ISSN: 0973-7510
E-ISSN: 2581-690X
Mycoplasma bovis is one of the important pathogens in mycoplasma types that cause disease in cattle. The young calves from one to four months of age are most likely to develop pneumonia caused by M. bovis. In this study, we isolated M. bovis from tracheal swabs of cattle which showed respiratory symptoms. A total of about 100 tracheal swab samples were collected from cattle in Kafer El-Shikh slaughterhouse, Egypt. The collected samples from cattle were between 3-12 months of age. Mycoplasma bovis was identification in tracheal swab samples by using 16S rDNA gene sequencing and biotyping by using rep-PCR, respectively. The microbiological method could not give positive results, while the PCR showed that M. bovis infections were positive in 16 different cattle samples with about 16%. The partial sequences of the 16S rRNA genes of the Mycoplasma isolates were obtained and phylogeny tree showed that Sixteen Mycoplasma isolates were identified into Mycoplasma bovis. the similarity to Mycoplasma bovis MYC 84, M. bovis L22 and M. bovis MYC 76 was 100, 99 and 95%, respectively. The ten Rep-PCR primers produced about 139 fragments, 53.3% of them consider as monomorphic and 46.7% of them consider as polymorphic bands. According to genetic similarity and intraspecies differentiation, the sixteen Mycoplasma isolates were grouped into two main different clusters with about 60% genetic similarity in genetics dendrogram. These results suggest that PCR technique is a specific molecular detection technique identified to determine Mycoplasma and It is easy and fast methods to detect and isolate infected animals.
Genotyping, 16S rDNA gene, rep-PCR, phylogeny and Mycoplasma bovis.
Mycoplasma bovis is one of the most pathogenic agents in the Mycoplasma species that cause disease in cattle. Mycoplasma bovis-associated pneumonia occurs in cattle, including dairy and beef calves, beef cattle after arrival at a feedlot, and adults at any age (Maunsel et al., 2011). Mycoplasma bovis is a particularly important cause of calf pneumonias (Nicholas, 2004; Soehnlen et al., 2012). Especially young calves under 4 months of age are at increased risk for pneumonia caused by M. bovis (Nicholas, 2004). Mycoplasma bovis infections can be explained as chronic and polymicrobial (Nicholas and Ayling, 2003; Arcangioli et al., 2008). Animals can be infected via the respiratory system (Nicholas, 2004). Respiratory tract and nasal secretions are important for epidemiology of infection (Maunsel et al., 2011; Caswell and Archambault, 2008). Beside this, the importance of aerosols in calf-to-calf transmission of M. bovis is unknown but recently Maunsell et al., (2011), reported that M. bovis has been isolated from air in shed containing diseased calves and calves may be experimentally infected by inhalation of M. bovis. Infected cattle spread M. bovis to environment through respiratory secretions for many months as reservoir (Nicholas and Ayling, 2003). The severity of pneumonia could be varied depending on the maintenance and environmental conditions. The effects of antibiotics and vaccines are not known to have negligible effects in calves (Soehnlen et al., 2012). Control of infection is difficult and economic loses is imminent (Sachse et al., 1993; Thomas et al., 2002). The first condition is to ensure a high level of hygiene in the prevention of M. bovis infection. Because of non-specific clinical manifestations, and a wide range of variation in the epizootiology and pathogenesis, all M. bovis infections are have to be had specific diagnostic and control procedures (Thomas et al., 2002). Mycoplasma bovis infections could be diagnosed by bacteriological culture and serological methods (Ghadersohi et al., 1997; Fulton, 2009). Nevertheless, these methods are time consuming and false-negative results could be common (Ghadersohi et al., 1997). Serological methods are less time consuming than the cultural methods and also more samples can be investigated. Recently, new molecular identification methods were improved and used in diagnosis of M. bovis infections worldwide by several authors (Hayman and Hirst, 2003; Sung et al., 2006; Mohamed et al., 2011). PCR is much shorter in comparison to the conventional culture method for the identification of M. bovis infection (Sung et al., 2006). In Egypt there are few reports about the M. bovis infections in cattle (Sung et al., 2006). In this study we investigated M. bovis from tracheal swabs of cattle that showed respiratory symptoms for the situation of M. bovis in Egypt. Also we aimed to compare the efficiency of molecular and bacteriological methods for detection of M. bovis infections.
