ISSN: 0973-7510
E-ISSN: 2581-690X
Methicillin-resistant Staphylococcus aureus (MRSA) is a major cause of Nosocomial, community acquired infections and neonatal sepsis. The Glycopeptide vancomycin was the drug of choice for treating infections. Aim: Identifying the vancomycin- resistance phenotypically and genotypically among the MRSA isolates from Shibin El-Kom teaching Hospital. Materials and Methods: All samples were collected from Neonatal intensive care unit (NICU) of Shibin El-Kom teaching hospital in Minoufiya, Egypt and identified by conventional methods. S aureus and MRSA were isolated and identified from different clinical samples using conventional methods confirmed by antibiogram of the isolates and mec A gene detection. vancomycin MIC and Vancomycin screening agar were determined following CLSI guidelines. Van A was amplified by PCR using standard primers. Out of the 200 neonates included in this study, 85% were positive growth and 15% were negative growth. Among them, 25% isolates were staphylococci, 42 isolates had nuc gene. Out of 42 S. aureus, 80.95% had mecA gene and 19.05% had not Mec A gene. The VRSA isolates had not van A gene. Conclusions: Vancomycin was still the most effective drug against S. aureus infection. All MRSA in Shibin El-Kom Teaching Hospital had not vanA gene.
MRSA, VRSA, Nuc gene, Antibiotic resistance
Over the past few decades, there has been an alarming increase in the prevalence of antibiotic-resistant pathogens and strains in serious infections. The occurrence of bacterial infection had decreased with the discovery of penicillin in 1940 until Staphylococcus aureus began producing B-lactamase, which destroys the penicillin B-lactam core ring (Khan, et al., 2013). This increase in resistance towards penicillin drove the development of methicillin drugs, which are virtually resistant against many genetic variations of the B-lactamase enzyme. Since then, MRSA has become endemic in hospitals and nursing homes worldwide (Noor & Munna, 2015). The treatment of the S. aureus infections has become problematic because of the increase of resistance to methicillin, vancomycin and other antibiotics (Mathews et al., 2010).
The glycopeptide vancomycin was considered to be the best alternative for the treatment of multi drug-resistant MRSA. However, there are increasing numbers of reports indicating the emergence of vancomycin-resistant S. aureus (VRSA) strains exhibiting two different resistance mechanisms. Initially, vancomycin-intermediate S. aureus (VISA) noted to be due to the thickened cell wall (Hiramatsu et al., 1997), where many vancomycin molecules were trapped within the cell wall. The trapped molecules clog the peptidoglycan meshwork and finally form a physical barrier towards further incoming vancomycin molecules (Cui et al., 2006) The second was identical to the mechanism seen in vancomycin-resistant Enterococcus. Vancomycin-resistant Enterococcus faecium harbours the vanA operon, which contains five genes, VanS, -R, -H, -A and -X8. But Tiwari and Sen have reported a VRSA which is van gene-negative. (Tiwari and Sen 2006) Subsequent isolation of VISA and VRSA isolates from other countries has confirmed that the emergence of these strains is a global issue. Vancomycin was the drug of choice for treatment of infections caused by MRSA. High vancomycin MIC for MRSA which are susceptible to vancomycin may indicate the drug resistance to many antibiotics (Kshetry et al., 2016). The vancomycin has been a high molecular weight antibiotic, does not diffuse in concentration gradient manner while diffusing through the agar medium when the disc susceptibility test is employed. Moreover, it does not differentiate between vancomycin-susceptible isolates from vancomycin-intermediate isolates of S. aureus and hence disc diffusion testing is not used for detection of VRSA (CLSI 2012).
The aim of the present study was to identify the emergence of vancomycin-resistant MRSA among S. aureus isolates from patients with Neonatal sepsis attending the Intensive Care Unit in Shibin El-Kom Teaching Hospital.
