ISSN: 0973-7510
E-ISSN: 2581-690X
Streptococcus agalactiae, an important etiological agent of bovine mastitis is poorly studied as a cause of caprine mastitis in India. Thus, the present study aimed at studying the antibiogram and virulence profile of S. agalactiae isolates from cases of caprine subclinical mastitis. In the present study, prevalence of subclinical mastitis in goats was estimated to be 51% by SLS test. Out of a total of 100 milk samples tested, seven S. agalactiae isolates (7%) were obtained and all isolates were obtained from mastitic milk samples. The S. agalactiae isolates were screened for ten virulence genes, of which only six were detected viz., cyl, glnA, cfb, hylB, fnbB and scaA gene. To our knowledge, this is the first report on detection of cyl, glnA, hylB, fnbB and scaA genes in caprine S. agalactiae isolates from India. Out of six antibiotic resistance genes tested, only one gene, i.e. tetM gene was detected by PCR. Antibiotic sensitivity results revealed highest resistance to tetracycline and ampicillin. Moreover, 71.4% of isolates were found to be multidrug-resistant and all MDR isolates exhibited a MAR index more than 0.2 which is reflective of the wide antibiotic usage in goats across Punjab.
Antibiotic Resistance, Caprine Subclinical Mastitis, MALDI-TOF, S. agalactiae, Virulence Genes
Small ruminants, particularly goats, greatly cater to the socioeconomic and nutritional needs of the country as goats are often referred to as the poor man’s cow. Goat milk is recommended for those who are allergic to cow milk as well as in certain diseases like dengue. One of the major diseases affecting lactating goats is mastitis, an inflammation of parenchyma of mammary gland which is endemic in India and affects all lactating animals.1 The disease affects both quality and quantity of milk and is therefore of great economic importance to the poor farmers.2 Subclinical mastitis in goats reduces the income of underprivileged rural people by reducing goat farming profits and lowering milk output. A wide array of infectious pathogens is attributed as etiological agents for subclinical mastitis. One major pathogen that causes subclinical mastitis is Streptococcus.
Streptococci are Gram-positive, facultatively anaerobic, catalase and oxidase-negative bacteria that belong to the family Streptococcaceae and tend to grow in pairs or chains as cell division occurs along a single axis. More than 60 species of Streptococci have been recognized so far, many of which have been associated with important diseases in animals. The species of Streptococcus most associated with subclinical mastitis in small ruminants include S. agalactiae and S. dysgalactiae. Streptococcus agalactiae, an important pathogen, has often been associated as a cause of udder fibrosis and reduction in milk production.3 When Streptococci invade a tissue or organ, they produce a wide range of virulence associated factors which permit their survival as well as replication in different tissues in host.4 S. agalactiae produces different virulence factors that lead to its pathogenicity viz., gene lmb, which codes for the laminin-binding protein, the cyl gene which encodes for a hemolysin, glnA which encodes for the glutamine synthetase and scaA gene which is an aggregation factor among others.5 All the resistance determinants as well as virulence factors help in initiation, development, and persistence of Streptococcal infections in hosts. Their study is therefore essential to understand the pathogenesis of the disease.
While treating subclinical mastitis of bacterial origin in small ruminants, antibiotic susceptibility testing is routinely carried out to obtain the best possible therapeutic result.6 Antibiotics have been used for therapy and prevention of Streptococcal subclinical mastitis in goats globally and this abuse of antibiotics has led to the menace of antimicrobial resistance which is becoming more and more alarming. The rise in number of different multidrug-resistant bacterial species pose a serious hazard to both human and animal health.7 S. agalactiae, an important pathogen responsible for subclinical mastitis in bovines, is highly contagious and has the ability to degrade milk quality. However, this pathogen is poorly studied as a cause of caprine subclinical mastitis, particularly in India. To the best of our knowledge, this study is the first report on determining virulence genes and detection of antimicrobial resistance, phenotypically and genotypically from caprine isolates of S. agalactiae in India.