Isolation of Mycoplasma
The tracheal swabs were randomly collected from about 100 cattle in Kafer El-Shikh slaughterhouse, Egypt from October, 2017 to September, 2018. The swabs were cultured in pleuropneumonia-like organisms (PPLO) broth media at 37°C for 24 h., and then inoculated in PPLO media agar plate at 37°C for 1 week to examine the presence of Mycoplasma colonies (Evans et al., 2009).
DNA extraction
Genomic DNA was extracted from the Mycoplasma sample by using bacterial DNA extraction kit (Promega, USA), according to the manufacturer’s instructions. DNA samples were stored at -20°C until used.
Amplification of 16S-rRNA gene
For 16S-rRNA gene analysis, two pair primers were used for molecular detection of mycoplasma isolates. PCR amplification of 16S-rRNA primer was designed according to (Lauerman et al., 1995) with about 425 bp length. Universal primer was used to target the conserved region of 16S rDNA in Mycoplasma and Thermal profiles were done according Mohamed et al., (2018).
Sequencing of 16S-rRNA gene
About 425 bp 16S rDNA fragments were purified using QIAquick PCR purification kit (QIAGEN, Valencia, CA, USA) according to the manufacturer’s instructions and sequenced with the same primers using the sequencer (Gene analyzer 3121) by Macrogen co., Korea. The Mycoplasma 16S-rDNA sequences obtained were then aligned with known 16S-rDNA sequences in Genbank using the basic local alignment search tool (BLAST) at the National Center for Biotechnology Information, and percent homology scores were generated to identify Mycoplasma species. The deduced sequence was aligned using Molecular Evolutionary Genetics Analysis (MEGA) version 7.10 and drawn phylogeny tree (Kumar et al., 2016).
Rep-PCR analysis
For repetitive sequence analysis, PCR conditions for Mycoplasma isolates in the present investigations were standardized. Ten repetitive sequence primers (BOX-A1, (GTG)5, Rep-2, Rep-8, Rep-10, Rep-12, Rep-16, Rep-18, Rep-28 and Rep-29), were used to amplify genomic DNA of the Mycoplasma isolates according to (Hassan et al. 2014; Gaber et al., 2015). For DNA amplification, the C1000TM Thermo Cycler Bio-Rad, Germany, was programmed under the conditions involving denaturation at 94°C for 5 min; 30 cycles of denaturation at 94°C for 1 min, primer annealing at 52°C for 45 Sec and primer extension at 72°C for 2.5 min; final extension step at 72°C for 10 min.
Data analysis
In order to determine the genetic relationship among studied bacteria, Rep-PCR data were scored for presence (1) or absence (0) of the bands using Gene Tools software from Syngene. A simple matching coefficient was estimated by means of the Jaccard’s coefficient to construct a similarity matrix. Cluster analysis and dendrogram were produced on the basis of the unweighted average pair group method (UPGMA) using the NTSYS-PC Statistical Package (Rohlf, 2000).
Bacteriological method
The study was conducted in two stages. Traditional culture method and PCR method to identify mycoplasma colonies. Initially, a total of about 100 samples were taken from tracheal swabs of cattle animals. With culture method, mycoplasma isolation was not achieved from any of these samples in a tube with PPLO liquid culture medium and/or PPLO solid culture media. Traditionally, culture methods was used to identify mycoplasma colonies on agar medium, but no positive results obtained from this methods (Atalla et al., 2015; McAuliffe et al., 2004). So, we used PCR with universal primers (Delgado et al., 2001; Chen et al., 2014), for identification of mycoplasma from swabs culture. This is in agreement with findings of other researchers who showed that PCR provides a rapid diagnosis and identification of mycoplasma, and also showed that PCR assay has several features that simplify the diagnosis of mycoplasma infections (Chen et al., 2014). To increase sensitivity of mycoplasma detection, PCR based on specific sequences of nucleotides has been used (Atalla et al., 2015). Multiplex PCR, PCR-RFLP also developed for mycolpasma detection and identification (Chen et al., 2014; Barghash et al., 2016).