Study Site and Population: The study was done at department of Microbiology, Shibin El-kom Teaching Hospital and Department of Microbial Biotechnology, Genetic Engineering and Biotechnology Research Institute, University of Sadat City. The present study was carried out among the Neonatal sepsis attending intensive care unit (ICU) at Shibin El-kom Teaching Hospital.
Isolation and Identification of Staphylococcus aureus: A total of 200 blood clinical samples were inoculated on blood culture and incubated aerobically at 37°C for 48 hours. S. aureus was identified by colony morphology, Gram stain, DNase, catalase and coagulase tests and fermentation of mannitol by conventional methods. (Oxoid, England) (De vos et al., 2009). Staph isolates were confirmed by detection of the nuc gene.
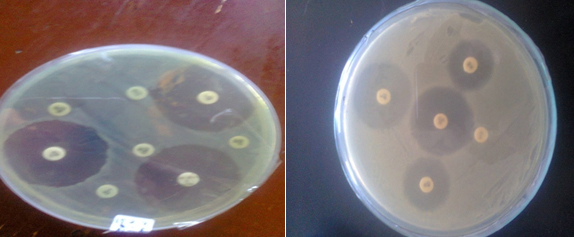
Antibiotic susceptibility testing: The antibiotic resistance profile was determined by the disc agar diffusion (DAD) technique using different antimicrobial agents; Amikacin (30µg), Gentamicin (10µg), Streptomycin (10µg), Clarithromycin (15µg), Cefoxitin (30µg), Ampicillin (10µg), Ciprofloxacin (5µg), Erythromycin (15µg), Oxacillin (1µg), Vancomycin (30µg), Meropenem (10µg), Imepenem (10µg) and Cephardine (30µg) (Oxoid, England) according to the guidelines recommended by Clinical and Laboratory Standards Institute (CLSI, 2014).
Determination of MIC: MIC of vancomycin was determined by agar dilution method using CLSI guidelines (CLSI 2014). Briefly, gradient plates of Mueller-Hinton agar (Oxoid, England) were prepared with vancomycin (2-32 mg/l, Oxoid, England). 0.5 McFarland equivalent inoculum prepared using 18-24h; old culture was spotted on to gradient plates. Plates were incubated overnight at 35°C for 24h before assessing the visible growth.
Vancomycin screening plate: Brain heart infusion (BHI) was used for vancomycin (VA) screening plate (6µg/ml). All plates were inoculated with 10µl of an inoculum suspension prepared with growth from an overnight blood agar plate, with a turbidity equivalent to 0.5 McFarland standard. The plates were incubated for 48h, and growth was reported after both 24 and 48h (Bhateja et al., 2005).
DNA Extraction: DNA of S. aureus isolated according to the Kit (Jena Bioscience. Germany) according to the manufacturer’s recommendations. The eluted DNA was stored at –20°C until use
PCR amplification for Nuc, MecA and VanA genes
Amplification was carried out according to the following thermal and cycling condition for Nuc gene ( table 1) using Gene Amp (Applied Biosystems, USA) The primers and the PCR conditions were as described by (Zhang et al., 2004):and primeres are mentioned in table 1. Amplification condition consists of Initial denaturation at 94oC for 5 minutes. Thirty cycles of amplification consisting of Denaturation at 94oC for 60 seconds, annealing at 50oC for 60 seconds and final extension at 72oC for 120 seconds. After the last cycle, a final extension of 10 minutes at 72 °C was done.
Table (1):
Primers used for amplification of MecA gene, VanA gene and Nuc gene.
| Primer | Sequence(5′-3′) | Reference | Size of Amplified Product (bp) |
|---|---|---|---|
| Nuc | 5′-GCGATTGATGGTGATACGGTT-3′ 3′-AGCCAAGCCTTGACGAACTAAAGC-5′ |
||
| Zhang et al., 2004) | 279 | ||
| VanA | 5′-ATGAATAGAATAAAAGTTGC-3′ 3′-TCACCCCTTTAACGCTAATA-5′ |
(Biswajit et al., 2008) | 1030 |
| MecA | 5′-CCAATTCCACATTGTTTCGGTCATA-3′ 3′-GTAGAAATGACTGAACGTCCGATAA-5′ |
Zhang et al., 2004 | 310 |
Amplification of MecA gene (Table 1) (Zhang et al., 2004):
- Initial denaturation at 94oC for 5 minutes.