Sample collection
Since in case of cattle, it has been reported that most infected cows do not exhibit overt signs of disease such as abnormal milk; therefore, we chose to screen milk samples from apparently healthy as well as mastitic milk samples from goats. A total of 100 milk samples were collected from both apparently healthy and mastitic goats from five districts of Punjab viz. Barnala (n = 34), Faridkot (n = 20), Ferozepur (n = 8), Jalandhar (n = 28) and Ludhiana (n = 10). The distance between Ludhiana and other districts ranged from 60 km to 180 km. The minimum distance was between Ludhiana and Jalandhar (60 km) while the maximum distance was 180 km between Ludhiana and Ferozepur. Milk samples were aseptically collected in sterile tubes for collection and transported to lab on ice. Before collecting milk samples, the teats were cleaned by using an antiseptic like 70% ethanol with initial strips of milk being discarded.
Confirmation of subclinical mastitis by Sodium Lauryl Sulphate (SLS) test
All the milk samples were tested by SLS (Sodium Lauryl Sulphate) test for confirmation of subclinical mastitis cases. In the test paddle, 5 ml of milk sample and 5 ml of SLS reagent were added followed by mixing immediately by swirling the paddle. Observations were made by recording a colour change and formation of gel in the paddle.
Isolation of S. agalactiae and its identification
All the milk samples were inoculated on Blood agar (BA), incubated for 24 hours at 37 °C for primary Streptococcal species isolation. The colonies were presumptively recognized as S. agalactiae based on beta-hemolysis on blood agar, by Gram’s staining and biochemical tests including catalase, oxidase and CAMP test. These colonies were additionally purified by streaking on BHI agar and Edward’s medium containing 5% defibrinated blood obtained from sheep.
MALDI-TOF
All S. agalactiae isolates were subjected to MALDI-TOF for confirmation. BHI plates were streaked by presumptive S. agalactiae isolates, incubated for 24 hrs at 37 °C and MALDI-TOF was performed by following the extended direct transfer (cDT) method. A single pure bacterial colony was spotted on the MALDI target plate as a thin film by using sterile toothpick followed by addition of 1 µl of 70% formic acid on the spot. The plate was allowed to dry at room temperature and the spot was covered by 1 µl solution of α-Cyano-4-hydroxycinnamic acid (HCCA) matrix followed by drying the plate and proteomic identification of bacteria.
DNA extraction
Genomic DNA from S. agalactiae isolates was extracted by hot cold lysis method. Briefly, 5-6 pure bacterial colonies were mixed in 200 µl of nuclease free water in a microcentrifuge tube and placed in a dry bath at 100 °C for 10 mins followed by immediately placing the tubes at -20 °C for at least 2 hours. The suspension was centrifuged at 10,000 g for 5 mins, supernatant was kept at -20 °C for further use as template DNA in PCR.
Molecular detection of virulence genes
All confirmed S. agalactiae isolates were tested by PCR for presence of virulence genes by use of primers as described previously.5,8 PCR consisted of 12.5 µL 2X Green Taq mastermix (Promega), 20 pM each of 1 µL forward primer, 1 µL reverse primer and 4 µL template DNA in a 25 µL reaction volume. The oligonucleotide sequence of primers for detecting virulence genes have been presented in Table 1. The amplification was carried out in a thermocycler (ABI, Thermo) as per the PCR program described in Table 2. Analysis of PCR amplicons was done in 1.5% agarose gel having 0.5 µg/mL ethidium bromide followed by visualization in UV transilluminator.
