ISSN: 0973-7510
E-ISSN: 2581-690X
Enterococci species are known commensals of the gastrointestinal flora; however, in recent years, they have emerged as important nosocomial pathogens that possess many virulence factors that are attributed to the pathogenesis of diseases caused by them. The study evaluated and compared the virulence factors of Enterococci isolated from fecal and clinical samples. From the obtained isolates, the clinical enterococcal isolates produced 35%, 20%, and 50%, and fecal isolates produced 23%, 13%, and 13% gelatinase, hemolysin, and biofilm, respectively. Biofilm production determined by the Congo Red agar, tube, and microtiter plate methods was 23%, 39%, and 49%, respectively. The sensitivity of the Congo Red agar and tube method compared to the microtiter plate method was 27% and 46%, respectively, whereas the specificity of both tests was 79%. This study showed that biofilm production plays a significant role in the pathogenesis of diseases caused by Enterococci. Detection of biofilm production using the microtiter plate method is more sensitive and specific than the Congo Red agar and tube method.
Biofilms, Enterococci, Hemolysin, Gelatinase, Virulence Factors
Enterococci, commensals of the genital tract, oral cavity, and gastrointestinal tract, have recently emerged as nosocomial pathogens that cause serious infections such as bloodstream infections (the incidence of which is steadily increasing), urinary tract infections (UTI), catheter-associated UTI (CAUTI), and intra-abdominal or intrapelvic abscesses.1-3 Among the Enterococcus species, E. faecalis is responsible for human enterococcal infections in 80–90% of cases, followed by E. faecium.3 Enterococci are intrinsically resistant to antibiotics due to the acquisition of genetic sequences responsible for drug resistance in other bacteria by transferring plasmids, transposons, or chromosome mutations.4-6
In addition to studies analyzing the trend of increased drug resistance in these organisms, several virulence factors have been studied. A few hemolysins, gelatinase, can form biofilms (enterococcal surface proteins). In contrast, other putative virulence factors, such as hyaluronidase, are not considered to be the disease-causing capability of Enterococcus strains.2 Several studies have reported the prevalence of Enterococci in India.7 However, few studies have focused on the prevalence of virulence factors in Enterococci. This study evaluated and compared the virulence factors of Enterococci isolated from fecal and clinical samples.
The study was conducted in the Department of Microbiology, R L Jalappa Hospital, and Research Center for over one year. Seventy-five clinical isolates of Enterococcus species were included in this study. The isolates were obtained from blood, sterile body fluids (cerebrospinal fluid, peritoneal fluid, and pleural fluid), urine, and pus/wound swabs. Routine bacteriological methods were used to isolate and identify Enterococcus species.8,9
Thirty isolates of Enterococcus species were collected from stool samples to compare virulence factors. The institutional ethics committee approved this study. The isolated clinical Enterococcus strains and commensal strains (from stool samples) of Enterococcus species were subjected to phenotypic methods to detect virulence factors. Hemolysin was detected using brain-heart infusion agar supplemented with 5% human blood. Gelatinase production was detected using peptone yeast extract agar containing 30 g/L gelatin. Biofilm production was detected using Congo Red agar, tube, and microtiter plate methods.2,3,10,11
Seventy-five Enterococcus clinical isolates and 30 fecal isolates were tested for virulence factors. Of the 75 clinical Enterococci, 41 (55%) were E. faecalis and 34 (45%) were E. faecium. In contrast, among the 30 fecal isolates of Enterococci, 60% were E. faecium and 40% were E. faecalis.
Table (1):
Gelatinase, hemolysin and biofilm production in pathogenic and fecal isolates.
