ISSN: 0973-7510
E-ISSN: 2581-690X
A highly efficient and reproducible tissue culture regeneration system is an important prerequisite for achieving successful transformation. However, most indica rice varieties remain recalcitrant to genetic transformation and require refinement of the tissue culture protocol for the generation of embryogenic calli or regeneration of fertile plants from transformed cells. In this study, standardized regeneration system for indica rice cultivars was carried out. Further the expression of miR164c was investigated in the different rice genotypes. The expression of miR164c was proportional to the percentage of regeneration of the rice genotypes. These results provide interesting insights that will significantly improve our understanding of direct regeneration of rice from callus tissue.
miRNA, tissue culture, Rice regeneration, Genotypes.
In vitro tissue culture is widely applied for experimental breeding and biotechnological and genetic manipulation of plants in basic and applied research. Plants can be readily regenerated from cultured tissue; this characteristic is termed totipotency and is a robust tool for genetic engineering (Larkin and Scowcroft, 1981). Although tissue culture techniques are well known with respect to monocotyledonous plants, Graminae species are recalcitrant during in vitro manipulation and hence cannot be cultured easily. Moreover, the competency of tissues or cells for regenerating a whole plant is a prerequisite of in vitro culture.
Fujiwara and Ojima (1955) cultured excised roots and immature embryos (Amemiya et al., 1956, Li-na et al., 2010) for the in vitro culture of rice. Furthermore, different explants were cultured in different nutrient media augmented with different concentrations of phytohormones (Vasil and Vasil, 1982, Vasil, 1988, and Pipatpanukul et al., 2004). As a result, rice became the first cereal that was regenerated into a whole plant (Vasil, 1983). Strikingly, the tissue culture of the japonica variety of rice showed a higher callus yield and frequency of regeneration than those of indica (Abe and Futstuhara, 1986).
Embryogenesis is initiated by altered development during the in vitro regeneration of plant tissues effectuated by differential expression of genes in somatic cells. However, only a few such molecular pathways have been established in rice. Thus, a map-based cloning strategy was utilized to isolate a major quantitative trait locus (QTL) that encoded a ferredoxin-nitrite reductase, which estimated the regeneration ability in rice (Nishimura et al., 2005). This regeneration ability was enhanced by introgression of QTL (Takeuchi et al., 2000 and Kwon et al., 2001& 2002), indicating the requirement of key loci or genes. Oryza sativa L., or rice, has two ecotypes, indica and japonica. Hitherto, plant regeneration has been reproducible in japonica rice. Although a few indica varieties have also been reported to be regenerated, this ecotype is primarily recalcitrant to genetic manipulation (Ignacimuthu et al., 2000). Both rice cultivars are regenerated as follows. Briefly, 2,4-D medium is used for the inoculation of dehusked mature seeds, which helps in the formation of embryogenic calli from the scutellum within 3-4 weeks. The culture is supplemented with auxin and cytokinin and incubated in the light. Subsequently, the calli differentiate into green sectors that later form into shoot bud primordia. Additionally, plant regeneration and genotype-dependent variations in in vitro callus formation have been detailed (Ge et al., 2006). The transcripts that are produced by members of the MIR gene family are processed to the identical or almost identical mature miRNA molecules. Different members of the MIR gene family are expressed in a developmental and tissue-specific manner and in response to various biotic and abiotic stimuli (Zhao et al., 2007, 2011; Moldovan et al., 2010; Kruszka et al., 2014).
Present study was aimed to standardize the callus induction and regeneration protocols for the selected indica rice varieties. Additional experiments were carried out to identify the expression pattern of miRNA 164C key molecular regulator underlying the regeneration of rice plants.
Plant materials used
Rice varieties Pusa Basmati 1, IR64, IRBB60, N22, Lalat, and Basmati 370 were used to carry out tissue culture experiments.
Establishment of culture and explant preparation
The dehusked rice seeds were washed with sterile distilled water to remove the dust and other visible floating particles, followed by surface sterilization with 70% (v/v) ethanol 3 min, rinsed with sterile distilled water 3 times, resterilized with 2% NaOCl for 20 min, and finally rinsed again with sterile distilled water 5 times.
Subsequently, approximately 20–25 seeds were dried using sterile filter papers and inoculated on callus induction medium such that half the embryo was in contact with the media while the other half was exposed to air. The seeds plated on Petri plates (90 mm diameter x 16 mm height) were incubated in either the dark or light at 26°C for 21 days.
In vitro regeneration of plantlets from calli
The diverse combinations of auxin and cytokinins in the MS basal medium would increase the efficiency of the in vitro regeneration of rice varieties. The cytokinin BAP (3 mg/L), the synthetic auxin naphthalene acetic acid (NAA) (0.5 mg/L), sucrose (30 g/L), phytagel (0.4%), casein hydrolysate (500 mg/L), and L-proline (500 mg/L) at concentrations comprised the media, which were adjusted for pH and sterilized.