Table (1):
Details of primers used for the detection of virulence genes in S. agalactiae isolates
| No. | Gene | Oligonucleotide Sequence 5’-3’ | Amplicon size | Ref. |
|---|---|---|---|---|
| 1 | β-hemolysin (cyl) | F: ACG GCT TGT CCA TAG TAG TGT TTG | 345 | 5 |
| R: AAC GAC ACT GCC ATC AGC AC | ||||
| 2 | Glutamine synthetase (glnA) | F: ACG TAT GAA CAG AGT TGG CTA TAA | 471 | |
| R: TCC TCT GAT AAT TGC ATT CCA C | ||||
| 3 | CAMP factor (cfb) | F: ATG GGA TTT GGG ATA ACT AAG CTA G | 193 | |
| R: AGC GTG TAT TCC AGA TTT CCT TAT | ||||
| 4 | Aggregation (scaA) | F: ACG GTA TCA ACC TTG AAA CTG G | 256 | |
| R: TCA GTG TTG ATT TCC CAG ATG TA | ||||
| 5 | Hyaluronidase (hylB) | F: ACA AAT GGA ACG ACG TGA CTA T | 346 | |
| R: CAC CAA TTG GCA GAG CCT | ||||
| 6 | Fibrinogen bindingprotein (fnbB) | F: TGA TGC TGC AAA AGA ATT GC | 629 | 8 |
| R: TTA CAG CCC CTT TTT GAG GA | ||||
| 7 | Fibronectin-binding protein (pavA) | F: TTC CCA TGA TTT CAA CAA CAA G | 495 | |
| R: AAC CTT TTG ACC ATG AAT TGG TA | ||||
| 8 | Laminin-binding protein (lmb) | F: AGT CAG CAA ACC CCA AAC AG | 397 | |
| R: GCT TCC TCA CCA GCT AAA ACG | ||||
| 9 | Capsule, C5a peptidase (scpB) | F: AGT TGC TTC TTA CAG CCC AGA | 567 | |
| R: GGC GCA GAC ATA CTA GTT CCA | ||||
| 10 | β-C protein (cba) | F: TGC ACC GAA GTT ACC AGA TG | 149 | |
| R: CAG CCA ACT CTT TCG TCG TT |
Molecular detection of antibiotic resistance genes
All S. agalactiae isolates obtained were subjected to PCR for determination of antibiotic resistance genes as per primers described previously9,10 and presented in Table 3. PCR reaction mixture consisted of 2X Green Taq mastermix (12.5 µL), 20 pM forward primer (1 µL), 20 pM reverse primer (1 µL) and 4 µL of template DNA in a 25 µL reaction volume. The PCR cycle consisted of: initial denaturation at 94 °C for 10 mins, 35 cycles each of denaturation for 1 min at 94 °C, annealing at temperatures between 46 °C to 56 °C depending on the primer pair (Table 3) for 1 min, extension for 1 min at 72 °C. Final extension was done for 10 mins at 72 °C. The PCR amplicons were analyzed in 1.5% agarose gel containing 0.5 µg/mL ethidium bromide followed by visualizing in UV transilluminator.
Table (2):
PCR cycling conditions for amplifying virulence genes in S. agalactiae isolates
| No. | Gene | Initial Denaturation | Denaturation | Annealing | Extension | No. of cycles | Final extension |
|---|---|---|---|---|---|---|---|
| 1 | cyl | 94 °C for 5 min | 94 °C for 30 sec | 52 °C for
30 sec |
72 °C for 90 sec | 35 | 72 °C for 10 min |
| 2 | glnA | ||||||
| 3 | cfb | ||||||
| 4 | scaA | ||||||
| 5 | hylB | ||||||
| 6 | lmb | 94 °C for 4 min | 94 °C for 30 sec | 53 °C for 55 sec | 72 °C for 55 sec | 72 °C for 8 min | |
| 7 | pavA | 58 °C for 55 sec | |||||
| 8 | scpB | 57 °C for 55 sec | |||||
| 9 | fnbB | 58 °C for 55 sec | |||||
| 10 | cba | 56 °C for 55 sec |
Table (3):
Details of primers for detecting antibiotic resistance genes in S. agalactiae isolates
| No. | Gene | Oligonucleotide Sequence (5’-3’) | Annealing temp. (°C) | Product size (bp) | Ref. |
|---|---|---|---|---|---|
| 1 | ermB | F: GAA AAG GTA CTC AAC CAA ATA | 55 | 640 | 9 |
| R: AGT AAC GGT ACT TAA ATT GTT TAC | |||||
| 2 | ermA | F: TCT AAA AAG CAT GTA AAA GA | 52 | 640 | |
| R: CTT CGA TAG TTT ATT AAT ATT AGT | |||||
| 3 | mefA | F: AGT ATC ATT AAT CAC TAG TGC | 52 | 348 | |
| R: TTC TTC TGG TAC TAA AAG TGG | |||||
| 4 | mefE | F: CGT AGC ATT GGA ACA GC | 52 | 513 | 10 |
| R: TCG AAG CCC CCT AAT CTT | |||||
| 5 | tetO | F: AAC TTA GGC ATT CTG GCT CAC | 55 | 519 | |
| R: CGG CGG GGT TGG CAA ATA | |||||
