ISSN: 0973-7510
E-ISSN: 2581-690X
Diabetic foot ulcer (DFU) is one of the most serious and costly complications of diabetic patients. Enterococcus faecalis (E. faecalis) represents one of the most virulent microorganisms in diabetic foot infections (DFIs). We therefore aimed to study the frequency and precise identification of E. faecalis in DFU. Six hundred thirty specimens collected from diabetic foot patients were used in the current investigation. Biochemical identification was carried out by the Vitek® 2 system. Proteomic analysis was implemented by MALDI-TOF MS and confirmed by SYBER Green real-time polymerase chain reaction (SYBER Green qPCR). According to the results, the overall frequency of E. faecalis in patients with DFU was 8.25% (52/630). Out of 52 E. faecalis strains, 40 isolates were isolated from males and 12 from females. The results of biochemical identification revealed that 92.30% (48/52) of E. faecalis isolates were properly recognized at the species level. Whereas 100% (52/52) of E. faecalis isolates were properly recognized by MALDI-TOF MS as 44.23% (23/52), 51.92% (27/52) and 3.84% (2/52) with a score value ranging from 2.300 to 3.000, 2.000-2.299 and 1.700-1.999 Da, respectively. Seven E. faecalis virulence genes, including asa1, GelE, cylA, esp, hy1, VanA, and VanB, were detected by SYBER Green RT-PCR. In conclusion, E. faecalis was the utmost predominant single organism isolated from the DFIs. MALDI-TOF mass spectrometry is considered a fast, trustworthy and economic detection method for various significant microorganisms. E. faecalis isolates were also found to carry several virulence genes. Our findings may serve as an urgent issue for supplementary investigations of contagions caused by E. faecalis.
Diabetic foot infections, E. faecalis, Incidence, Identification, MALDI-TOF MS
Diabetes mellitus (DM) is a chronic deteriorating disease associated with many complications. Diabetic foot ulcer (DFU) is considered one of the major problems of DM with a yearly incidence of 10% among diabetic patients. It is calculated that 15% of diabetic patients are suffering from ulcers through their lifetime, and 10%–30% of these cases ultimately progress to amputation 1. The infection rate is a significant causative issue in this case, according to this review of literature; about 60% of these amputations are preceded by the presence of infected ulcers. Saudi Arabia is one of the top ten Middle Eastern/Arab countries with the highest diabetes prevalence in adults, which will certainly lead to high rates of foot ulcers along with increasing morbidity and costs 2. Most diabetic foot infections (DFIs) present with a polybacterial etiology caused by enterococcal strains, which are part of the multifaceted diabetic foot infection. Former researches stated that the genus Enterococcus is one of the utmost virulent microorganisms in DFIs 3. Numerous microbes, can lead to DFIs, of which Enterococci are the common type of bacteria 4. Former researches indicated that the genus Enterococcus genus is the most public organisms in DFIs, leading to increase the severity of infection and even death 5,6. Recently, the genus Enterococcus is composed of thirty-eight species; Enterococcus faecalis (E. faecalis) represents the common significant species of this genus 7. Their identification is established mostly on morphological and biochemical and molecular techniques 8,9. This diversity of techniques permits a consistent detection of Gram-positive cocci. Nevertheless, phenotypic and genetic approaches have many limitations such as laborious, expensive and labor-intensive, and the molecular methods do not usually exist in numerous investigative test centers.
In recent times, numerous studies assessed the application of matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) for the detection of various microorganisms in the diagnostic laboratories 10,11,12,13,14,15,16. Throughout this technology, microorganisms are recognized by means of either whole cells or cell extracts. The procedure is quick, delicate, and inexpensive. This skill has been willingly approved by specialists who have described the application of MALDI-TOF for a quantity of determinations such as microbiological identification, characterization, and discrimination 17,18. In Germany, Werner et al. 19 indicated that MALDI TOF MS detected 64 E. faecalis and 37 E. faecium isolates, which were established by PCR. In Zagreb, Dobranic et al. 20 stated that MALDI-TOF MS identification presented 100% concordance with API 20 Strep in the identification of E. faecalis.
Therefore, the possibility for MALDI-TOF to rapidly describe a broad range of microbes, comprising fungi, bacteria, and viruses increase its possible use in various parts of biological identification. Consequently, this equipment can be applied for quick identification of numerous types of pathogens at comparatively low cost and also represents another powerful technique for traditional and molecular methods. The main impact of this technique consists of a significant improvement in limiting the time consumed from preparing the samples till identification.
From the previously mentioned date, the current study was delineated to examine several parameters: (i) Determine the colonization rate of E. faecalis in DFI in Saudi Arabia; (ii) Biochemical identification of E. facials using the VITEK 2 System; (iii) Proteomic identification of E. faecalis using MALDI-TOF MS confirmed by qPCR.