- Thirty cycles of amplification consisting of:
- Denaturation at 94oC for 30 seconds.
- Annealing at 54oC for 30 seconds.
- Extension at 72oC for 30 seconds.
-
- After the last cycle, a final extension of 5 minutes at 72 °C was done.
Amplification of Van A gene (table 1) (Biswajit et al., 2008):
- Initial denaturation at 94oC for 5 minutes.
- Thirty cycles of amplification consisting of:
- Denaturation at 94oC for 60 seconds.
- Annealing at 50oC for 60 seconds.
- Extension at 72oC for 120 seconds.
- After the last cycle, a final extension of 10 minutes at 72 °C was done.
Amplified fragments were visualized on a 1% agarose gel electrophoresis stained with ethidium bromide. A DNA ladder (100–1500bp) was used to estimate allele sizes in base pairs (bp) for the gel.
Table (2):
Distribution of 200 Neonates with suspected sepsis according to Gram positive, Gram negative, Candida and No growth.
| Type of growth | Gram positive | Gram negative | Candida | No growth |
|---|---|---|---|---|
| No % | No % | No % | No % | |
| Number | 70(35%) | 75 (37.5%) | 25 (12.5%) | 30 (15%) |
Table (3):
Identification of 50 staphylococci according to biochemical test.
S. aureus |
CoNS |
|
|---|---|---|
Coagulase Test |
42(84%) |
8(16%) |
MSA |
45(90%) |
5(10%) |
DNase |
42(84%) |
8(16%) |
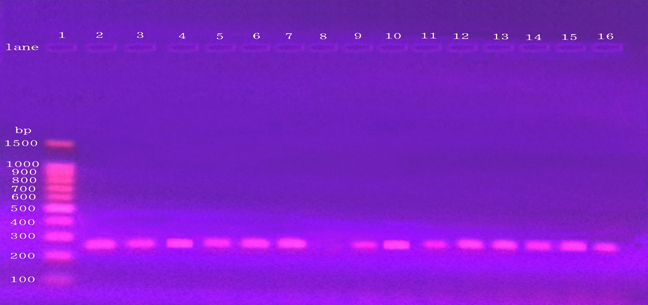
Out of the 200 neonates had neonatal sepsis, 85% were positive growth and 15% were negative growth. Out of 85% positive growth, 35% were Gram Positive, 37.5% were Gram-Negative and (12.5%) were Candida (Tab 2). Among 70 of Gram-Positive, 50 isolates were confirmed phenotypically as S. aureus. PCR amplification confirmed the presence of nuc gene in 42(84%) of the 50 isolates (table 4). The PCR product appeared a single DNA band with a size equal to 279 bp fragments (Fig 3).
Table (4):
Detection of Nuc gene among 50 staphylococcal isolates.
Positive No (%) |
Negative No (%) |
|
|---|---|---|
Nuc gene |
42 (84%) |
8(16%) |
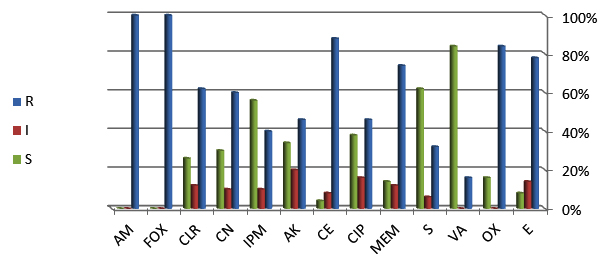
Antibiotic susceptibility tests were evaluated by the disk diffusion method for all isolates of S. aureus (Table 5 & Fig 2). Isolates of S. aureus showed different level of resistance against some antibiotics (Fig 1).
