ISSN: 0973-7510
E-ISSN: 2581-690X
In this study, sixty specimens were collected of Staphylococcus aureus isolation from several sources but found twenty-five specimens of Staphylococcus aureus isolated from skin from External laboratories in Baghdad, Iraq, during the period of December to May 2018. Then the samples were inoculated on culture media (Mannitol agar). The results of the sequencing showed congruence with isolation Staphylococcus aureus of amplified product of 16S rRNA gene appeared 99%compatibility. The results as shown After alignment of product amplification of 16S rRNA having seven Transition(A>G, A>G, T>C, T>C, G>A, C>T and A>G, and having three Transversion (T>G, G>C and A>T) from the Gene Bank.Registration of Iraqi isolate for Staphylococcus aureus bacteria in National Center for Biotechnology Information and under the accession number MH 145371.1. and it is available for download at NCBI: https://www.ncbi.nlm.nih.gov/nuccore/ MH 145371.1. The aim of the study was to determine the phylogenetic tree of Staphylococcus aureus based on 16S rRNA sequencing.The local Iraq isolate was registered after the correspondence of (NCBI), and obtained accession number and became a reference to Iraq and the Middle East and the world found variations of the local strain on the world.
Staphylococcus aureus, 16S rRNA gene, Skin infections
Staphylococcus aureus is a ubiquitous organism responsible for widespread infections both in community as well as in hospital settings1. These infections include pneumonia, skin and soft tissue infections, osteomyelitis, endocarditis, toxic shock syndrome etc.2. Skin and soft tissue infections ranged from furuncle, carbuncle, boil, impetigo, erysipelas, cellulitis, and abscesses to surgical site infections etc. Incision and drainage is the primary therapy for abscesses3. Staphylococcus aureus is a Gram-positive coccus bacterium, facultative anaerobic, catalase-positive, with single cells aggregate in clusters. Its responsible for many infection in human for example skin lesions (impetigo, furunculosis, folliculitis, hidradenitis-suppurativa, mastitis and Staphylococcal scalded skin syndrome.), in addition to infections of the subcutaneous tissue4. its colonizes the mucous membranes of nasopharynx, it is presented in apportion 50-60% in human as carriers5. The carrier state is a major risk factor for staphylococcal food poisoning, folliculitis and furunculosis6. S. aureus is transmitted mostly via contaminated hands of patients and medical staff and this lead to nosocomial infection. S. aureus has multiple factors and mechanisms of antibiotics resistance thereby is able to cause health and life-threatening infections4. The major virulence factors in this bacteria include enzymes, peptidoglycan and toxins. Other factors which lead to damaged tissue include (protein A and clumping factor)7. Hemolysin and Panton-Valentine leukocidin cause killing of white blood cells and red blood cells, whereas hyaluronidase, lipases, proteases, DNA, coagulase and fibrinolysin facilitate spreading the toxins within the human body8. PCR amplification from protected regions of the bacterial genome, especially the 16S rRNA gene followed by sequence analysis, is a steadfast technique for the identification of Enterobacter cloacae9. 16SrRNA gene sequence are considered as perfect standard for conclusion the phylogenetic relationship of microorganism10,11. Aim of the study was to determine the phylogenetic tree of Staphylococcus aureus based on 16S rRNA sequencing.
Bacterial isolates
In this study, sixty specimens were collected of Staphylococcus aureus isolation from several source but found twenty five specimens of Staphylococcus aureus isolation from skin. Then the samples were inoculated on culture media (Mannitol agar). Microscopic examination was done by using Gram stain 12. Mannitol salt agar was used as selective medium13.