Sample collection
The current investigation was conducted from June 2016 to January 2017 at King Saud Medical City, Riyadh region and Bukyriah General Hospital (BGH) Qassim region. The King Saud Medical City was opened in 1970 with a capacity of 1500 beds and provides hospital services to the province of Riyadh city. BGH opened in 1985 and accommodates 150 beds and covers hospital services to the province of Bukayriyah and surrounding areas. It is a patient-referral hospital that covers more than 20 health centers in addition to three small (30 beds) hospitals. A total of 630 samples were surveyed during June 2016 to January 2017. Swabs were collected from patients’ DFI deep wound and abscess specimens. Pus from the lesions should be put in a sanitary sterilized bottle and/or an anaerobic transport bottle (Procedures Guidelines for the Microbiology Laboratory). Swabs were microscopically examined and classified. Before sample collection, a sterile cotton swab was soaked with sterile saline solution. Bacterial description and proof of identity procedures were accomplished by standard protocols. From air dried and heat fixed smears, gram staining and cell morphology were implemented. Epidemiological data were recorded with respect to gender, age, and weight.
Isolation of E. faecalis
Fifty-two strains of E. faecalis were isolated from 630 diabetic foot patients. From the primary routine cultures, all specimens were cultured into blood agar and MacConkey agar plates (both from Sigma-Aldrich, USA). After incubation of all plates at 37oC for 24 to 48 h, E. faecalis colonies appeared as small, round, white colonies. The purified isolates were then subjected to presumptive identification tests, including Gram staining, catalase and bile esculin, growth at 45°C, and 6.5% of NaCl. Catalase-negative and bile esculin-positive, Gram-positive cocci arranged in pairs and/or chains forming alpha-hemolytic or non-hemolytic colonies on blood agar media were recognized as Enterococci and then they were spectated with the API Rapid Strep system.
Biochemical identification of E. faecalis
The identification- gram positive card (ID-GPC) of the VITEK® 2 system (Biomérieux, France) was used in this study for detection of E. faecalis isolates according to the manufacturer’s instructions. Briefly, 2-3 fresh colonies were inoculated with 5 ml of salt solution and thoroughly mixed. The bacterial turbidity was tested in the range from 0.50 and 0.63 using DensiChekTM (BioMe2 rieux, France). A test tube filled with 5 ml of this suspension was loaded into the machine. The filled cassette was finally sited in the device, and the data from an unknown bacterium were compared to the particular catalogue to define a quantitative value for proximity to each of the database taxa. Both ATCC 19433 E. faecalis and ATCC 25923 S. aureus were utilized as quality control isolates.
Proteomic investigation of E. faecalis by MALDI-TOF MS
Preparation of bacterial samples
We then applied the MALDI biotyper device (Bruker Daltonik, Bremen, Germany) for fast and precise detection of E. faecalis isolates. A single colony from freshly streaked agar incubated for 18-24h at 37°C, was utilized for each isolate and inoculated onto two spots of the target plate. Each colony was then enclosed with one microliter of matrix solution. The microbial bands were directly produced by applying compass software, and evaluation was conducted with a MALDI biotyper machine. We used Escherichia coli as a positive control (bacterial test standard) according to the recommendations of Bruker Corporation.
Data analysis and clustering
The score value of unidentified spectrum ranging from 0 to 3 was determined by comparing the corresponding unknown spectrum with the known spectrum in the Bruker library. The accuracy of strain recognition was detected by Bruker Daltonik, Bremen, Germany. The precise detection of species was performed using the Microflex LT device when the score value ranges from 2.3 to 3.0. Species and genus levels were also recognized in the range from 2.00 to 2.29 and 1.700 to 1.999, respectively. Furthermore, a score of 0.00 to 1.69 means that the proof of identity is not reproducible. The diverse spectra created with the Microflex LT compass software were measured in a m/z range from 2, 000 to 20, 000 Da. The official standards depended on fifty laser shots per spot. According to the Microflex LT library, which contains 5989 bacterial and fungal sub-species, a dendrogram was generated from the minimal spanning tree (MSP) data set. The MSP dendrogram was generated based on evaluation of the main spectra of the various species.
Molecular identification of E. faecalis
Genomic DNA isolation
Extraction of bacterial DNA was done by QuickGene-810 (Fujifilm, Tokyo, Japan) with a special DNA isolation kit (QuickGene DNA Tissue Kit S). All procedures were carried out based on the manufacturer recommendations. The purity and concentration of extracting DNA were detected by NanoDrop™ 2000 UV Spectrophotometer.