Table (5):
Percentage of antibiotic resistant isolates of staphylococci (n=50).
| Antibiotic Name |
Number of isolates % | ||
|---|---|---|---|
| R | I | S | |
| E | 39 (78%) | 7 (14%) | 4 (8%) |
| OX | 42 (84%) | — | 8(16%) |
| VA | 8(16%) | — | 42(84%) |
| S | 16(32%) | 3(6%) | 31(62%) |
| MEM | 37(74%) | 6(12%) | 7(14%) |
| CIP | 23(46%) | 8(16%) | 19(38%) |
| CE | 44(88%) | 4(8%) | 2(4%) |
| AK | 23(46%) | 10(20%) | 17(34%) |
| IPM | 20(40%) | 2(4%) | 28(56%) |
| CN | 30(60%) | 5(10%) | 15(30%) |
| CLR | 31(62%) | 6(12%) | 13(26%) |
| FOX* | 50(100%) | — | — |
| AM | 50(100%) | — | — |
R: Resistant, I: Intermediate, S: Sensitive
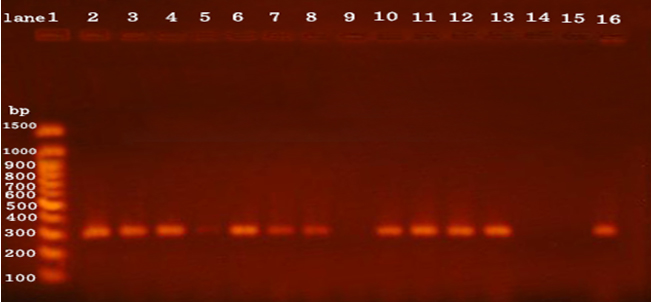
Among the 50 clinical isolates of S. aureus, 50(100%) were identified as methicillin- resistant S. aureus (MRSA) by disc diffusion method. This was confirmed by detection of mecA gene. The PCR product appeared a single DNA band with a size equal to 310 bp fragment corresponding to the mec A gene for 34 (68%) isolates of S. aureus and 16(32%) had not mec A gene (Fig 4).
MIC of vancomycin was determined by agar dilution method using CLSI guidelines. MIC of Vancomycin for 50 staphylococci isolates were 48 isolates sensitive for vancomycin and two isolates resistant for vancomycin in concentration 2µg/mL while 49 isolates sensitive for vancomycin and one isolate resistant for vancomycin in concentration 4 and 8µg/mL while 50 isolates sensitive for vancomycin in concentration 16 and 32µg/mL (Tab 5). By Vancomycin Screening Agar for 50 staphylococci isolates were 49 isolates sensitive for vancomycin and one isolate resistant for vancomycin (Table 6).
Table (6):
Detection of MecA gene among 50 staphylococcal isolates.
Positive No (%) |
Negative No (%) |
|
|---|---|---|
MecA gene |
34 (68%) |
16 (32%) |
Table (7):
MIC of vancomycin for 50 isolates of Staphylococci.
MIC ( μg/mL) |
No. of sensitive strains |
Percentage |
|---|---|---|
≤ 2 |
48 |
(96%) |
4-8 |
49 |
(98%) |
16-32 |
50 |
(100%) |
Table (8):
Vancomycin Screening Agar for 50 staphylococci.
Sensitive |
Resistant |
|
|---|---|---|
Vancomycin 6μg/ml |
49 |
1 |
Table (9):
Detection of VanA gene among 50 staphylococcal isolates.
Positive |
Negative |
|
|---|---|---|
VanA gene |
0 |
50 |
The PCR amplification did not appear band corresponding to van A gene for S. aureus and 50 had not van A gene (Fig 5).