| 6 | tetM | F: TTA TCA ACG GTT TAT CAG G | 46 | 397 | |
| R: CGT ATA TAT GCA AGA CG |
In vitro antibiotic susceptibility testing
All the S. agalactiae were tested for antibiotic sensitivity in vitro by using Kirby Bauer’s disc diffusion method11 on blood Mueller-Hinton Agar plates as per CLSI guidelines.12
A panel of 14 antibiotics viz., ampicillin (AMP) (10 µg/disc), azithromycin (AZM) (15 µg/disc), cefotaxime (CTX) (10 µg/disc), ceftriaxone (CTR) (30 µg/disc), chloramphenicol (C) (30 µg/disc), ciprofloxacin (CIP) (5 µg/disc), cotrimoxazole (COT) (25 µg/disc), erythromycin (E) (15 µg/disc), gentamicin (GEN) (10 µg/disc), meropenam (MRP) (5 µg/disc), ofloxacin (OF) (5 µg/disc), penicillin G (P) (2U/disc), tetracycline (TE) (30 µg/disc) and vancomycin (VA) (30 µg/disc) were used. The diameter of inhibition zone was measured using a ruler, the results were noted in millimeters (mm) and isolates were designated as susceptible (S), intermediate (I) or resistant (R) to antibiotics as per CLSI (Clinical and Laboratory Standards Institute) guidelines.12
Determination of multidrug-resistance and MAR index
The multidrug-resistance in the S. agalactiae isolates was identified. Isolates exhibiting resistance to minimum one antimicrobial drug in three/more antimicrobial classes were designed as MDRs (multidrug-resistant). MAR index (Multiple antibiotic resistance) was also calculated; as ratio between the number of drugs to which an isolate is resistant and total number of antibiotics tested, as it is a practical tool for identifying the sources of antibiotic resistant organisms.13 A MAR index >0.2 indicates high risk source of contamination where antibiotics are frequently used.14
Statistical analysis
Chi-square test was used to examine the effect of breed and age of goats on S. agalactiae colonization. All S. agalactiae isolates in present study were obtained from mastitic milk samples and the goats housed in semi-intensive housing system.
Of the 100 milk samples collected, 51 were mastitic by SLS test and thus prevalence of caprine subclinical mastitis in Punjab was found as high as 51% by SLS test. Similar results demonstrating a high prevalence of caprine subclinical mastitis has been reported by various researchers across different geographical locations. A high prevalence of caprine subclinical mastitis (41.2%) has been observed in J & K, India, by the California mastitis test.15 Similar results have been obtained in Nigeria where an overall subclinical mastitis prevalence in lactating goats was estimated by CMT and clinical examinations of the udder as 40.4%.16 Similarly, higher prevalence of S. agalactiae is reported in other countries like 40.5% in Brazil,17 43.33% in Bangladesh,18 48.1% in Egypt19 and 61% in Kenya.20
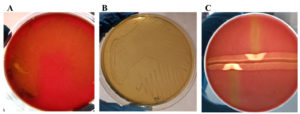
Out of these 100 samples, a total of 100 isolates were obtained, out of which only seven isolates exhibited characteristic beta-hemolysis on blood agar, stained as Gram-positive cocci, in chains of varying length, were catalase and oxidase negative and demonstrated a typical arrow head zone of hemolysis at intersection of S. aureus and S. agalactiae streaks (CAMP test positive). These seven isolates exhibited grey to colourless colonies with absence of aesculin hydrolysis on Edwards medium while on BHI agar yielded small and translucent colonies and were therefore presumptively identified as S. agalactiae. The colonies of S. agalactiae observed on different media have been shown in Figure 1. All the seven presumptive isolates were confirmed as S. agalactiae by MALDI-TOF.