PCR amplification of 16S rRNA genes
Partial 16S rRNA gene sequences were amplified by PCR using primers 16S rRNA Forward strand primers 5’- AGAGTTTGATCCTGGCTCAG -3’ and Reverse strand primers 5’- GGTTACC-TTGTTACGACTT -3’ 14. The PCR reaction mixture included 1.5 ml of bacterial DNA, 16.5 of ddH2O, 5 ml Master Mix PCR (intron, korea), and l ml each primer in a final reaction volume of 25 ml. Amplifications were performed as follows: initial denaturation at 5 min at 95°C, followed by 40 cycles of denaturation 95°C for 45 sec, annealing at 52°C for 1 min, extension at 72°C for 1 min and a final extension of 72°C for 7 min. The PCR products were separated on 1% agarose gel. The gel is left to run for 90min with a 70volt/65Am current. Following electrophoresis, visualization was conducted with a UV trans illuminator after red stain staining.
Genetic Detection of Staphylococcus aureus by 16S-rRNA
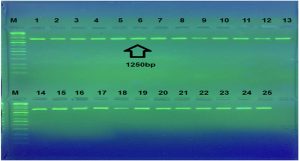
The present research findings pertain to the isolation of Staphylococcus aureus , samples were confirmed as on the basis of morphological, and molecular characterization . PCR is a rapid, sensitive, specific and inexpensive method. After gradient PCR has been performed ,bands has been obtained as presents in Figure 1. Showing that the best annealing a temperature to give 16S-rRNA was (52°C, 45s) depending on results of gradient PCR amplification procedure Several studies have used PCR for the detection of 16S rRNA by PCR method more rapidly and reliably. The results of current study agree with result of a same study in Iran reporte15. Who noticed that amplification of 16s-rRNA confirmed all the staphylococcal isolates as Staphylococcus aureus. Utilizing the 16S rRNA gene instead from whole genome information is not only computational efficient but also economical16 however, will provide an analysis that demonstrates that at least in the context of oral microbial communities, the 16S rRNA gene retains sufficient information to allow us detect unknown bacteria17. Figure 1 showed PCR amplification of the 16SrRNA where a specific product at 1250bp was observed.
Fig. 1. Agarose gel electrophoresis for 16SrRNA gene (1250bp) amplification of Staphylococcus aureus. electrophoresis on a 2% agarose gel and visualized under U.V. light after staining with red safe staining. Lane: 1 (M: 100bp ladder), Lane: 1to 25 product PCR positive.
Sequencing
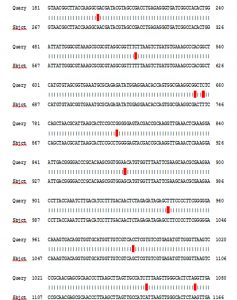
The sequencing and BLAST analysis of partial 16S rRNA gene figure 2 and 3. The results of the sequencing showed congruence with isolation Staphylococcus aureus of amplified product of 16SrRNA gene appeared 99% compatibility. (87 to 1178) number of nucleotide from gene of gene bank, and have number score (1925) bits, expect (0), and corresponds to the global number ID: NR_118997.2. The results as shown After alignment of product amplification of 16S rRNA having seven Transition (A>G, A>G, T>C, T>C, G>A, C>T and A>G, and having three Transversion (T>G, G>C and A>T) from the Gene Bank. 16SrRNA gene was successfully amplified using specific PCR primers for gene.
Table (1):
Represent type of polymorphism of 16S rRNA gene from Staphylococcus aureus isolate.
| Sequence ID | Nucleotide | Location | Type of substitution | No. of sample |
|---|---|---|---|---|
| ID: NR_118997.2 | A>G | 285 | Transition | isolates |
| T>C | 599 | Transition | ||
| A>G | 741 | Transition | ||
| T>C | 744 | Transition | ||
| T>G | 892 | Transversion | ||
| G>A | 955 | Transition | ||
| C>T | 1031 | Transition | ||
| G>C | 1078 | Transversion | ||
| A>T | 1143 | Transversion | ||
| A>G | 1161 | Transition |
Staphylococcus aureus strain ATCC 12600 16SrRNA, complete sequence Sequence ID:NR_118997.