SYBER Green Real time PCR identification
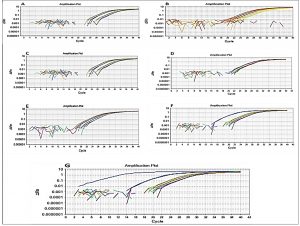
The SYBER Green RT-PCR for E. faecalis was then carried out by 7500 Fast RT-PCR System (Applied Biosystems, USA). Seven different virulence genes of E. faecalis, including asa1, GelE, cylA, esp, hy1, VanA and VanB. E. faecalis were detected as described by the manufacturer’s instructions of Thermo Fisher Scientific, USA. Lists of selected primers, qPCR conditions, and amplified products are presented in Table 1. Briefly, a total of 20 µl reaction volume containing 10 µl of SYBER Green PCR master mix, 1 µl each of forward and reverse primer, 1 µl target DNA, and 7 µl of RNAse/DNase free water was used. All reactions were performed in duplicate. Preparation of a standard curve dilution series was done by adding 90 µl of template preparation buffer to five tubes and labeling them. The positive control template (E. faecalis [ATCC 19433]) was considered as tube 1, and 10 µl was transferred from this tube to tube 2 and mixed thoroughly. Ten microliters from tube 2 were then transferred into tube 3, continuing in the same manner up to the 9th tube. Finally, for the standard curve, 5 µl of the standard template was transferred to each well, making the final volume in each well 20 µl. Amplifications were then completed in the RT-PCR system. Thermal cycling consisted of 37 °C for 15 min with AmpErase® uracil N-glycosylase (UNG) in order to prevent PCR carryover contamination and 95 °C for 2 min for enzyme activation followed by 40 cycles with every denaturation step at 95 °C for 10 s. Annealing and extension was performed at 60 °C for 60 s. Data were analyzed using sequence detection system (SDS) software. Amplification findings were expressed by plotting Delta Rn (DRn) versus cycle number for the interpretation of infection 21.
Table (1):
Primers utilized in the RT-PCR for E. faecalis detection.
Target gene |
Virulence factor |
Primer |
Sequence (59–39) |
Product size (bp) |
|---|---|---|---|---|
asa1 |
Aggregation substance |
ACA11 ACA12 |
GCACGCTATTACGAACTATGA TAAGAAAGAACATCACCACGA |
375 |
GelE |
Gelatinase |
Gel11 Gel12 |
TATGACAATGCTTTTTGGGAT AGATGCACCCGAAATAATATA |
213 |
cylA |
Cytolysin |
CYT I CYT IIb |
ACTCGGGGATTGATAGGC GCTGCTAAAGCTGCGCTT |
688 |
esp |
Enterococcal surface protein |
ESP 14F ESP 12R |
AGATTTCATCTTTGATTCTTGG AATTGATTCTTTAGCATCTGG |
510 |
hyl |
Hyaluronidase |
HYL n1 HYL n2 |
ACAGAAGAGCTGCAGGAAATG GACTGACGTCCAAGTTTCCAA |
276 |
VanA |
Vancomycin-resistant A |
vanA 1 vanA2 |
GGGAAAACGACAATTGC GTACAATGCGGCCGTTA |
175–191 907–891 |
VanB |
Vancomycin-resistant B |
Van B1 vanB 2 |
ATGGGAAGCCGATAGTC GATTTCGTTCCTCGACC |
173–189 807–791 |
Incidence of E. faecalis in diabetic foot patients
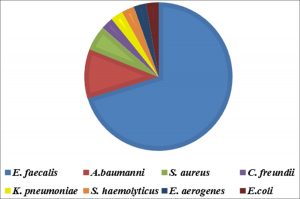
The incidence of E. faecalis was investigated in 630 DFI samples. The frequency of the organisms in the DFI is shown in fig. 1. According to the results, of the 630 DFI samples, 74 samples were positive as 52 E. faecalis, 8 Acinetobacter baumanni (A. baumanni), 4 Staphylococcus aureus (S. aureus), 2 Citrobacter freundii (C. freundii), 2 Klebsiella pneumoniae (K. pneumoniae), 2 Staphylococcus epidermidis (S. epidermidis), 2 Enterobacter aerogenes (E. aerogenes) and 2 Escherichia coli (E. coli).
Distribution of E. faecalis and its associated organisms among males & females
As presented in fig. 2, 40 E. faecalis isolates were recovered from males and 12 from females. Six isolates of A. baumanni were recovered form males and 2 from females. All S. aureus, C. freundii, K. pneumoniae, S. epidermidis and E. coli isolates were recovered from males only, whereas E. aerogenes isolates were isolated from females only.