Figure 1. Colonies of S. agalactiae observed on different media after aerobic incubation at 37 °C after 24 hrs. A: Colourless colonies of S. agalactiae on Edward’s media indicating absence of aesculin hydrolysis. B: S. agalactiae colonies on BHI agar depicting small and translucent colonies. C: Positive CAMP test between S. agalactiae (vertical streak) and S. aureus (horizontal streak) depicted by formation of arrowhead of hemolysis at intersection of S. aureus and S. agalactiae streaks
Prevalence of S. agalactiae in caprine milk samples was estimated as 7%. Interestingly, all S. agalactiae isolates were obtained from mastitic goats. Therefore, prevalence of S. agalactiae from caprine mastitic cases in Punjab, India was 7%. Similarly, high prevalence has been reported from other countries viz. Malaysia where a prevalence of 6.5% has been reported21 and from Nigeria where a prevalence of 11% has been reported.16 The S. agalactiae isolates were obtained from different districts. Although there is the possibility of spread of bacteria causing subclinical mastitis between districts, inter-district transmission of subclinical mastitis causing bacteria appears limited in this study, as large distances between districts led to lack of migration thereby reducing the risk of bacterial spread. However, spread between different villages within the same district is likely.
Molecular detection of virulence genes
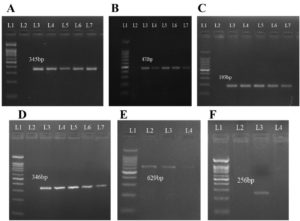
The S. agalactiae isolates were tested for the presence of ten virulence genes viz., cyl (codes for b-Hemolysin), glnA (Glutamine synthetase), cfb (CAMP factor), scaA (Aggregation), hylB (Hyaluronidase), fnbB (binding protein for Fibrinogen), pavA (binding protein for Fibronectin), lmb (Laminin-binding protein), scpB (Capsule, C5a peptidase), and cba (β-C protein). Out of these, only six genes were detected (Figure 2). The cyl, glnA, cfb and hylB genes were detected in five out of seven isolates while fnbB gene was observed in four isolates. The scaA gene was detected in only one S. agalactiae isolate.
Figure 2. Molecular detection of Virulence genes in S. agalactiae isolates (L1 = 100 bp Molecular weight marker, L2 = negative control, L3 onwards = positive samples). A: Molecular detection of cyl gene (345 bp amplicon), B: Molecular detection of glnA gene (471 bp), C: Molecular detection of cfb gene (193 bp), D: Molecular detection of hylB gene (346 bp), E: Molecular detection of fnbB gene (629 bp), F: Molecular detection of scaA gene (256 bp)
To the best of our knowledge, this paper represents the first report on detecting cyl, glnA, hylB, fnbB and scaA genes in caprine S. agalactiae isolates. Previous studies have extensively shown these virulence genes in bovine S. agalactiae isolates, but none of the published literature was available on presence of these genes in caprine isolates. In one of the previous studies conducted in Iran, none of the caprine S. agalactiae isolates carried the cyl gene,22 even though cyl gene has been detected in bovine S. agalactiae isolates in various studies.23,24 Similarly, glnA gene which encodes for enzyme glutamine synthetase, enables bacteria to assimilate ammonium from glutamate and is involved in nitrogen metabolism, thus contributes to the virulence of the bacterium, has not been detected in caprine Streptococcal isolates from Iran,22 but has been widely detected in bovine S. agalactiae isolates.25 The hylB gene codes for hyaluronidase enzyme which degrades the polysaccharides such as chondroitin, chondroitin sulphate, N-acetylglucosamine thus facilitating S. agalactiae spread during infection. This gene has been detected from bovine S. agalactiae isolates but none of the previous studies have reported its prevalence in caprine S. agalactiae isolates.22 The fnbB gene is another important virulence gene which encodes for fibrinogen binding protein that helps the Streptococci to bind to fibrinogen which enables bacterial adhesion to host surfaces and also helps the bacteria to avoid the immune system. In contrast to the results obtained in the present study, the caprine S. agalactiae isolates have not been found to harbour this gene.22 The scaA gene which codes for aggregation factor has also remained undetected in caprine isolates22 but bovine Streptococcal isolates have shown the presence of this gene.26
Cfb gene which encodes for CAMP factor has been detected from bovine as well as caprine S. agalactiae isolates. Similar results have been reported from 68.75% caprine S. agalactiae isolates in a study conducted in Iran.22 The cfb gene has also been detected in S. agalactiae isolates from dairy goats of China,27 in 73.3% isolates in Egypt.28 This gene has been observed in S. agalactiae isolates obtained from bovines.26
Molecular detection of antibiotic resistance genes
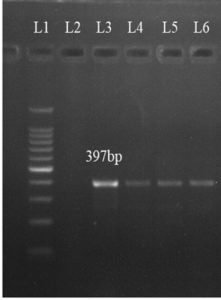
Out of six antibiotic resistance genes tested, only one antibiotic resistance gene, viz. tetM gene could be detected in S. agalactiae isolates. The tetM gene was detected in four isolates (Figure 3).