The present study a total of 200 samples tested, 50 isolates were staphylococci. S. aureus was identified and classified by PCR using specific primers for housekeeping genes in these bacteria, such as the nuc gene. The PCR depending on a diagnostic protocol was used to detect the different genes (Mehrotra et al., 2000). Nuc gene is base line in identification and classification of S. aureus and in the present study, the nuc gene fragment was equal to 279 bp. Some reports indicated that the nuc gene was encoded to enzyme thermo nuclease and the length fragment of nuc gene was equal to 279 bp (Brakstad et al., 1992). By applying PCR method, among the 126 clinical samples that were identified as S. aureus with phenotypic methods, 101 (80.2%) isolates were found to be nuc positive (Roxana et al., 2014), this agrees with our results.
Infections caused by methicillin-resistant S. aureus have been associated with high morbidity and mortality rates. MRSA is one of the common causes of hospital-acquired infections and 30 to 80 percent methicillin resistance in S. aureus based on antibiotic sensitivity tests has been reported from different hospitals Anupurba et al., (2003).
Antibiotic susceptibility testing has been found to be a good epidemiological marker for MRSA phenotyping. The present study, among MRSA isolates high degree of resistance was encountered for Ampicillin (100%), Cefoxitin (100%), Cephardine (88%), Oxacillin (84%), and Meropenem (74%). Our study similar to Mundhada et al., which also found a high level of resistance to Erythromycin (Mundhada et al., 2016). In our study, the isolates of S. aureus revealed increasing resistance against many antibiotics, such as Cefoxitin while they appeared sensitive to other antibiotics like Vancomycin. 84% isolates of S. aureus were Oxacillin-resistant, 78% were Erythromycin-resistant and 16% were Vancomycin-resistant. In other reports, the percentage of Tetracycline-resistant S. aureus isolates was equal to 47.4% or 60.4% (Shittu et al., 2011) (Duran et al., 2012). Our results did not agree with the reports of Duran et al., and Brakstad et al., which showed that all clinical samples of S. aureus were resistant to Vancomycin (Brakstad et al., 1992) (Duran et al., 2012), and agree with Kruzel et al., Holland and Fowler showed that Vancomycin as a susceptible drug for treating S. aureus infection (Kruzel et al., 2011) (Holland & Fowler, 2011).
The cefoxitin disc diffusion method was more sensitivity and specificity than other routinely methods used for detection of MRSA. This method can be preferred in clinical microbiology laboratories, because it is easy to prepare (Mundhada et al., 2016).
S. aureus is a major causes of community-acquired and nosocomial infections .They are also among the most frequently isolated bacteria in clinical microbiology laboratories (Gosbell et al., 2001). Simple and rapid identification and discrimination of S. aureus is detection of MET resistance is essential for prompt institution of effective antimicrobial chemotherapy and for limiting the unnecessary use of certain classes of antibiotics. Since MET resistance (encoded by mec A) are mediated by the expression of PBP2a, PCR detection of mec A is considered the “gold standard” for the detection of MET resistance (Zhang et al., 2004,
Our results confirmed the presence of mec A gene among 34 (68%) isolates of S. aureus and 16(32%) had not mec A gene.
Other investigators have shown that the presence of mecA gene correlates 100% with the detection of methicillin resistance in S.aureus when it is compared with the other methods (Thompson et al., 1982). From 126 S.aureus isolates, 87 (69%) isolates harbor the mec A gene and identified as methicillin-resistant S.aureus (MRSA) and the remaining 39 (31%) isolates were methicillin-susceptible (MSSA) (Roxana et al., 2014). Sixty seven (83.8%) of the 80 MRSA isolates and 26.8% of the total 250 Staphylococcus aureus isolates which were tested, were found to be multidrug resistant MRSA (Rashedul et al., 2016).