The effect of age on the prevalence of bacterial DFIs
To assess the effects of age on the prevalence of DFIs, patients enrolled in this study were categorized into different age groups varying from ≤1 year to >80 years. As illustrated in Table 3, patients in the age group ≤1- 10, 21–30 years, and 41–50 years showed the highest prevalence of bacterial DFIs at frequencies of 20.2%, 18.9%, and 17.5%, respectively, whereas patients in the age groups 11–20 years, 31–40 years, and 61–70 years exhibited the lowest risk of bacterial DFIs with a frequency of 5.4%.
Table (2):
Distribution of E. faecalis and its associated organisms among males & females.
| Isolates | Total isolates = 74 | ||
|---|---|---|---|
| Males | Females | Total | |
| Enterococcus faecalis | 40 | 12 | 52 |
| Acinetobacter baumanni | 6 | 2 | 8 |
| Staphylococcus aureus | 4 | 0 | 4 |
| Citrobacter freundii complex | 2 | 0 | 2 |
| Klebsiella pneumoniae | 2 | 0 | 2 |
| Staphylococcus epidermidis | 2 | 0 | 2 |
| Enterobacter aerogenes | 0 | 2 | 2 |
| Escherichia coli | 2 | 0 | 2 |
Table (3):
Distribution of recovered DFIs bacteria in relation to age groups.
| Age Group | Total bacteria isolates (n=74) |
Total gram positive (n= 58) |
Total gram Negative (n=16) |
|||
|---|---|---|---|---|---|---|
| N | % | N | % | N | % | |
| ≤1 – 10 y | 15 | 20.2 | 13 | 22.4 | 2 | 12.5 |
| 11 – 20 y | 4 | 5.4 | 2 | 3.4 | 2 | 12.5 |
| 21 – 30 y | 13 | 17.5 | 2 | 3.4 | 0 | 0 |
| 31 – 40 y | 4 | 5.4 | 2 | 3.4 | 0 | 0 |
| 41 – 50 y | 14 | 18.9 | 10 | 17.2 | 4 | 25 |
| 51 – 60 y | 12 | 16.2 | 8 | 13.7 | 4 | 25 |
| 61 – 70 y | 4 | 5.4 | 4 | 6.8 | 0 | 0 |
| 71 – 80≥ y | 10 | 13.5 | 6 | 10.3 | 4 | 25 |
Identification of E. faecalis and associated organisms Biochemical identification
A total of 52 E. faecalis and other 22 organisms recovered from DFI were assessed by the VitekTM 2 system for biochemical identification. As shown in Table 2, a total of 70 out of 74 isolates were correctly identified as 49/52 (94.23%) E. faecalis, 7/8 (87.2%) A. baumanni, 4/4 (100%) S. aureus, 2/2 (100%) C. freundii complex, 2/2 (100%) K. pneumoniae, 2/2 (100%) S. haemolyticus, 2/2 (100%) E. aerogenes, and 2/2 (100%) E. coli.
Proteomic identification of E. faecalis
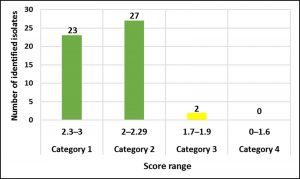
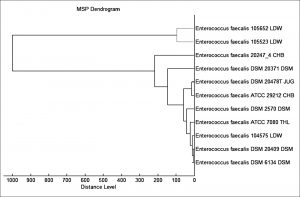
According to our findings, 52 E. faecalis strains recovered from patients suffering from DFI were properly recognized by MALDI Biotyper (MBT). In the current study, all isolates were investigated using the MBT device, and the obtained spectra were matched with the storing spectra in the Bruker taxonomy of compass software. As seen in fig. 2, it was noticed that 52 E. faecalis strains were sufficiently well recognized at the species level as 44.23% (23/52), 51.92% (27/52) and 3.84% (2/52) with a score value ranging from 2.300 to 3.000 and 2.000-2.299 Da, respectively. In contrast, only two isolates (3.84%) were identified at the genus level with a log score ranging from 1.7 to 1.99. Ion peaks were observed in the line spectra of 2,000-16,000 Da. The strong peaks were concentrated in between 3,000 and 5,000 Da, which corresponded with different types of bacterial strains assessed by the library of compass software (fig. 3). A current gel spectral image detected 52 E. faecalis strains. Numerous spectra were scattered within the range from 2, 000 to 15, 000 Da, and most peaks were condensed between 4, 400 and 5,000 Da (fig. 4). Furthermore, to clarify if the Microflex LT compass software could distinguish clonally-associated strains at the species level, spectra from 52 well-identified E. faecalis strains were investigated as shown on the gel. The various spectra were then developed to generate a novel cross-wise minimal spanning tree (MSP) dataset (fig. 5). According to the results, the dendrogram exhibited that analyzed E. faecalis strains were closely related to 11 E. faecalis reference strains in the Bruker taxonomy at a distance of 200.
