ISSN: 0973-7510
E-ISSN: 2581-690X
This study aimed to characterize, validate, and evaluate the plant growth potential of bacterial isolates (E-2, T-2, and T-1) to determine their suitability for application as biofertilizers and/or plant-biostimulants. The plant growth-promoting potential of bacteria (E-2, T-2, and T-1) has been validated in a hydroponic study on paddy plants by inoculating bacterial isolates and monitoring the phenotypic and plant growth responses. The applicability of bacteria was tested based on their tolerance to salinity, susceptibility to antibiotics, and identification based on 16S rDNA sequencing. The isolates E-2, T-2, and T-1 improved plant growth variably and significantly (P < 0.05 at 95% confidence interval) when inoculated into the plant growth matrix, ensuring nutrient availability to the plants grown under a nutrient (nitrate or phosphate) deprived growth matrix. Isolates E-2, T-2, and T-1 grew at salt (NaCl) concentrations of 7%, 6%, and 6%, respectively, and were tolerant to saline conditions. Although these three isolates exhibited resistance to certain antibiotics, they were susceptible to a large number of readily available antibiotics. Isolates E-2, T-2, and T-1 were identified as Klebsiella sp. strain BAB-6433, Citrobacter freundii strain R2A5, and Citrobacter sp. DY1981 respectively, and all of these may be assigned to Risk-Group-2 and hence are safe in view of their susceptibility to readily available antibiotics. Hence, these isolates are promising for extensive evaluation as bioinoculants to ecologically improve soil quality, fertility, crop growth, and yield.
PGPR, Hydroponic study, Root length, Salinity, Shoot length, Plant-biostimulant, Citrobacter, Klebsiella
Human society is largely dependent on agricultural crop vegetation for their requirement of food, nutrition, and other livelihoods. A rise in agricultural products is the need of today and tomorrow to meet the food requirements of the growing human population. The extensive use of chemical fertilizers/pesticides to meet the increased demand poses many threats to various life forms and environment.1,2 Plant growth-promoting rhizobacteria (PGPR) serve as sustainable and environment friendly approach for improving crop yield and offer an alternative to the extensive application of chemical fertilizers and pesticides.3-7 The potential of PGPR in the development of sustainable agriculture systems has been investigated earlier and is still being explored in the search for PGPR that can be applied under various environmental conditions.8-10,3,4,5,11
PGPR colonize plant roots in different manners and enhance their growth by a wide variety of mechanisms, including providing plant nutrient and exogenous plant growth hormones, protecting against plant pathogens, and helping plants tolerate many biotic and abiotic stresses.12-15,9,10 Plant roots exude various compounds of nutritional and regulatory importance, such as carbohydrates, organic acids, amino acids, proteins, enzymes, flavonoids, and indole compounds, into the rhizosphere, which interact with microbes in soil to establish the microbial community (some of these microbes are PGPR) around the plant root system.16-19 PGPR enhance plant growth and crop yield via many different mechanisms, such as the formation of a suitable soil structure, decomposition of organic matter, mineralization and recycling of essential elements, and mineral nutrients such as nitrogen fixation, phosphate solubilization, and siderophore production and release of plant growth regulators such as indole acetic acid (IAA), which influences root system architecture, reduction of plant ethylene-mediated stress via microbial ACC (aminocyclopropane carboxylate) deaminase, biocontrol of soil and seed-borne plant pathogens, and others.20,8,15
Nitrogen (N) and phosphorus (P) are essential nutrients for plant growth. Naturally, it is available in various combined organic and inorganic forms that plants cannot take directly because of their complexity. Many microorganisms in the rhizosphere have the potential to convert these complex forms, through nitrogen fixation and phosphate mobilization, to forms suitable for plants and hence promote plant growth and yield.21-25. In addition, PGPR may improve plant growth by producing phytohormones and iron chelating compounds.21,26 Iron chelating compounds, such as bacterial siderophores, which help in efficient iron acquisition, can be easily taken up by plants for their own growth. Thus, PGPR improves the growth of plants, which is measured and represented by growth parameters such as length, number, and biomass of whole plants or parts such as roots, shoots, and others.27-29
The use of PGPR has also been studied in many crops, such as rice, potato, sugar beet, radish, and sweet potato, where increases in product quality, growth, and yields are evident.30-35 Many bacteria, such as Agrobacterium, Arthrobacter, Azospirillum, Azotobacter, Bacillus, Burkholderia, Caulobacter, Chromobacterium, Enterobacter, Erwinia, Flavobacterium, Gluconacetobacter diazotrophicus LMG7603, Herbaspirillum seropedicae LMG6513, Micrococcous, Pseudomonas, Rhizobium and Serratia have been found to enhance plant growth.36,37,15 Identification of efficient PGPR as potentially useful microbes for application in agriculture to improve plant growth, health, and crop yields has received attention over the past several years, and research has frequently been carried out successfully in field experiments.30,38,39 Many biofertilizers are in application across the globe, but their selection is subjective to their performance, which may be influenced by various environmental factors (temperature, salinity, pH, and many others that influence the survival and growth of microorganisms) that vary across geographically distinct locations and with time.40,41 Genetic variation is an ongoing process, which may be spontaneous or induced, leading to the emergence of efficient and new PGPR genera or species. Thus, the isolation of efficient PGPR strains at par application environments is a prime concern for eco-friendly and sustainable agriculture. Safety issues are of utmost importance, especially when studies must be carried out under field conditions to explore the in-field performance of PGPR isolates. The present work has been undertaken to validate and evaluate the plant growth-promoting potential, tolerance to saline environment, and preliminary concern about safety issues of rhizospheric bacteria in lab conditions, which can be further investigated under field conditions for application as a biofertilizer/plant-biostimulant.