Virulence factors |
Clinical isolates n = 75 (%) |
Fecal isolates n = 30 (%) |
P value |
|---|---|---|---|
Gelatinase |
26 (35%) |
7 (23%) |
0.2593 |
Hemolysin |
15 (2 %) |
4 (13.3%) |
0.4228 |
Biofilm |
35 (50%) |
4 (13.3%) |
0.0303 (<0.05) (statistically significant) |
Table 1 shows that out of 75 strains of Enterococci isolated from clinical samples, 26 (35%), 15 (20%), and 35 (50%) were gelatinase, hemolysin, and biofilm producers, respectively. In contrast, out of 30 fecal isolates of Enterococci, 7 (23%), 4 (13.3%), and 4 (13.3%) were gelatinase, hemolysin, and biofilm producers, respectively. Biofilm production was observed significantly more frequently in clinical isolates (p <0.05).
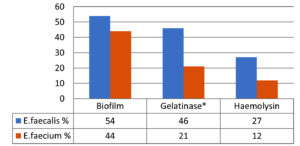
Figure 1 shows the virulence factors in Enterococcus species of the clinical isolates. In total, 54%, 46%, and 27% of isolates of E. faecalis were biofilm, gelatinase, and hemolysin producers. Among E. faecium isolates, 44%, 21%, and 12% were biofilm, gelatinase, and hemolysin producers, respectively. (* Gelatinase production: statistically significant, p = 0.0281)
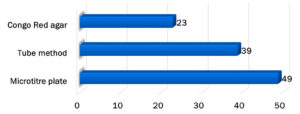
Figure 2 shows the biofilm detection using different methods. The microtiter plate method (49%) was more sensitive than the Congo Red agar (23%) and tube methods (39%). The sensitivity of the Congo Red agar and tube method compared to the microtiter plate method was 27% and 46%, respectively, whereas the specificity of both tests was 79%. Among the virulence factors, higher biofilm production was observed in Enterococci isolated from UTI 7 (70%), CAUTI 4 (50%), sepsis 5 (45%), postoperative wound infection 8 (44%), and diabetic wound infection 3 (43%) (Table 2).
Table (2):
Virulence factors and associated clinical conditions.
Clinical conditions (N) |
Biofilm production |
Gelatinase production |
Hemolysin production |
|---|---|---|---|
Urinary tract infection (UTI) (10) |
7 (70%) |
3 (30%) |
– |
Catheter-associated UTI (8) |
4 (50%) |
2 (25%) |
2 (25%) |
Sepsis (11) |
5 (45%) |
2 (18%) |
3 (27%) |
Post-op wound infection (18) |
8 (44%) |
7 (39%) |
3 (17%) |
Diabetic wound infection (7) |
3 (43%) |
2 (29%) |
3 (43%) |
Wound infection (12) |
5 (42%) |
8 (67%) |
3 (25%) |
Perforation (3) |
1 (33%) |
– |
1 (33%) |
Enterococci are emerging as important nosocomial pathogens due to their ability to acquire and spread genes responsible for antibiotic resistance. However, the role of other virulence factors of Enterococci in causing the disease cannot be neglected.2
Enterococci are well-adapted to areas with low redox potential, such as the oral cavity, gut, and genitourinary tract.9,12 Enterococci first adhere to specific host tissues, invade, and exert their pathogenic effects. In unfavorable environments, the expression of various enterococcal traits ultimately contributes to virulence.13
With the emergence of new virulence factors in Enterococci, they evolved like other pathogenic organisms. Antibiotic resistance has been the best studied in E. faecium. In contrast, virulence traits have been studied in E. faecalis. Increased virulence is assumed to be directly associated with increased antibiotic resistance. However, cost–benefit analyses have shown that virulence and antibiotic resistance are two completely different aspects of bacterial cells; hence, increased virulence may or may not be associated with increased antibiotic resistance in bacterial cells.13-15 Therefore, intrinsic virulence and antibiotic resistance play roles in the pathogenesis of diseases caused by Enterococci; however, in a complementary way.7,16
In our study, of the 75 clinical Enterococci isolates, the predominant species isolated was E. faecalis (55%), followed by E. faecium (45%). The distribution ratio of E. faecalis to E. faecium is similar to that reported in other studies.17-20