Figure 3. Molecular detection of tetM gene in S. agalactiae isolates at 397 bp (L1 = 100 bp Molecular weight marker, L2 = negative control, L3-L6 = positive samples)
In a Spanish study, tetM gene was detected in only 1.0% of the caprine isolates.29 In contrast to results of present study, 63% of isolates obtained in China contained at least one tetracycline resistance gene, with 13 isolates harbouring the tetM gene.30 69.64% of the S. agalactiae isolates obtained from Iran exhibited high resistance to tetracycline and erythromycin22 while very low erythromycin resistance has been found in present study. In a study of Malaysia, 33.3% of S. agalactiae isolated have been observed to be penicillin-resistant21 while 57.1% penicillin resistance has been observed in Pakistan.2 All S. agalactiae isolates obtained in present study were completely resistant to tetracycline but only 57.14% isolates had the tetM gene. One of the plausible explanations for this could be that tetracycline resistance can be caused by various genes, but in this study, we screened the isolates only for tetM and tetO. This suggests that tetracycline resistance may be caused by some different resistance genes. Interestingly, 14.28% of the S. agalactiae isolates were found to be erythromycin resistant, but no resistance gene was detected. This phenomenon has been explained as phenotypic resistance where resistance is observed without any genetic alteration and is generally associated with specific bacterial processes like growth in biofilms, stationary growth phases or persistence.31 The resistance may also be attributable to different resistance genes like ermTR, msrA, mefG for which the screening was not performed.32
In vitro Antibiotic susceptibility assay
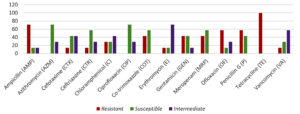
Among the S. agalactiae isolates obtained in the present study, resistance was highest for tetracycline and ampicillin, followed by penicillin and ofloxacin. The detailed AST results have been depicted in Figure 4.
Figure 4. Antibiotic resistance profile of S. agalactiae isolates: the bars in red indicate resistance, the bars in green indicate susceptible and in violet indicate intermediate resistance
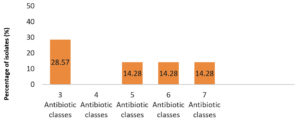
The S. agalactiae isolates which had resistance to three/more than three antibiotic classes were identified as multidrug-resistant (MDR). Five out of seven isolates were multidrug-resistant with 28.57% isolates exhibiting resistance to three and 14.28% isolates showing resistance to five, six and seven classes of antibiotics each (Figure 5).
Figure 5. Multidrug-resistant S. agalactiae isolates. X-axis represents number of antibiotic classes to which an isolate was resistant to, Y-axis denotes percentage of isolates
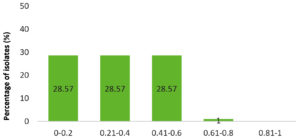
It has been observed that 71.4% of the S. agalactiae isolates had a MAR index >0.2 (Figure 6). Higher MAR index reflects wide antibiotic usage in small ruminants across Punjab.
Figure 6. Multiple Antibiotic Resistance (MAR) of S. agalactiae isolates as shown by MAR index. X-axis represents MAR index value and Y-axis represents percentage of isolates. A MAR index >0.2 indicates higher antimicrobial usage in the geographical area
Statistical analysis
Chi-square test was used to assess effect of age and breed on S. agalactiae colonization. No significant difference was observed between age and breed of the animals to S. agalactiae colonization.