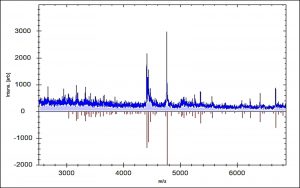
Fig. 3. Comparison of mass spectrum protein profiles of unknown samples (blue) with reference strain E. faecalis 20371 DSM (red).
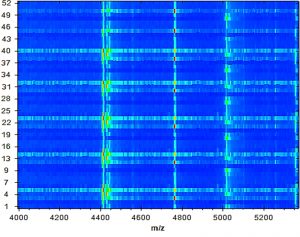
Fig. 4. A gel protein spectrum for 52 E. faecalis strains demonstrates that most peaks are condensed between 4, 400 and 5, 000 Da.
Molecular identification of E. faecalis
The qPCR method using the fast qPCR 7500 (Applied Biosystems) was used for detection of E. faecalis isolates. SYBR®Green qPCR was performed on DNAs extracted from seven reference strains as positive controls for the seven possible genes, including asa1, GelE, cylA, esp, hy1, VanA, and VanB. Specific primers designed using a reference for seven gene sequences representing asa1, GelE, cylA, esp, hy1, VanA, and VanB were collected from the GenBank database. As shown in fig. 6, all seven genes of E. faecalis were detected by qPCR.
Fig. 5. The MSP dendrogram for 52 E. faecalis strains exhibited a strong relationship with 11 E. faecalis reference strains in the Bruker taxonomy
Foot ulcer represents one of the major complications in diabetic patients and currently represent the most public diabetes-related cause of hospitalization. Patients suffering from diabetes have approximately a 25% chance of rising a foot ulcers in their life. DFIs lead to high morbidity rates and a lower extremity amputation may be resulted. Recently, DFIs represent the major cause of lower extremity amputations in many parts of the world 22.
In our study, 52 (8.25%) E. faecalis strains were recovered from 630 pus swabs from DFI. Similar results were obtained by 23,24, who stated that E. faecalis are rarely encountered in DFIs. Spichler et al. 22 established that the primary bacterial genera in DFIs were Staphylococcus (29.7%), Peptoniphilus (6.9%), Rhodopseudomonas (6.9%) and Enterococcus (6.4%). The incidence of E. faecalis in DFIs was also studied in Malaysia by Raja 25 who found that Gram-negative bacteria were the major isolated pathogens followed by S. aureus (44%), Group B Streptococci (25%), and Enterococcus spp. (9%). Enterococci are among the most common Gram-positive bacteria, and they are also characterized as significant nosocomial pathogens. Moreover, Edmonds 26, Mathangi and Prabhakaran 27, Roudbary et al. 28 and Sousa Lages et al. 29 reported that the frequency of E. faecalis in DFIs was 2%, 6.4%, 13.5% and 8.97%, respectively.
Recently, traditional techniques applied for microbial detection in the clinical biological laboratories have been based mainly on morphological and biochemical analyses, but these methods are still laborious, need multifaceted measures, and require huge quantities of biological agents, which is predominantly hard to accomplish for problematic microorganisms with unusual biochemical features 30.Different molecular techniques, including 16S rRNA gene sequencing, have been established as a compromise; nevertheless, they are not routinely applied as a result of their high price and high workload on technicians. Although, cultivation and other conventional methods have been established for identification of E. faecalis, numerous restrictions have been detected. Therefore, we used MBT as a rapid technique for detection of E. faecalis recovered from DFIs in Saudi Arabia. This technology is recently used as a potent technique for recognition of various pathogens 16,18,17,31.
In our study, recognition and clustering of E. faecalis strains were performed by proteomic fingerprinting and the occurrence of accurate identûcation at the species level was 52/52 (100%) with the score levels of 2,000-3,000 Da. Similar findings were documented formerly by Werner et al. 19 who detected 64 E. faecalis and 37 E. faecium strains recovered from milk samples of mastitic cows by MBT. They established that the maximum identûcation rate was 99.3%. Another study was conducted by Benagli et al. 32, who identified E. faecalis by MALDI-TOF MS with a confidence level of 100%. In addition, Griffin et al. 33 identified vancomycin resistant E. faecalis (VRE) stains by MBT with high identification rate of 96.7% and 98.1%, correspondingly.