We studied biofilm formation, hemolysin, and gelatinase production in fecal and clinical isolates. We observed no significant differences in the production of hemolysin and gelatinase between the fecal and clinical isolates. However, for biofilm production, a statistically significant difference (P <0.05) was observed between the clinical and fecal isolates, similar to studies by Upadhyaya et al. and Jett et al.3,11 Nosocomial strains are known to develop different mechanisms of colonization and cause infection. One of these mechanisms is the production of biofilms that facilitate both surface adherence and invasion of host cells by the organism causing the infection. Our results concord with those of Ira et al., who reported that gelatinase production was statistically significant among clinical isolates.2
Gelatinase production was considerably higher than biofilm and hemolysin production in E. faecalis isolates (46%) than in E. faecium (21%) (p = 0.0281). Thus, biofilm production is responsible for more infections by E. faecalis than by E. faecium. Gelatinase and hemolysin production help Enterococci spread infection, thus increasing the severity of the infection. The production of gelatinase and hemolysin helps acquire and meet Enterococci’s nutritional needs. Our results are similar to those reported by Fernandes et al. and Sood et al.1,19
In this study, biofilm detection using the microtiter plate method was more sensitive (49%) than the Congo Red agar (23%) and tube methods (39%). In contrast, a study by Ruchi et al.20 showed that the Congo Red agar method was more sensitive (40%) for biofilm detection than the tube method (37%) and microtiter plate method (27%).
The microtiter plate method is preferred because it is easy to perform, cost-effective, and assesses the biofilm-forming capacity of an isolate both qualitatively and quantitatively. The subjective error in interpreting biofilm production findings was overcome using an enzyme-linked immunosorbent assay reader. Other methods, such as epifluorescence microscopy and microscopic biofilm formation assays, have been used to study the biofilm-forming abilities of bacteria. The Congo Red agar method is a reproducible and rapid technique that is sensitive to the added advantage of test colonies on the medium remaining viable. Moreover, subjective errors during reporting were observed more frequently with the tube method than with the Congo Red agar method.2,21
In this study, biofilm production was higher in clinical isolates than in fecal isolates. This likely plays a key role in the pathogenesis of diseases caused by Enterococci. These results are similar to those reported by Fernandes et al., Hemalatha et al., and Marothi et al.1,16,21
Enterococci have been associated with biofilm production on various types of indwelling devices, such as prosthetic heart valves, urinary catheters, and artificial hip prostheses, and this capability to produce biofilms is considered an important virulence factor of this organism.2
This study showed that biofilm production plays a significant role in the pathogenesis of diseases caused by Enterococci. Detection of biofilm production using the microtiter plate method is more sensitive and specific than the Congo Red agar and tube method. Clinical isolates from patients with UTI, CAUTIs, and septicemia showed higher rates of biofilm production than fecal isolates. Mechanisms or interventions to stop biofilm production could have added advantages in managing patients suffering from nosocomial infections caused by multidrug-resistant Enterococci.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
SK, UPCR and PMB conceptualized and designed the work. UPCR, AN, SM and SK performed data analysis. AN, SM and SK performed data interpretation. SK and SM wrote the manuscript. PMB supervised the study. AN approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee, Sri Devaraj URS Medical College, Tamaka, Kolar, India, with reference number DMC/KLR/UDOME/IEC-CER/117/2014-15.
- Fernandes SC, Dhanashree B. Drug resistance & virulence determinants in clinical isolatesof Enterococcus species. Indian J Med Res. 2013;137(5):981-985. PMCID: PMC3734693
- Ira P, Sujatha S, Chandra PS. Virulence factors in clinical and commensal isolates of Enterococcus species. Ind J Patho Gyn Micro. 2013;56(1):24-30.
Crossref - Upadhyaya PMG, Umapathy BL, Ravikumar KL. Comparative study for the presence of enterococcal virulence factors gelatinase, hemolysin and biofilm among clinical and commensal isolates of Enterococcus faecalis. J Lab Physicians. 2010;2(2):100-104.