In the current study, all S. agalactiae isolates were isolated from mastitic milk samples and none could be detected from healthy caprine milk samples. Therefore, it can be concluded that, S. agalactiae, is a significant subclinical mastitis causing pathogen in goats. Furthermore, multidrug-resistance and a wide range of virulence factors have been detected in S. agalactiae isolates. The isolates had a MAR index >0.2 which indicates wide antimicrobial usage in caprines in Punjab. Since S. agalactiae isolates obtained in the current study have been found to be resistant to common antimicrobials, thus, proper antimicrobial susceptibility testing is warranted prescribing antibiotic for treatment.
ACKNOWLEDGMENTS
The authors greatly acknowledge the facilities and funding provided by the Guru Angad Dev Veterinary and Animal Sciences University, Ludhiana, India, for the present investigation.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
JB and PK conceptualized and designed the study. SG performed sample collection. JS performed sampling. SG performed experiments. PK performed data validation. SG wrote the manuscript. PK, AKA and TSR reviewed the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
ETHICS STATEMENT
This study was approved by the Institutional Animal Ethics Committee (IAEC) of Guru Angad Dev Veterinary and Animal Science University, Ludhiana, India, vide Reference no. GADVASU/2021/IAEC/61/22 dated 13/10/2021 and from the Institutional Biosafety Committee (IBSC), Guru Angad Dev Veterinary and Animal Sciences University, Ludhiana, India, vide reference no. IBSC/2021/1763-64 dated 13/10/2021.
- Rinaldi M, Li RW, Capuco AV. Mastitis associated transcriptomic disruptions in cattle. Vet Immunol Immunopathol. 2010;138(4):267-279.
Crossref - Najeeb MF, Anjum AA, Ahmad MUD, Khan HM, Ali MA, Sattar MMK. Bacterial etiology of subclinical subclinical mastitis in dairy goats and multiple drug resistance of the isolates. J Anim Plant Sci. 2013;23(6):1541-1544.
- Bergonier D, De Cremoux R, Rupp R, Lagriffoul G, Berthelot X. Subclinical mastitis of dairy small ruminants. Vet Res. 2003;34(5):689-716.
Crossref - Kabelitz T, Aubry E, Van Vorst K, Amon T, Fulde M. The role of Streptococcus spp. in bovine subclinical mastitis. Microorganisms. 2021;9(7):1497.
Crossref - Dmitriev A, Shakleina E, Tkaeikova L, Mikula I, Totolian A. Genetic heterogeneity of the pathogenic potentials of human and bovine group B streptococci. Folia Microbiol. 2002;47(3):291-295.
Crossref - Landfried LK, Barnidge EK, Pithua P, et al. Antibiotic use on goat farms: an investigation of knowledge, attitudes, and behaviors of Missouri goat farmers. Animals. 2018;8(11):198.
Crossref - Parra SA, Rather MI, Para PA, Ganguly S. The emergence of drug resistant bacteria: effects on human health. J Environ Life Sci. 2017;2(3):77-79.
- Pires R, Rolo D, Gama-Norton L, et al. Group A streptococci from carriage and disease in Portugal: evolution of antimicrobial resistance and T antigenic types during 2000-2002. Microb Drug Resist. 2005;11(4):360-370.
Crossref - Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother. 1996;40(11):2562-2566.
Crossref - Lopardo HA, Vidal P, Jeric P, Centron D, Paganini H, Facklam RR, Elliott J. Six-month multicenter study on invasive infections due to group B streptococci in Argentina. J Clin Microbiol. 2003;41(10):4688-4694.
Crossref - Bauer AW, Kirby WMM, Sherris JC and Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45(4):493-496.
Crossref - Clinical And Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. CLSI. 2020. https://clsi.org
- Sandhu R, Dahiya S, Sayal P. Evaluation of multiple antibiotic resistance (MAR) index and Doxycline susceptibility of Acinetobacter species among inpatients. Indian J Microbiol Res. 2016;3(3):299-304.
Crossref - Rotchell D, Brown PD. Multiple Antibiotic Resistance Index. Fitness and Virulence Potential in Respiratory Pseudomonas aeruginosa from Jamaica. J Med Microbiol. 2016;65(4):251-271.
Crossref - Taku A, Badroo GA, Gazal S, et al. Prevalence and bacteriological examination of clinical and sub-clinical cases of caprine subclinical mastitis in Jammu, India. J Pure Appl Microbiol. 2016;10(2):1585-1590.