In this context, the lower score values for two E. faecalis isolates at the species level (1.700 to 1.999) might be related to sample handling, and the amount of matrix in the sample were not accurate. Consequently, it is very significant to apply other standard methods such as PCR as an additional technique to support the suspected findings detected by MALDI-TOF MS 34. The data of the present investigation were evaluated by MALDI-TOF MS, and it was confirmed that the furthest spectral peaks for the examined E. faecalis strains ranged from 3,000 to 10,000 Da. A similar range was obtained in numerous other studies on bacteriological recognition using MALDI-TOF MS 35,36,37. Benagli et al. 32 indicated that MALDI-TOF MS is considered a trustworthy, economic, and rapid identification technique for clinically significant microorganisms, including Enterococci. In addition, Pavlovic et al. 38 demonstrated that this technology has the ability to identify various pathogens through the generation of fingerprints of extremely plentiful proteins, followed by association with the reference spectra in the Bruker taxonomy. Other studies conducted by Pavlovic et al. 38 and Quintela et al. 39 stated that there are incomplete data on the application of this technique for identification of various strains of Enterococci. Moreover, Chabros et al. 40 showed that MBT may be a valuable instrument to discriminate between the species E. faecium and E. gallinarum. Although MALDI-TOF is considered an innovative method compared with the other biochemical methods, they found some complications with the identification of the Enterococcus species.
The cost and maintenance of this technique still represent the most important disadvantage of this method 13,38,41. However, this technology is considered a potent, innovative method that has the possibility to replace classical identûcation methods for common routine isolates in the hopes of solving the problem of DFIs using an accurate and primary diagnostic technique.
In our investigation, we also applied an easy-to-use qPCR technique based on the amplification of fragments of asa1, GelE, cylA, esp, hy1, VanA, and VanB genes with SYBER green probe qPCR. This specific method permits identifying of the seven possible genes for E. faecalis. The seven fluorescence results are readable on a 7500 Fast Applied Biosystems. Based on our findings, the six E. faecalis genes were detected. The developed sensitivity of the qPCR technique could be explained by the fact that it possibly targets free-floating DNA and DNA from non-viable and cultivable viable cells 42. In addition, our results were matched with the results obtained by Rocas et al. 43, who used a PCR method to detect E. faecalis in endodontic infections.
This study showed that several bacterial species were recovered from diabetic foot patients. E. faecalis was the most predominant single organism isolated from the lesions. These findings show the significance of E. faecalis in diabetic foot infections. E. faecalis isolates were found to carry several virulence genes, suggesting that additional researches on the dissemination and the possibility of virulent strains to carry these genes may be necessary. In addition, MALI Biotyper is a fast, trustworthy and a beneficial detection method for various microorganisms. Our outcomes may serve as a source for supplementary investigation studies of infections caused by this bacterium.
Acknowledgements
The authors gratefully acknowledge the King Abdulaziz City for Science and Technology, Saudi Arabia for funding this research (Project number 1-17-01-001-0054).
Conflict Of Interest
The authors declare that there is no conflict of interest.
- Doria M, Rosado V, Pacheco LR, Hernández M, Betriu A, Valls J, Franch-Nadal J, Fernández, Mauricio D. Prevalence of Diabetic Foot Disease in Patients with Diabetes Mellitus under Renal Replacement Therapy in Lleida, Spain. Biomed Res Int 2016;8:1-8.
- Al Dawish MA, Robert AA, Braham RA, Hayek AA, Saeed A, Ahmed RA, Al Sabaan FS. Diabetes Mellitus in Saudi Arabia: A Review of the Recent Literature. Curr Diabetes Rev 2016;12(4):359-368.
- Semedo-Lemsaddek T, Mottola C, Alves-Barroco C, Cavaco-Silva P, Tavares L, Oliveira M. Characterization of multidrug-resistant diabetic foot ulcer enterococci. Enferm Infecc Microbiol Clin 2016; 34(2):114–116.
- Nair N, Biswas R, Götz F, Biswas L. Impact of Staphylococcus aureus on pathogenesis in polymicrobial infections. Infect Immun 2014; 82(6):2162-2169.
- Lipsky BA, Richard JL, Lavigne JP. Diabetic foot ulcer microbiome: one small step for molecular microbiology One giant leap for understanding diabetic foot ulcers. Diabetes 2013; 62(3):679-681.
- Karmaker M, Sanyal SK, Sultana M, Hossain MA. Association of bacteria in diabetic and non-diabetic foot infection – An investigation in patients from Bangladesh. J Infect Public Health 2016; 9(3):267-77.
- John Vu and Carvalho J. Enterococcus: review of its physiology, pathogenesis, diseases and the challenges it poses for clinical microbiology. J Front Biol 2011; 6: 357.
- Bosshard PP, Abels S, Altwegg M, Böttger EC, Zbinden R. Comparison of conventional and molecular methods for identification of aerobic catalase-negative Gram-positive cocci in the clinical laboratory. J Clin Microbiol 2004;42:2065–2073.