The MIC value of vancomycin indicated that all isolates were sensitive to vancomycin. The genetic mechanism of vancomycin resistance in VRSA is not well understood. Several genes have been proposed as being involved in certain clinical VRSA strains (Jansen et al., 2007). The experimental transfer of the vanA gene cluster from E. faecalis to S. aureushas raised fears about the occurrence of such genetic transfer in clinical isolates of methicillin resistant S. aureus. In this study, all the phenotypically VRSA isolates contained mecA, but no isolates contained vanA. Tiwari & Sen (2006) have also reported a van gene-negative VRSA.Results of Rana-Khara et al., showed that all strains sensitive for vancomycin (Rana-Khara et al., 2016), which agree with our result.
In conclusion, the results of this study showed the occurrence of vanA gene-negative VRSA in Egypt. Though resistance to Vancomycin among Staphylococcus aureus isolates is not found in our study, the MRSA prevalence is higher and calls for measures to control its spread in hospital and infections due to it. Moreover regular testing to detect vancomycin resistance should be carried out so that the emergence of any resistant strain is detected, especially among the MRSA strains. A large proportion of these MRSA was found to be multidrug resistant, which takes attention to strict antibiotic policy should be enforced to curtail the irrational use of antibiotics.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
- Anupurba S., Sen M.R., Nath G., Sharma B.M., Gulati A.K., Mohapatra T.M. Prevalence of methicillin resistant Staphylococcus aureus in a tertiary referral hospital in eastern Uttar Pradesh. Indian J Med Microbiol.; 2003; 21:49–51
- Bhateja, P., Mathur, T., Pandya, M., Fatma, T., and Rattan, A., Detection of vancomycin resistant Staphylococcus aureus: A comparative study of three different phenotypic screening methods. Indian J Med Microbial. 2005; 23(1): 52-55.
- Biswajit, S., Anil, K.S., Abhrajyoti, G., and Manjusri, B., Identification and characterization of a vancomycin-resistant Staphylococcus aureus isolated from Kolkata (South Asia). J MedMicrobial; 2008; 57: 72-9.
- Brakstad, O.G., Aasbakk, K., and Maeland, J.A., Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J ClinMicrobiol. 1992; 30(7):1654–60.
- Clinical and Laboratory Standards Institute., Performance standards for antimicrobial susceptibility testing, 22nd informational supplement (M100-S22). Wayne, Pa: Clinical and Laboratory Standards Institute 2012.
- Clinical and Laboratory Standards Institute., Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement. CLSI document M100 S24. Wayne, PA: Clinical and Laboratory Standards Institute 2014.
- De vos, P., et al., Bergey’s manual of systematic bacteriology, the firmicutes. Springer, 2nd Edition, 2009; 19-124, 392-420.
- Duran, N., Ozer, B., Duran, G.G., Onlen, Y., and Demir, C., Antibiotic resistance genes & susceptibility patterns in staphylococci. Indian J Med Res 2012; 135: 389–96.
- Gosbell, I.B., Neville, S.A., Mercer, J.L., Fernandes, L.A., and Fernandes, C.J., Evaluation of the MRSA-Screen test in detecting oxacillin resistance incommunity and hospital isolates of Staphylococcus aureus. Pathology. 2001; 33: 493–5.
- Holland, T. L., and Fowler, Jr. VG., Vancomycin minimum inhibitory concentration and outcome in patients with Staphylococcus aureus bacteremia: pearl or pellet? J Infect Dis, 2011; 204(3):329–31.
- Jansen A, Turck M, Szekat C, Nagel M, Clever I, Bierbaum G. Role of insertion elements and yycFGin the development of decreased susceptibility to vancomycin in Staphylococcus aureus. Int J Med Microbiol. 2007; 297:205–15
- Khan, S.A., Feroz, F. and Noor, R., Study of extended spectrum b-lactamase producing bacteria from urinary tract infection in Dhaka city, Bangladesh. Tzu Chi Med J, 2013; 25: 39-42.