- Danmallam FA, Pimenov NV. Study on prevalence, clinical presentation, and associated bacterial pathogens of goat subclinical mastitis in Bauchi, Plateau, and Edo states, Nigeria. Vet World. 2019;12(5):638.
Crossref - Oliveira AA, Melo CB, Seixas L, et al. Mastitis and milk composition in first partum Santa Ines ewes. J Vet Adv. 2013;3(8):220-231.
Crossref - Ferdous J, Rahman MS, Khan MI, Khan M, Rima UK. Prevalence of clinical and subclinical caprine subclinical mastitis of Northern region in Bangladesh. Progress Agric. 2018;29(2):127-138.
Crossref - Mbindyo CM, Gitao CG, Bebor LA. Cross-sectional study on the prevalence of subclinical subclinical mastitis and antimicrobial susceptibility patterns of the bacterial isolates in milk samples of smallholder dairy goats in Kenya. Am J Res Commun. 2014;2(8):30-51.
- El-Shymaa AA, Mohamed IE, Afaf MM. The prevalence and etiology of subclinical subclinical mastitis in sheep and goats. Zagazig Vet. J. 2018;46(2):96-104.
Crossref - Ariffin SMZ, Hasmadi N, Syawari NM, et al. Prevalence and antibiotic susceptibility pattern of Staphylococcus aureus, Streptococcus agalactiae and Escherichia coli in dairy goats with clinical and subclinical subclinical mastitis. J Anim Health Prod. 2019;7(1):32-37.
Crossref - Momtaz H, Eliyasi M, Soleimani R, Jazayeri A. Prevalence of Virulence Factors and Antimicrobial Resistance of Streptococcus agalactiae and Streptococcus uberis in Ruminant Sub-clinical Mastitic Milk in Iran. Int J Med Lab. 2017;4(1):34-47.
- Tian XY, Zheng N, Han RW, et al. Antimicrobial resistance and virulence genes of Streptococcus isolated from dairy cows with subclinical mastitis in China. Microb Pathog. 2019;131:33-39.
Crossref - Shen J, Wu X, Yang Y, et al. Antimicrobial resistance and virulence factor of Streptococcus dysgalactiae isolated from clinical bovine subclinical mastitis cases in northwest China. Infect Drug Resist. 2021;2021(14):3519-30.
Crossref - Zhao Y, Shao W, Wang F, et al. Antimicrobial resistance and virulence genes of Streptococcus agalactiae isolated from subclinical mastitis milk samples in China. J Vet Res. 2022;66(4):581-590.
Crossref - Zhang Z, Yang F, Li XP, et al. Distribution of serotypes, antimicrobial resistance and virulence genes among Streptococcus agalactiae isolated from bovine in China. Acta Sci Vet. 2019;47(1):97254.
Crossref - Shi H, Zhou M, Zhang Z, et al. Molecular epidemiology, drug resistance, and virulence gene analysis of Streptococcus agalactiae isolates from dairy goats in backyard farms in China. Front Cell Infect Microbiol. 2023;12:1915.
Crossref - Abed AH, Menshawy AMS, Zeinhom MMA, et al. Subclinical subclinical mastitis in selected bovine dairy herds in North Upper Egypt: Assessment of prevalence, causative bacterial pathogens, antimicrobial resistance and virulence-associated genes. Microorganisms. 2021;9(6):1175.
Crossref - Petrocchi-Rilo M, Martinez-Martinez S, Aguaron-Turrientes A, et al. Anatomical site, typing, virulence gene profiling, antimicrobial susceptibility and resistance genes of Streptococcus suis isolates recovered from pigs in Spain. Antibiotics. 2021;10(6):707.
Crossref - Huang J, Shang K, Kashif J and Wang L. Genetic diversity of Streptococcus suis isolated from three pig farms of China obtained by acquiring antibiotic resistance genes. J Sci Food Agric. 2015;95(7):1454-1460.
Crossref - Corona F, Martinez JL. Phenotypic Resistance to Antibiotics. Antibiotics. 2013;2(2):237-55.
Crossref - Varaldo PE, Montanari MP, Giovanetti E. Genetic elements responsible for erythromycin resistance in streptococci. Antimicrob Agents Chemother. 2009;53(2):343-353.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.