Score |
Expect |
Identities |
Gaps |
Strand |
|---|---|---|---|---|
1925 bits(2134) |
0.0 |
1082/1092(99%) |
0/1092(0%) |
Plus/Plus |
Phylogenetic tree structuring
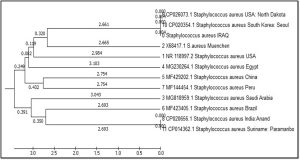
The phylogenetic tree diagrammatic by (MEGA) software version 6.0 is shown in figure 3. This study take care for phylogenetic analysis for geographic genetic distances determination The isolated sequences have shown 99% compatibility. Neighbor-joining tree was constructed for phylogenetic analysis. These alignments appeared the genetic distance and other global strains by partial sequence similarity in 16S ribosomal RNA gene for translating specific region. The genetic dimension between Iraq and the isolates of the world is detailed according to the Phylogenetic tree and the comparison table. hierarchical cluster analysis determine the following clusters: large Cluster divided into several neck: first root USA: North Dakota, South Korea: Seoul and IRAQ the genetic dimension was by 2.661, second root Muenchen the genetic dimension was by 2.665, third root USA the genetic dimension was by 2.984, fourth root Egypt the genetic dimension was by 3.103, and small Cluster divided into two root: China and Peru the genetic dimension was by 2.754. The last cluster is divided into two branches the first branch Saudi Arabia the genetic dimension was by 3.043), the second branch divided into two neck : India, Brazil and Suriname: Paramaribo the genetic dimension was by 2.693).
Table 2 represents comparison between strain of Staphylococcus aureus isolated from skin, recorded in the National Center Biotechnology Information (NCBI) and isolated from different source Avicenniae marina (white mangrove), buffalo milk, Minas cheese surface, sputum, milk, fatal septicaemia and septic arthritis in a 16-month-old American-Indian girl who had not risk agents associated with health care, blood and Perineum , and throat/groin but the same host (Homo sapiens) have under sequence(ID: NR_118997.218,
ID: X68417.118., ID: MG818959.119.,
ID: MG230264.120., ID: MF429202.121.,
ID:MF423405.122,
ID: MF144454.123.,ID: CP020656.124.,
ID: CP026073.125., ID: CP020354.126.,and
ID: CP014362.127. respectively with source of isolation and showed compatibility 99% and score (1925 and 1920), and expect 0.0 with gene bank.
Table (2):
Sequencing ID in gene bank, and compatibility of DNA sequences obtained from National Center Biotechnology Information (NCBI).
ACCESSION |
isolate |
country |
Source |
Compatibility |
|---|---|---|---|---|
ID: NR_118997.2 |
strain=”ATCC 12600″ |
USA |
Staphylococcus aureus |
99% |
ID: X68417.1 |
strain=”ATCC 12600″ |
Muenchen |
Staphylococcus aureus |
99% |
ID: MG818959.1 |
strain=”FA-1″ |
Saudi Arabia |
Staphylococcus aureus |
99% |
ID: MG230264.1 |
strain=”AM” |
Egypt |
Staphylococcus aureus |
99% |
ID: MF429202.1 |
strain=”CAU1287″ |
China |
Staphylococcus aureus |
99% |
ID: MF423405.1 |
strain=”SABRC56″ |
Brazil |
Staphylococcus aureus |
99% |
ID: MF144454.1 |
strain=”FQXIII” |
Peru |
Staphylococcus aureus |
99% |
ID: CP020656.1 |
strain=”K5″ |
India : Anand |
Staphylococcus aureus |
99% |
ID: CP026073.1 |
strain=”Mw2″ |
USA: North Dakota |
Staphylococcus aureus |
99 % |
ID: CP020354.1 |
strain=”FORC59″ |
South Korea: Seoul |
Staphylococcus aureus |
99 % |
ID: CP014362.1 |
strain USA300-SUR1 |
Suriname: Paramaribo |
Staphylococcus aureus |
99 % |
Submission of local Iraq isolate in NCBI
Broad-range PCR targets the 16S rRNA gene which is the most common housekeeping genetic marker because the ribosomal small subunit is present universally in all bacteria28. Registration of Iraqi isolates for Staphylococcus aureus bacteria in National Center for Biotechnology Information and under the accession number MH 145371.1. and it is available for download at NCBI: https://www.ncbi.nlm.nih.gov/nuccore/ MH 145371.1.