The embryogenic calli were placed in regeneration media and incubated in the dark at 26 ± 2°C for 4-5 days and under fluorescent light at the same temperature for 3-4 weeks. The regeneration efficiency was calculated as follows:
![]()
RNA isolation
Dehusked and sterilized rice seeds were grown on 2,4-D medium for 2 weeks. Subsequently, the calli were transferred to the regeneration medium. The callus and regeneration tissue samples were collected and pooled from four independent experiments and stored at -80°C until RNA isolation. Total RNA was isolated using the SpectrumTM Plant Total RNA Kit according to the manufacturer’s instructions (Sigma, USA). DNase treatment of the samples removed any remaining trace DNA. The purity of the RNA was evaluated on 1.2% denatured agarose gels and quantified by a NanoDrop 1000 Spectrophotometer (Thermo Scientific, USA).
Expression analysis using semiquantitative RT-PCR
After subculturing once on the maintenance medium for 15 days, the embryogenic calli were transferred to the regeneration medium to initiate differentiation and harvested after 14 days. Total RNA was extracted, and 5 µg was synthesized into cDNA using the Superscript III cDNA Synthesis Kit (Invitrogen, USA). RT-PCR was performed in a 20 µL reaction for the selected genes using Kappa PCR MasterMix (Kappa Biosystems, USA). The primers (Sigma, USA) are listed in Table 1. For miRNA primers were designed according to Wan et al., 2010. PCR was performed as follows: initial denaturation at 94°C for 2 min; 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72ºC for 30 s; and a final extension at 72°C for 5 min. The PCR products were analyzed by 2.5% agarose gel electrophoresis, and images were captured using a gel documentation system (Alpha Innotech, USA).
Table (1):
List of primers used in the study.
S.No |
Primer Name |
Sequence |
|---|---|---|
1 |
U6 forward |
5′ CGATAAAATTGGAACGATACAGA3′ |
2 |
U6 reverse |
5′ ATTTGGACCATTTCTCGATTTGT3′ |
3 |
Mir164c RT |
5′ CACCGTTCCCCGCCGdUCGGTGGCACGT3′ |
4 |
Mir164c qrt F |
5′ CCCGCCTGGAGAAGCAGG3′ |
5 |
Mir164c qrt R |
5′ GCCGTCGGTGTGCACGTAC3′ |
Most indica rice varieties are recalcitrant to hereditary control. Accordingly, enhanced tissue culture is a standard practice for embryogenic calli or for the recovery of fruitful plants from a changed cell. Although MS medium is widely used, factors such as explant, carbon source, development controllers, natural enhancements, and gelling operators are necessary for callus growth and plant recovery, particularly in indica rice varieties. In addition, a widespread culture medium is not characterized for a random genotype/assortment. Therefore, in this study, we refined the medium by changing the carbon source and expanding the development controllers, amino acids, and gelling operator to produce embryogenic calli for indica rice cultivars.
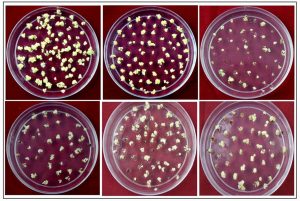
Seed-derived embryogenic calli were transferred to the regeneration medium and maintained under fluorescent light. Consequently, small shoots were produced within 10 days, and after 2-3 weeks, calli were observed to induce a large number of green shoots (Figure 1 and Table 2). The mean frequency of regeneration was found to be highest 82% in the case of Pusa Basmati 1 and lowest 34.3 in lalat variety. Based on the above findings, it can be deduced that an efficient regeneration system is imperative before undertaking any transformation study. A majority of regeneration studies have been conducted in japonica because indica cultivars are not conducive to tissue engineering (Pandey et al., 1994, Seraj et al., 1997 and Nishimura et al., 2005). Additionally, the optimal media composition and culture conditions for efficient plant regeneration are currently under intensive research focus (Khanna and Raina, 1997, Datta et al., 1992, Kishore et al., 1999 and Lin and Zhang, 2005).
Fig. 1. Rice Regeneration images of different indica genotypes A)Pusa basmati 1, B) N22, C) Lalat, D) IRBB60, E) Basmati 370, F) IR64
Table (2):
Percentage of Regeneration in different indica rice genotypes.