PCR method
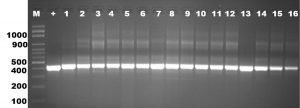
Mycoplasmal DNA was successfully extracted from cells in the liquid medium. These samples were tested using universal primer, 16 of 100 samples given PCR-products as positive results, including the sample from the tracheal swabs. The 16S rDNA PCR amplicon from the 16 positive samples was obtained, showing that they had the same length as Mycoplasma sp. (Figure 1), a single 425 bp PCR amplicon band. The 16S rDNA PCR products confirmed that is Mycoplasma. Then we used the 16S rDNA sequencing methods and rep-PCR methods to identify and characterize these Mycoplasma isolates.
Fig. 1. Analysis of PCR products from sixteen Mycoplasma isolates obtained by amplification with universal primer of 16S rDNA. Molecular-size was applied from lane 2 to 18 and a positive control in lane 1. The approximate sizes of the amplicons are given in 425 bp.
Sequencing of 16S-rRNA gene
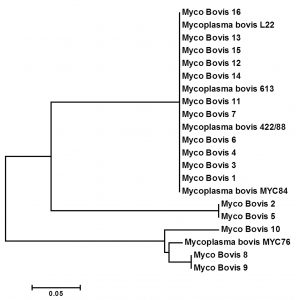
The partial sequences of the 16S rRNA genes of the Mycoplasma isolates were obtained, and Phylogenetic analysis of these sequences that deposited in the Gene Bank database were studied, together with related sequences in GenBank. Sixteen Mycoplasma isolates were identified into Mycoplasma bovis. For isolates Myco-Bovis 1, 3, 4 and 6 identified as Mycoplasma bovis and the similarity to Mycoplasma bovis MYC 84 was 100%. Moreover isolates Myco-Bovis 12, 13, 14, 15 and 16 were identified as M. bovis and it was 99% similarity to M. bovis 613 and M. bovis L22. On the other hand, isolates Myco-Bovis 8, 9 and 10 were identified as M. bovis and similarity to M. bovis MYC 76 was ranged from 95 to 98% (Figure 2). The 16S rDNA sequence data play an important role in classification of mycoplasmas as more species are described (Heldtander et al., 1998), because conventional methods provide less discriminatory information. The number of deposited 16S rRNA sequences from mycoplasmas is close to 100, and it well, therefore, is possible to determine the phylogenetic cluster and close relatives to a new mycoplasma. There is a constant debate concerning the construction of phylogenetic trees, and it is impossible to cover all aspects in this study. The 16S rDNA sequence of the mobile Mycolpasma strains deposited in GenBank was identical to that determined in this work. Ribosomal operons are of great relevance for the study of Mycoplasma evolution and phylogeny (Zarei et al., 2009). Sequencing of 16S rDNA has been widely used to re-construct phylogenetic relationships of microorganisms (Hassan and Ismail, 2014).
Fig. 2. Phylogenetic tree based on 16S rRNA sequences of mycoplasmas representing all groups and clusters. Representatives of the closely related genera Mycoplasma bovis. The tree was constructed by neighbor-joining from a distance matrix corrected by the one parameter nucleotide substitution model by using the MEGA 7.1 program.
Rep-PCR analysis
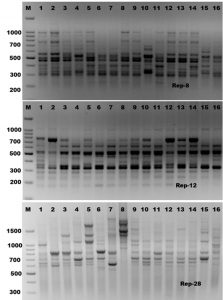
Rep-PCR is a new typing method that differentiates microbes by using primers complementary to interspersed repetitive consensus sequences that enable amplification of diverse-sized DNA fragments consisting of sequences between the repetitive elements (Healy et al., 2004, Hassan and Belal, 2016). The Rep-PCR fingerprinting method, utilizing repetitive sequence oligonucleotides, is particularly a powerful tool for genetic studies and it is useful as a screening genotyping method (Healy et al., 2004). Rep-PCR can generate various fingerprint patterns with unlimited number of fragments (Hassan et al., 2014). In the present study, ten Rep-PCR primers were used for estimating of genetic diversity of Mycoplasma bovis isolates. Rep-PCR reactions were performed in duplicate and all amplification products were found to be reproducible (Figure 3 and Table 2). The Rep-PCR results using primer (GTG)5 has showed a total of 18 bands