The efficiency of genetic methods in the detection of accurate and rapid bacterial isolates thus shortening the time and cost and giving the correct treatment and accurate treatment of disease without error and time loss. After obtaining the global number of Iraqi isolates proved near the Iraqi isolates of the isolates USA :North Dakota , South Korea: Seoul.
Acknowledgements
Praise be to Allah, the Lord of words. First of all, I owe the utmost sincere gratitude to my creator, Allah, for giving me strength and patience to face the difficulties of completion of this Research. I would like to thank the Al-maarif University College, Department of Medical Laboratory Techniques , and all the staff of Biotechnology Research Center, Al-Nahrain University.
Conflict Of Interest
The authors declare that there is no conflict of interest.
- Tianming, Li, Yan Song, Yuanjun Zhu, Xin Du, Min LI. Current status of Staphylococcus aureus infection in a central teaching hospital in Shanghai, China. BMJ Microbiol., 2013; 13: 153.
- Dennis, L., Stevens, et al. Practice Guidelines for the Diagnosis and Management of Skin and Soft-Tissue Infections. CID, 2005; 41: 1373–406.
- Swati Rahul Dhope., Dnyaneshwari Purushottum Ghadage et al. Study of Antibiotic Susceptibility Pattern in Staphylococcus aureus Isolated from Skin and Soft Tissue Infections in a Tertiary Care Hospital. Int.J. Curr. Microbiol .App. Sci., 2017; 6(7): 1776-1779.
- Pawel Kwiatkowski1., Magdalena Mnichowska Polanowska., et al. Activity of essential oils against Staphylococcus aureus strains isolated from skin lesions in the course of staphylococcal skin infections, 2017; 63(1): 43-52.
- Verma RS, Padalia RC, Chauhan A. Rose-scented geranium (Pelargonium sp.) oils. In: Preedy VR, ed. Essential oils in food preservation, flavor and safety. 1st ed. London. Academic Press, 2015 ;697-704.
- Toshkova K, Annemüller C, Akineden O, Lämmler C. The significance of nasal carriage of Staphylococcus aureus as risk factor for human skin infections. FEMS Microbiol Lett., 2001; 202(1):17-24. doi: http://dx.doi.org/10.1111/j.1574-6968.2001.tb10774.x
- Kaneko J, Kamio Y. Bacterial two-component and hetero-heptameric pore-forming cytolytic toxins: structures, pore-forming mechanism, and organization of the genes. Biosci. Biotechnol Biochem, 2004; 68(5):981-1003.
- Skaar EP, Schneewind O. Iron-regulated surface determinants (Isd) of Staphylococcus aureus: stealing iron from heme. Microbes Infect., 2004; 6(4):390-7. doi: http://dx.doi.org/10.1016/j.micinf.2003.12.008.
- Mignard S, Flandrois JP.16S rRNA sequencing in routine bacterial identification: a 30-month experiment. J. Microbiol. Methods, 2006; 67:574-581.
- Nayak, B . S.; Badgley, B. and Harwood , V.J.Comparison of genotypic and phylogenetic relationships of environmental Enterococcus isolates by Box-PCR typing and 16SrRNA gene sequencing. Appl. environ. Microbiol., 2011; 77(14):5050-5055.
- Ciccareli, F.D.; Doerks. T.; Von Mering, C.; Creevey, C.J.; Snel, B. and Bork, P. Toward automatic reconstruction of a highly resolved tree of life Science, 2006; 311(5765):1283-1287.
- Shaymaa Khudhr Al–Alak, Daremkadhim Qassim. Molecular Identification of 16S rRNA gene in Staphylococcus aureus Isolated from Wounds and Burns by PCR Technique and Study Resistance of Fusidic acid. Iraqi Journal of Cancer and Medical Genetics, 2016.
- Anderson and Cindy. Great Adventures in the Microbiology laboratory Pearson. 2013; pp:175-176.