Variety |
% of regeneration |
|---|---|
IRBB60 |
59.4±1.84 |
IR64 |
64.2±0.95 |
N22 |
72.3±1.91 |
Basmati 370 |
73±1.78 |
Pusa Basmati 1 |
82±1.97 |
Lalat |
34.3±1.8 |
Values are means of five replications. ± is SE
The present results suggest that the regeneration of rice is dependent on the type of calli and the concentrations of auxin and cytokinin in the media (Jain, 1997, Saharan et al., 2004, Aygun and Dumanoglu, 2015, Saha et al., 2017, Bekircan 2018 and Stevens and Pijut, 2018). The strategies for improving the frequency of plant regeneration in rice have steadily evolved for two decades (Kyozuka et al., 1988 and Datta et al., 1992). Reportedly, partial desiccation of calli is beneficial for embryogenesis and regeneration in several plant species, including rice.
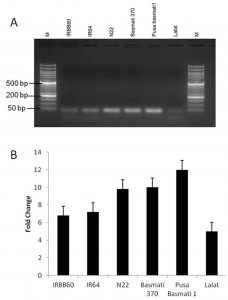
MIRNA (MIR) genes play a vital role in the regulation of the expression of the transcription factor miRNAs. miRNAs are single-stranded RNA molecules 21–24 nucleotides long that regulate the expression of the genes involved in plant development (Bartel, 2009 and Rubio-Somoza and Weigel, 2011). To study the role of miRNA 164c in rice regeneration semiquantitative RT-PCR and realtime PCR were used. RNA from different rice regenerating tissues of different genotypes were analysed for miRNA 164c expression levels. Subsequently, the expression of miR164c was found to be directly proportional to the regeneration percentage of different rice genotypes (Figure 2).
Fig. 2. expression of miRNA 164c in regenerating tissue. A) semiquantitative RT-PCR, B) Realtime PCR analysis
In conclusion the differential regulation of miR164c is effecting the percentage of rice regeneration from callus tissue. These results provide interesting insights that will significantly improve our understanding of direct regeneration in rice from callus tissue.
Conflict Of Interest
The authors declare that there is no conflict of interest.
- Abe, T., Futsuhara Y. Genotypic variability for callus formation and plant regeneration in rice. Theoretical and Applied Genetics, 1986; 72, 5-10.
- Amemiya, A., Akemine, H., Toriyama, K. Culture conditions and growth of immature embryo in rice plant (Studies on the embryo culture in rice plant 1). Bulletin of the National Institute of Agricultural Sciences, 1956; D6, 1-40.
- Aygun, A., Dumanoglu, H. In vitro shoot proliferation and in vitro and ex vitro root formation of Pyrusel aeagrifolia Pallas. Frontiers in Plant Science, 2015; 6, 225.
- Bartel, D. P. MicroRNAs: target recognition and regulatory functions. Cell, 2009; 136, 215–233.
- Bekircan, T., Yaþar, A., Yýldýrým, S., Sokmen, M., Sokmen, A. Effect of cytokinins on in vitro multiplication, volatiles composition and rosmarinic acid content of Thymus leucotrichus Hal. Shoots. 3 Biotech, 2018; 8, 180.
- Datta, S. K., Datta, K., Donn, G., Potrykus, I. Herbicide resistant indica rice plants from IRRI breeding lines after PEG- mediated transformation of protoplast. Plant Molecular Biology, 1992; 20, 619-692.
- Fujiwara, A., Ojima, K. Physiological studies of plant roots (part-1). Influence of some environmental conditions on growth of isolated roots of rice and wheat. Journal of Soil Science, 1955; 28, 9-12.
- Ge, X. J., Chu, Z. H., Lin, Y. J., Wang, S. P. A tissue culture system for different germplasms of indica rice. Plant Cell Reports, 2006; 25, 392–402.
- Ignacimuthu, S., Arockiasamy, S., Terada, R. Genetic transformation of rice: current status and future prospects. Current Science, 2000; 79, 186–195.
- Jain, R. K. Effects of some factors on plant regeneration from indica rice cells and protoplasts. A review. Indian journal of experimental biology, 1997; 35, 323-331.
- Khanna, H. K., Raina, S. K. Enhanced in vitro plantlet regeneration from mature embryo-derived primary callus of Basmati rice cultivar throughout modification of nitrate-nitrogen and ammonium-nitrogen concentrations. Journal of Biochemistry and Biotechnology, 1997; 16, 85-89.
- Kishore, P. B. K., Sangan, S., Naidu, K. P. Sodium, potassium, sugar, alcohol and proline mediated somatic embryogenesis and plant regeneration in recalcitrant rice callus. Plant Tissue Cult. Biotech. Emerging Trends. Proc. Symposium held at Hyderabad, India. 1999; 78-85.
- Kruszka, K., Pacak, A., Swida-Barteczka, A., Nuc, P., Alaba, S., Wroblewska, Z., Karlowski, W., Jarmolowski, A., Szweykowska-Kulinska, Z. Transcriptionally and post-transcriptionally regulated microRNAs in heat stress response in barley. Journal of Experimental Botany, 2014; 65, 6123–6235.