Crossref - Vergis EN, Shankar N, Chow JW, et al. Association between the presence of Enterococcal virulence factors gelatinase, haemolysin and enterococcal surface protein and mortality among patients with bacteremia due to Enterococcus faecalis. Clin Infect Dis. 2002;35(5):570-575.
Crossref - Mollering RC Jr. Emergence of Enterococcus as significant pathogen. Clin Infect Dis. 1992;14(6):1173-1178.
Crossref - Manavalan J, Kannaiyan K, Velayutham A, Vadivel S, Kuthalaramalingam S. Phenotypic speciation of enterococci with special reference to prevalence, virulence and antimicrobial resistance. Int J Res Med Sci. 2015;3(10):2623-2629.
Crossref - Facklam RR, Collins MD. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J Clin Microbiol. 1989;27(4):731-734.
Crossref - Murray PK, Baron EJ, Landry ML, Jorgensen JH, Pfaller MA. Manual of clinical microbiology. 9th ed. Washington DC: ASM Press; 2007.
Crossref - Mishra RK, Saraf G, Patidar V, Tiwari Y, Pundir S, Pawan K. A study for the presence of enterococcal virulence factors gelatinase, haemolysin among clinical isolates in a tertiary care hospital. Research & Reviews: A Journal of Microbiology and Virology. 2017;7(3):14-18.
- Umamageswari SSM, Prabhakaran N, Mohanram K, Banu ASS. A Study to Compare the Presence of Virulence Factors Gelatinase, Haemolysin, Enterococcal Surface Protein (esp) and Biofilm Formation Among Clinical and Commensal Isolates of Enterococcus Species. J Pure Appl Microbiol. 2016;10(4):3195.
Crossref - Jett BD, Huycke MM, Gilmore MS. Virulence of enterococci. Clin Microbiol Rev. 1994;7(4):462-478.
Crossref - Banerjee T, Anupurba S. Prevalence of Virulence Factors and Drug Resistance in Clinical Isolates of Enterococci: A Study from North India. J Pathog. 2015:692612.
Crossref - Mundy M, Sahm DF, Gilmore M. Relationships between enterococcal virulence and antimicrobial resistance. Clin Microbiol Rev. 2000;13(4):513-522.
Crossref - Creti R, Imperi M, Bertuccini L et al. Survey for virulence determinants among Enterococcus faecalis isolated from different sources. J Med Microb. 2004;53(1):13-20.
Crossref - Murray BE, Weinstock GM. Enterococci: new aspects of an old organism. Proc Assoc Am Physicians. 1999;111(4):328-334.
Crossref - Hemalatha G, Bhaskaran K, Sowmiya M, Howlader A, Sethumadhavan K. A study on virulence factors and antimicrobial resistance pattern among enterococci isolated from various clinical specimens from a tertiary care hospital. Int J Res Med Sci. 2017;5(7):2969-2974.
Crossref - Salem-Bekhit MM, Moussa I, Muharram MM, Alanazy FK, Hefni HM. Prevalence and antimicrobial resistance pattern of multidrug-resistant enterococci isolated from clinical specimens. Indian J Med Microbiol. 2012;30(1):44-51.
Crossref - Nasaj M, Mousavi SM, Hosseini SM, Arabestani MR. Prevalence of virulence factors and vancomycin resistant genes among Enterococcus faecalis and E. faecium isolated from clinical specimens. Iranian J Public Health. 2016;45(6):806.
- Sood S, Malhotra M, Das BK, Kapil A. Enterococcal infections & antimicrobial resistance. Indian J Med Res. 2008;128(2):111-121. PMID: 19001673
- Ruchi T, Baveja S,De A. Comparison of Phenotypic Methods for the Detection of Biofilm Production in Uro-Pathogens in a Tertiary Care Hospital in India. Int J Curr Microbiol App Sci. 2015;4(9):840-849.
- Marothi YA, Agnihotri H, Dubey D, Enterococcal Resistance-An Overview. Indian J Med Microb. 2005;23(4):214-219.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.