- Ramya Srinivasan, Ulas Karaoz, Marina Volegova, Joanna MacKichan, Midori Kato Maeda, Steve Miller, Rohan Nadarajan, Eoin L. Brodie, Susan V. Lynch. Use of 16S rRNA Gene for Identification of a Broad Range of Clinically Relevant Bacterial Pathogens. Journal. pone. 2015; 0117617,1-22.
- Malihe, Heidari.; Hassan ,Momtaz and Mahboobeh Madani. Detection of the antibiotic resistance genes in Staphylococcus aureus isolated from human infections and bovine mastitis. African Journal of Microbiology Research, Vol. 2011; 5(28), pp. 5132-5136.
- Lindner MS, Renard BY. Metagenomic profiling of known and unknown microbes with microbegps. PloS ONE, 2015; 10(2):0117711.
- Quang Tran, Diem-Trang Pham and VinhthuyPhan. Using 16S rRNA gene as marker to detect unknown bacteria in microbial communities. Tran et al. BMC Bioinformatics, 2017; 18(Suppl 14):499.
- Ludwig, W., Kirchof, G., Klugbauer, N., Weizenegger,M., Betzl, D, et.al. Complete 23S ribosomal RNA sequences of Gram-positive Bacteria with a low DNA G+C content. 15, 487-501. Botany @ Microbiology, King Saud18- Ameen,F. University, King Khaled Road, Riyadh, Riyadh 11451, Saudi Arabia, 1992.
- Hagaggi,N.S. Botany, Faculty of Science, Aswan University, Sahary, Aswan 81528, Egypt, 2017.
- Ren,F. and Zhao, L. College of Food Science & Nutrition Engineering, China Agricultural University, No. 17 Qinghua Road, Beijing 100083, China, 2017.
- Nino-Arias,F.C., Pitondo-Silva,A., Frazilio, D.A., Goncalves, L.O.,et.al. Department of Clinical Analysis, Toxicology and Food Science, School of Pharmaceutical Sciences of Ribeira Preto, University of Sao Paulo, Av. do Cafe S/N. Monte Alegr Ribeirao Preto, Sao Paulo 14040-903, Brazil, 2017.
- Aquino,R., Gonzales, E., Samaniego,S., Rivera,J., Cedeno,V., Urbina,Y. and Diringer,B. Lab. Molecular biotechnology, National University of Tumbes – INCABIOTEC SAC, CA.Filipinas 212, Tumbes, Tumbes, Tumbes 24001, Peru, 2017.
- Patel,K.J., Kunjadiya, A.P., Joshi, C.G., Nauriyal, D.S., Patel, A.B. and Pandit, R.J. Center for Interdisciplinary Studies in Science and Technology, Sardar Patel University Campus, Vallabh Vidyanagar, Gujarat 388 120, India, 2017.
- Utter,B., Tallon,L.J., DeShong Sadzewicz,L., Sengamalay,N.,Nagaraj,S., McCracken,C.L., Daugherty,S. and Sichtig,H. Institute for Genome Sciences, Universit of Maryland School of Medicine, 801 W. Baltimore St, Baltimore, MD21201, USA, 2015.
- Chun, J. and Ryu, S. Food Biotechnology, College of Agriculture and Life Sciences, Seoul National University, Gwanak-ro , Gwanak-guSeoul 08826, Republic of Korea, 2017.
- Sabat, A.J., Hermelijn, S.M., Akkerboom, V., Juliana, A., Degener,J.E. Grundmann, H. and Friedrich, A.W. Complete-genome sequencing elucidates outbreak dynamics of CA-MRSA USA300 (ST8-spa t008) in an academic hospital of Paramaribo,Republic of Suriname Sci Rep., 2017; 7: 41050 .
- Shin SH, Kim S, Kim JY, Lee S, Um Y et al. Complete Genome Sequence of Enterobacter aerogenes KCTC 2190. J Bacteriol., 2012; 194: 2373-2374.
- Taghavi S, van der Lelie D, Hoffman A, Zhang YB, Walla MD et al. Genome sequence of the plant growth promoting endophytic bacterium Enterobactersp.. PLOS Genet., 2010; 638: 6: e1000943.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.