- Kwon, Y. S., Kim, K. M., Eun, M. Y., Sohn, J. K. Quantitative trait loci mapping associated with plant regeneration ability from seed derived calli in rice (Oryza sativa L.). Molecules and Cells, 2001; 11, 64-67.
- Kwon, Y. S., Kim, K. M., Eun, M. Y., Sohn, J. K. QTL mapping and associated marker selection for the efficacy of green plant regeneration in anther culture of rice. Plant Breeding, 2002; 121, 10-16.
- Kyozuka, J., Otoo, E., Shimamoto, K. Plant regeneration from protoplast of indica rice: genotype differences in culture response. Theoretical and Applied Genetics, 1988; 76, 887-890.
- Larkin, P. J., Scowcroft, W. R. Somaclonal variation. A novel source of variability from the cell cultures for plant improvement. Theoretical and Applied Genetics, 1981; 60, 197-214.
- Lin, Y. J., Zhang, Q. Optimizing the tissue culture conditions for high efficiency transformation of indica rice. Plant cell Reports, 2005; 23, 540-547.
- Li-na, Y., Xia, L., Dan, W. The comparison in tissue culture ability of mature embryo in different cultivars of rice. Agricultural Sciences in China, 2010; 9(6), 840-846.
- Moldovan, D., Spriggs, A., Yang, J., Pogson, B. J., Dennis, E. S., Wilson, I. W. Hypoxia-responsive microRNAs and trans-acting small interfering RNAs in Arabidopsis. Journal of Experimental Botany, 2010; 61, 165-177.
- Nishimura, A., Ashikari, M., Lin, S., Takashi, T., Angeles, E. R., Yamamoto, T., Matsuoka, M. Isolation of a rice regeneration quantitative trait loci gene and its application to transformation systems. Proceedings of the National Academy of Sciences of the United States of America, 2005; 102, 11940-11944.
- Pandey, S. K., Ramesh, B., Gupta, P. K. Study on effect of genotype and culture medium on callus formation and plant regeneration in rice (Oryza sativa L.). Indian Journal of Genetics, 1994; 54, 293-211.
- Pipatpanukul, T., Bunnag, S., Theerakulpisut, P., Kosittrakul, M. Transformation of indica rice (Oryza sativaL.) cv. RD6 mediated by Agrobacterium tumefaciens. Songklanakarin Journal of Science and Technology, 2004; 26, 1–13.
- Rubio-Somoza, I., Weigel, D. MicroRNA networks and developmental plasticity in plants. Trends in Plant Science, 2011; 16, 258–264.
- Saha, D., Bhavya, C., Ashok, T. H. Effect of genotypes and different concentration of growth regulator on callus induction and plant regeneration through anther culture of rice. Journal of Pharmacognosy and Phytochemistry, 2017; 6(6), 1354-1358.
- Saharan, V., Yadav, R. C., Yadav, N. R., Chapagain, B. P. High frequency plant regeneration from desiccated calli of indica rice (Oryza sativa L.). African Journal of Biotechnology, 2004; 3, 256-259.
- Seraj, Z., Islam, Z., Omar Faruque, M., Devit, Ahmed, S. Identification of the regeneration potential of embryo derived callus from various indica rice varieties. Plant Cell Tissue and Organ Culture, 1997; 48, 9-13.
- Stevens, M. E., Pijut, P. M. Rapid in vitro shoot multiplication of the recalcitrant species Juglansnigra L. In Vitro Cellular & Developmental Biology – Plant, 2018; 01-09.
- Takeuchi, Y., Abe, T., Sasahara, T. RFLP Mapping of QTLs influencing shoot regeneration from mature seed-derived calli in rice. Crop Science, 2000; 40, 245–247.
- Vasil, I. K. Isolation and culture of protoplasts of grasses. International Review of Cytology, 1983; 16, 79-88.
- Vasil, I.K. Progress in the regeneration and genetic manipulation of cereal crops. Nature Biotechology, 1988; 6, 397.
- Vasil, V. L., Vasil, I. K. Characterization of an embryogenic cell suspension culture derived from cultured inflorescence of Pennisetum americanum (Pearl millet,). American Journal of Botany. 1982; 69, 1441-1449.
- Zhao, B. T., Liang, R. Q., Ge, L. F., Li, W., Xiao, H. S., Lin, H. X., Ruan, K. C., Jin, Y. X. Identification of drought-induced microRNAs in rice. Biochemical and Biophysical Research Communications,2007; 354, 585–590.
- Zhao, M., Ding, H., Zhu, J. K., Zhang, F., Li, W. X. Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytologist, 2011; 190, 906–915.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.