- Kruzel, M.C., Lewis, C.T., Welsh, K. J., Lewis, E.M., Dundas, N.T., Moher, J. F., et al., Determination of vancomycin and daptomycin MICs bydifferent testing methods for methicillin-resistant Staphylococcus aureus. J ClinMicrobiol. 2011; 49(6):2272–3.
- Kshetry, A.O. Pant, N.D. Bhandari, R., et al., “Minimum inhibitory concentration of vancomycin to methicillin resistant Staphylococcusaureus isolated from different clinical samples at a tertiary care hospital in Nepal,” Antimicrobial Resistance &Infection Control, 2016; 5(1): article 27.
- Mathews, A.A. Thomas, M., Appalaraju, B., Jayalakshmi, J., Evaluation and comparison of tests to detect methicillin resistant Staphylococcusaureus. Indian J PatholMicrobiol., 2010; 53(1): 79-82.
- Mehrotra, M. Wang, G., and Johnson, W.M., Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxin, toxic shock syndrome toxin 1,and methicilin resistance. J ClinMicrobiol; 2000; 38(3):1032-5.
- Mundhada, G. S., Ingole, V. K., Bhise, P.M., and Powar, M.R., Comparision of four diagnostic phenotypic methods for detection of methicilin resistant staphylococcus aureus (MRSA). Int J Pharm Bio Sci. 2016; 7(2): (B) 243 – 247.
- Noor, R., and Munna, M. S., Emerging diseases in Bangladesh: current microbiological research. Tzu Chi Med J 2015; 27:49-53.
- Rana-Khara R, Lakhani SJ, Vasava S, Shah K, Panjwani D. Methicillin Resistant Staphylococcus aureus (MRSA) and Vancomycin Resistant Staphylococcus aureus (VRSA) from a rural based tertiary care and teaching hospital in Vadodara district, Gujarat. IAIM, 2016; 3(7): 187-195.
- Rashedul, Hasan., Mrityunjoy, Acharjee. and Rashed Noor: Prevalence of vancomycin resistant Staphylococcus aureus (VRSA) in methicillin resistant S. aureus (MRSA) strains isolated from burn wound infections Tzu Chi Medical Journal, 2016; 28; 49-53.
- Roxana, SAHEBNASAGH., Horieh, SADERI. And Parviz, OWLIA. The Prevalence of Resistance to Methicillin in Staphylococcus aureus Strains Isolated from Patients by PCR Method for Detection of mecA and nuc Genes. Iranian J Publ Health, 2014; 43(1): pp. 84-92.
- Shittu, A.O., Oken, K., Adesida, S., Oyedara, O., Witte, W., Strommenger, B. et al. Antibiotic resistance and molecular epidemiology of Staphylococcus aureus in Nigeria. BMC Microbiol, 2011; 11: 8.
- Thompson, R. L., Cabezudo, I. and Wenzel, R.P. Epidemiology of nosocomial infections caused by methicillin-resistant Staphylococcus aureus. Ann Intern Med, 1982; 97: 309–17.
- Tiwari H.K., Sen M.R., Emergence of vancomycin resistant Staphylococcus aureus(VRSA) from a tertiary care hospital from northern part of India. Infect Dis. ; 2006; 6:156
- Zhang, K., Sparling, J., Chow, B. L., Elsayed, S., Hussain, Z. and Church, D.L. New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus au-reus from coagulase-negative staphylococci. J Clin Microb, 2004; 42(11): 4947-55.
- Hiramatsu K., Hanaki H., Ino T., Oguri Y.T., Tenover F.C. Methicillin-resistant Staphylococcus aureusclinical strain with reduced susceptibility. J Antimicrob Chemother. 1997; 40:135–6. [PubMed]
- Cui L., Iwamoto A., Lian J.Q., Neoh H.M., Maruyama T., Horikawa Y., et al: Novel mechanism of antibiotic resistance originating in vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother. 2006; 50:428–38. [PMC free article] [PubMed]
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.