sized ranged from 150-2800 bp long in all Mycoplasma isolates. Six common bands were observed in all isolates which exhibited about 33.4% monomorphism, while the other twelve fragments have showed 66.6% polymorphism (Table 2). In case of (Box-A1) primer, a total of fourteen fragments have showed 50% polymorphism among the sixteen Mycoplasma bovis isolates. The molecular size of the amplicon products ranged from 150-2300 bp long. The ten Rep-PCR primers produced about 139 fragments, 74 of them consider as monomorphic bands with about 53.3%, and other 65 fragments consider as polymorphic bands with about 46.7%. According to genetic similarity and intraspecies differentiation, the sixteen Mycoplasma isolates were grouped into two main different clusters with about 60% genetic similarity. M. bovis-15 and M. bovis-16 isolates were found to be alone in the first cluster. While, the second cluster was divided into two sub-clusters. The first sub-cluster contained M. bovis-1 isolate only, and the other Mycoplasma isolates were grouped in the second sub-cluster (Figure 4). Rep-PCR technique was proved to be useful genetic markers used for fingerprinting of Mycoplasma strains isolated from cattle. Although major bands from Rep-PCR reactions are highly reproducible, minor bands can difficult to repeat due to repetitive sequence priming nature of this PCR reaction and potential confounding effects associated with co-migration with other markers (Patrizia and Paola, 2003; Manakant and Tanaya, 2012; Hassan and Belal, 2016). Because AP-PCR has its limitations for widespread use, another more reproducible PCR method should be considered. The Rep-PCR has been described for the molecular genotyping of bacteria (Alharthi, et al., 2016). It also generates strain-specific DNA fragments when DNA is used as an amplification template (Hassan et al., 2014: Gaber et al., 2015). Therefore, the use of multiple primer sets in rep-PCR analysis can be used as a rapid method for preliminary biotyping of Mycoplasma strains.
Fig. 3. Rep-PCR profile of 16 Mycoplasma isolates generated with three repetitive sequence primers, rep-8, rep-12 and rep-28, respectively. First lane on each panel is 100 bp molecular weight markers.
Fig. 4. Dendrogram analysis among sixteen Mycoplasma bovis isolates based on the ten of rep-PCR primers.
Table (1):
Names and sequences of the rep-PCR primers used in this study.
Primer |
5¢®3¢ sequence |
|---|---|
BOX-A1 |
CTACGGCAAGGCGACGCTG |
GTG5 |
GTGGTGGTGGTGGTG |
Rep-2 |
GAGAGAGAGAGAGAGAA |
Rep-8 |
GACAGAGAGAGAGAGAG |
Rep-10 |
CAGCACACACACACACA |
Rep-12 |
AGAGAGAGAGAGAGAGC |
Rep-16 |
TCTCTCTCTCTCTCTCC |
Rep-18 |
ACACACACACACACACG |
Rep-28 |
AGAGAGAGAGAGAGAGT |
Rep-29 |
GAGAGAGAGAGAGAGAT |
Table (2):
Polymorphic bands of each genetic primers and percentage of polymorphism in sixteen Mycoplasma bovis isolates based on the ten of rep-PCR primers.
Primers |
Total Bands |
No. of Monomorphic Bands |
No. Polymorphic Bands |
% Monomorphic bands |
% Polymorphic bands |
|---|---|---|---|---|---|
BOX-A1 |
18 |
6 |
12 |
33.4 |
66.6 |
GTG5 |
14 |
7 |
7 |
50.0 |
50.0 |
Rep-2 |
16 |
9 |
7 |
56.2 |
45.8 |
Rep-8 |
14 |
8 |
4 |
57.1 |
42.9 |
Rep-10 |
12 |
10 |
2 |
83.4 |
16.6 |
Rep-12 |
14 |
10 |
4 |
71.4 |
28.6 |
Rep-16 |
13 |
7 |
6 |
53.8 |
46.2 |
Rep-18 |
13 |
8 |
5 |
61.5 |
38.5 |
Rep-28 |
14 |
6 |
8 |
42.9 |
57.1 |
Rep-29 |
11 |
3 |
8 |
27.3 |
72.7 |
Total |
139 |
74 |
65 |
Conflict Of Interest
The authors declare that there is no conflict of interest.
- Alharthi, A. A., Gaber, A. and Hassan, M. M. Molecular Characterization of mecA and SCCmec Genes in Pathogenic Staphylococcus spp. Collected from Hospitals in Taif Region, KSA. Biotechnology, 2016; 15(1): 26-34.
- Arcangioli MA, Duet A, Meyer G, Dernburg A, Bezille P, Poumarat F, Le Grand D: The role of Mycoplasma bovis in bovine respiratory disease outbreaks in veal calf feedlots. Vet J, 177, 89-93, 2008.