- Zbinden A, Kohler N, Bloemberg GV. recA-based PCR assay for accurate differentiation of Streptococcus pneumoniae from other viridans streptococci. J Clin Microbiol 2011; 49:523–527.
- Bizzini A, Durussel C, Bille J, Greub G, Prod’hom G. Performance of matrix-assisted laser desorption ionization-time of fiight mass spec- trometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J Clin Microbiol 2010; 48:1549–1554.
- Cherkaoui A, Hibbs J, Emonet S, Tangomo M, Girard M, François P, Schrenzel J. Comparison of two matrix-assisted laser desorption ionization-time of fiight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J Clin Microbiol 2011; 48:1169–1175.
- van Veen SQ, Claas EC, Kuijper EJ. High-throughput identification of bacteria and yeast by matrix-assisted laser desorption ionization- time of fiight mass spectrometry in conventional medical microbiology laboratories. J Clin Microbiol 2010; 48:900–907.
- Neville SA, Lecordier A, Ziochos H, Chater MJ, Gosbell IB, Maley MW,van Hal SJ. Utility of matrix-assisted laser desorption ionization-time of fiight mass spectrometry following introduction for routine labo- ratory bacterial identification. J Clin Microbiol 2011; 49:2980–2984.
- Dubois D, Grare M, Prere MF, Segonds C, Marty N, Oswald E. Performances of the MALDI-TOF mass spectrometry system VITEK MS for the rapid identification of bacteria in routine clinical microbiology. J Clin Microbiol 2012; 50:2568–2576.
- Tan EK, Ellis BC, Lee R, Stamper PD, Zhang SX, Carroll KC. Prospective evaluation of a MALDI-TOF MS system in a hospital clinical microbiology laboratory for the identification of bacteria and yeasts: a bench-by-bench study to assess the impact on time-to-identification (TTI) and cost-effectiveness. J Clin Microbiol 2012; 50:3301–3308.
- Elbehiry A, Al-Dubaib M, Marzouk E, Osman S, Edrees H. Performance of MALDI biotyper compared with Vitek(™) 2 compact system for fast identification and discrimination of staphylococcus species isolated from bovine mastitis. MicrobiologyOpen 2016;5(6):1061-1070.
- Singhal N, Manish K, Pawan KK, & Jugsharan SV. MALDI-TOF mass spectrometry: an emerging technology for microbial identfication and diagnosis. Frontiers in Microbiology 2015;6:791.
- Elbehiry A, Marzouk E, Hamada M, Al-Dubaib M, Alyamani E, Moussa IM, AlRowaidhan A, Hemeg HA. Application of MALDI-TOF MS fingerprinting as a quick tool for identification and clustering of foodborne pathogens isolated from food products. New Microbiol 2017; 40(4):269-278.
- Werner G, Fleige C, Fessler AT, Timke M, Kostrzewa M, Zischka M, Peters T Kaspar H, Schwarz S. Improved identification including MALDI-TOF mass spectrometry analysis of group D streptococci from bovine mastitis and subsequent molecular characterization of corresponding Enterococcus faecalis and Enterococcus faecium isolates. Vet Microbiol 2012; 160(12):162-169.
- Dobranic V, Kazazic S, Filipovic I, Mikulec N, Zdolec N. Composition of raw cow’s milk microbiota and identification of enterococci by MALDI-TOF MS – short communication. Veterinarski arhiv 2016; 86(4):581-590.
- Brittain-Long R, Nord S, Olofsson S, Westin J, Anderson LM, Lindh M. Multiplex real-time PCR for detection of respiratory tract infections. J Clin Virol 2008; 41(1):53-56.
- Spichler A, Hurwitz B, Armstrong D, Lipsky B. Microbiology of diabetic foot infections: from Louis Pasteur to crime scene investigation. BMC Medicine 2015; 13:2.
- Edmonds ME, Foster AM, Sanders LJ. Aprctical manual of Diabetic foot care.2004; ISBN 14051-0715-4.
- Mantzoros CS. Obesity and diabetes. Human press inc. 2007; ISBN 978-1-59259-985-1.
- Raja NS. Microbiology of diabetic foot infections in a teaching hospital in Malaysia: a retrospective study of 194 cases. J Microbiol Immunol Infect 2007; 40(1):39-44.
- Edmonds M. The treatment of diabetic foot infections: focus on ertapenem. Vascular Health and Risk Management 2009; 5:949–963.
- Mathangi T and Prabhakaran P. Prevalence of Bacteria Isolated from Type 2 Diabetic Foot Ulcers and the Antibiotic Susceptibility Pattern. Int J Curr Microbiol App Sci 2013; 2(10):329-337.
- Roudbary M, Razi F, Mohammadi SR, Annabestani Z, and Esfahani EN. Distribution of mycotic and bacterial infections among the patients with diabetic foot ulcers referred to diabetes and metabolic disorder clinic, Tehran, Iran 2011-2012. European Journal of Experimental Biology 2014; 4(4):20-25.
- Sousa Lages A, Oliveira P, Costa e Silva N, Guelho D, Cardoso L, Vicente N , Martins D, Oliveira D, Ventura M & Carrilho F. Microbiological profile in diabetic foot infections: identification and susceptibility profile of bacteria isolated in 5 years in a Portuguese tertiary care hospital. Endocrine Abstracts 2016; 41:446.
- Dupont C, Sivadon-Tardy V, Bille E. Identification of clinical coagulase-negative staphylococci, isolated in microbiology laboratories, by matrix-assisted laser desorption/ionization-time of flight mass spectrometry and two automated systems. Clin Microbiol Infect 2012; 16:998-1004.
- Carbonnelle E, Beret JL, Cotyn S, Quesne G, Berche P, Nassif X, & Ferroni A. Rapid identfication of Staphylococci isolated in clinical microbiology laboratories by matrix-assisted laser desorpton ionization-tme of fiight mass spectrometry. Journal of Clinical Microbiology 2007; 45:2156–2161.
- Benagli CV, Rossi M, Dolina M, Tonolla O, Petrini. Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry for the identification of clinically relevant bacteria. PLoS ONE 2011; 6:e16424.
- Griffin PM, Price GR, Schooneveldt JM, Schlebusch S, Tilse MH, Urbanski T, Hamilton B, Venter D. Use of matrix-assisted laser desorption ionization-time of flight mass spectrometry to identify vancomycin-resistant enterococci and investigate the epidemiology of an outbreak. J Clin Microbiol 2012; 50(9):2918-31.
- CLSI (Clinical and Laboratory Standards Insttute). Performance Standards for Antmicrobial Disk and Diluton Susceptbility Tests for Bacteria Iso- lated from Animals”, Approved Standards – (4th ed.) 2016; CLSI document VET01-A4, Wayne, PA, USA.
- Lartigue MF, Héry-Arnaud G, Haguenoer E, Domelier AS, Schmit PO, van der Mee-Marquet N, Lanotte P, Mereghetti L, Kostrzewa M, Quentin R. Identification of Streptococcus agalactiae isolates from various phylogenetic lineages by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 2009; 47(7):2284-7.
- Barreiro JR, Ferreira CR, Sanvido GB, Kostrzewa M, Maier T, Wegemann B, dos Santos MV. Short communicaton: identfi- caton of subclinical cow mastts pathogens in milk by matrix-assisted laser desorpton/ionizaton tme-of-fiight mass spectrometry. Journal of Dairy Science 2010; 93:5661–5667.
- Silva MB, Ferreira F, Garcia LN, Silva-Carvalho MC, Botelho LA, Figueiredo AM, Vieira-da-Mota O. An evaluaton of ma- trix assisted laser desorpton ionizaton tme-of-fiight mass spectrom- etry for the identficaton of Staphylococcus pseudintermedius isolates from canine infectons. Journal of Veterinary Diagnostc Investigaton 2015; 27:231–235.
- Pavlovic, M., I. Huber, R. Konrad, U. Busch. Application of MALDI-TOF MS for the identification of food borne bacteria. Open Microbiol J 2013; 7:135-141.
- Quintela Baluja MK, Böhme IC, Fernández-no S, Morandi ME, Alnakip S, Caamaño-Antelo J, Baros-Velázquez P, Calo-Mata. Characterization of different food-isolated Enterococcus strains by MALDI-TOF mass fingerprinting. Electrophoresis 2013; 34:2240-2250.
- Chabros L, Robert T, Kuthan, Anna Sawicka-Grzelak, Gra|yna MBynarczyk. Comparison of identification methods of vancomycin-resistant enterococci in MALDI-TOF MS, Vitek 2 and API 20 STREP. Post’py Nauk Medycznych, t. XXVIII, 2015; 4:201.
- Chen YL, Lin YC, Chen YS, Chen SC, Liu YM, & Tseng IL. Characterization of predominant molecular paterns of Burkholderia pseudomallei in Taiwan. Transactons of the Royal Society of Tropical Med- icine and Hygiene 2013; 107:165–169.
- Sedgley C, Nagel A, Dahlén G, Reit C, Molander A. Real-time quantitative polymerase chain reaction and culture analyses of Enterococcus faecalis in root canals. J Endod 2006; 32:173-177.
- Rocas IN, Jung IY, Lee CY, Siqueira JF. Polymerase chain reaction identification of microorganisms in previously root-filled teeth in a South Korean population. J Endod 2004; 30:504-508.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.