- Caswell JL, Archambault M: Mycoplasma bovis pneumonia in cattle. Anim Health Res Rev, 8 (2): 161-186, 2008.
- Fulton RW: Bovine respiratory diseases research (1983- 2009). Anim Health Res Rev, 10(2): 131-139, 2009.
- Gaber, A., Hassan, M. M.., El-Desoky, S. E. and Attia, O. A. In vitro Antimicrobial Comparison of Taif and Egyptian Pomegranate Peels and Seeds Extracts. J App Biol Biotech., 2015; 3 (02): 012-017.
- Ghadersohi A, Coelen RJ, Hirst RG: Development of a specific DNA probe and PCR for the detection of Mycoplasma bovis. Vet Microbiol, 56, 87-98, 1997.
- Hassan M. M. and Belal B. E. (2016). Antibiotic resistance and virulence genes in Enterococcus strains isolated from different hospitals in Saudi Arabia, Biotechnol. Biotechnol. Equip., 30(4): 726-732.
- Hassan, M. M., Gaber,A. and El-Hallous, E.I. Morphological and Molecular characterization of some Trichoderma spp. from Egyptian soil. Wulfinia, 2014b; 21: 80-96.
- Hayman B, Hirst R: Development of a semi-nested PCR for the improved detection of Mycoplasma bovis from bovine milk and mucosal samples. Vet Microbiol, 91, 91-100, 2003.
- Healy, H., Reece, K., Walton, D., Huong, J., Shah, K., Kontoyiannis, D.P. Identification to the Species Level and Differentiation between Strains of Aspergillus Clinical Isolates by Automated Repetitive-Sequence-Based PCR. J. Clin. Microbiol., 2004; 42: 4016-4024.
- Karahan M, Kalin R, Atil E, Çetinkaya B: Detection of Mycoplasma bovis in cattle with mastitis and respiratory problems in eastern Turkey. Vet Rec, 166, 827-829, 2010.
- Manakant, I. and Tanaya, K. Antibiotics resistance and RAPD-PCR typing of multidrug resistant MRSA isolated from bovine mastitis cases in Thailand. Sci. Asia., 2012; 38: 30-35.
- Maunsel FP, Woolums AR, Francoz D, Rosenbuch RF, Step DL, Wilson DJ, Janzen ED: Mycoplasma bovis infections in cattle. J Vet Intern Med, 25, 772-783, 2011.
- Nicholas RAJ, Ayling RD: Mycoplasma bovis: Disease, diagnosis, and control. Res Vet Sci, 74, 105-112, 2003.
- Nicholas RAJ: Recent developments in the diagnosis and control of mycoplasma infections in cattle. WBC Congress, Quebec, Canada, 2004. www.ivis.org
- Ongor H, Kalýn R, Karahan M, Cetinkaya B, McAuliffe L, Nicholas RAJ: Isolation of Mycoplasma bovis from broiler chickens in Turkey. Avian Pathol, 37 (6): 587-588, 2008.
- Patrizia, S. and Paola, M. (2003). Evaluation of Repetitive Element Sequence based PCR as a Molecular Typing Method for Clostridium difficile. J. Clin. Microbiol., 41:2454-2457.
- Sachse K, Pfützner H, Hotzel H, Demuth B, Heller M, Berthold E: Comparison of various diagnostic methods for the detection of Mycoplasma bovis. Rev Sci Tech Off Int Epiz, 12 (2): 571-580, 1993.
- Soehnlen NK, Aydin A, Murthy KS, Lengerich EJ, Hattel AL, Houser BA, Fenton GD, Lysczek HR, Burns CM, Townsend AM, Brooks JW, Wolfgang DR, Jayarao BM: Epidemiology of Mycoplasma bovis in Pennsylvania veal calves. J Dairy Sci, 95, 247-254, 2012.
- Sung H, Kang SH, Bae YJ, Hong JT, Chung CB, Lee CK, Song S: PCR-based detection of Mycoplasma species. J Microbiol, 44, 42-49, 2006.
- Thomas A, Dizier I, Trolin A, Mainil J, Linden A: Comparision of procedures of isolating pulmonary Mycoplasmas in cattle. Vet Res Commun, 26(5): 333-339, 2002.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.