Source and information of the microbial isolates
Three bacterial isolates (E-2, T-2, and T-1) were obtained from Kumar’s laboratory at Central University of Jharkhand, India.9,10 All three isolates exhibited four plant growth-promoting characteristics: (1) nitrogen fixation, (2) phosphate solubilization, (3) siderophore production, and (4) Indole acetic acid production.9,10 These three isolates were also tolerant and grew in a broad pH range (pH 3.5–10).10 These three isolates were used in this study.
Surface sterilization of paddy seeds
All experiments were performed in the Department of Life Sciences, Central University of Jharkhand, India. Plant inoculation assays were performed according to the modified method of Majeed et al.28 A total of 200 paddy seeds of variety Gorakhnath-509 (hybrid paddy, minimum germination 80%, minimum genetic purity 95%, minimum physical purity 98%, recommended area of cultivation including Jharkhand, produced and marketed by Nath Bio-Genes (I) Ltd. Nath House-India) with an average weight of 16.6 ± 0.2 mg per seed were used in this study. Seed surface sterilization was performed aseptically.42-44 Briefly, paddy seeds were washed thrice with 50 mL autoclaved double-distilled water (DDW) with vigorous shaking at 250 rpm at room temperature for 5 min. Water was then decanted, and the seeds were treated with 70% ethanol with shaking at 250 rpm for 2 min. The seeds were then rinsed thrice with autoclaved DDW for 2, 5, and 15 min. Steps of ethanol treatment followed by rinsing with autoclaved DDW were repeated. Seeds were then treated with 0.1% mercuric chloride (HgCl2) for 2 min and 1% sodium hypochlorite for 15 min, respectively, with each treatment followed by rinsing thrice with autoclaved DDW. Five surface-sterilized seeds and 100µL of water from the last wash step were plated on sterile Luria-Bertani (LB) agar media plates separately to check for sterility. After the microbial sterility check, the remaining seeds were soaked aseptically overnight in 50 mL autoclaved DDW. Seed germination was assessed prior to the hydroponic experiment. Most of the seeds germinated (86-88% germination) 3 d after sowing in the sand matrix flooded with Hoagland media.
Hydroponics study: Growth of paddy plants under different nutritional and inoculant control
Overnight-soaked seeds were sown (buried approximately 1 cm deep) aseptically in six beakers (volume-250 mL, height 95 mm, outer diameter 70 mm, inner diameter 63 mm). Each beaker contained sterilized sand-medium (red sand- repeatedly washed and autoclaved, 300 g of sand attained height of 74 mm and volume 183.78 cm3, added with 85 mL respective growth media) and 15 paddy seeds. The beakers were labeled 1-6:1- nitrate deficient Hoagland media with bacterial inoculum as H-NO3¯(IN), 2- nitrate deficient Hoagland media without bacterial inoculum as H-NO3¯(UNIN), 3- Hoagland media without bacterial inoculum as H(UNIN), 4- Hoagland media with bacterial inoculum as H(IN), 5- phosphate deficient but tri-calcium phosphate (TCP)-supplemented Hoagland media without bacterial inoculum as H-PO4¯+TCP(UNIN), and 6- phosphate-deficient but TCP supplemented Hoagland media with bacterial inoculum as H-PO4¯+TCP(IN). All the beakers containing seeds were kept under similar growth conditions of light, temperature, and humidity and kept in the same chamber of an enclosed light rack, and hydroponics experiments were performed in months of April-July for germination and growth.
Following germination of seeds, 3 days after sowing, 10 mL of bacterial cells (O.D. 1 in normal saline) was added to beakers labeled as (IN), whereas others labeled as (UNIN) received 10 mL of normal saline. A 10 mL (approximate volume determined equivalent to volume of water loss) autoclaved DDW was added on alternate days to the plant growth-supporting matrix in every beaker to keep the sand media flooded (~ 10 mm). Various phenotypic parameters of plant growth were recorded over 21 days following inoculation. Plants were uprooted on the 21st day, and the length of root (longest root in the plant) and shoot (length with longest leaf in the plant), number of leaves and number of root fibers/branches, fresh weight of roots, and fresh weight of shoots, as used by Souza et al30 Kumar et al38 and Majeed et al28 were measured separately for each plant except for fresh weight (Supplementary file). The root weights of individual plants in some treatment sets were very low. Therefore, the shoots and roots of all plants were cut and separated. Fresh weight of shoot/root of all plants in a treatment set, at a time, was measured and the average was calculated by dividing it with the number of plants in the respective treatment set.
Data analysis
One-way analysis of variance (ANOVA) was performed to determine the effects of different treatment/test conditions on plant growth. Plant growth promoting potential of all the three isolates E2, T2 and T1 was evaluated under the mentioned conditions except ‘Hoagland media without bacterial inoculum’ condition which was omitted for bacterial isolate T1. For each trait (shoot length, root length, ratio of shoot length to root length, number of leaves in each plant, and number of roots in each plant), data were analyzed using GraphPad Prism Software 5.0 (GraphPad, San Diego, CA, USA) through one-way analysis of variance (ANOVA) (Tukey’s post-hoc test: compared all pairs of columns). The results are expressed as mean ± standard deviation (SD). Replicates of plants (individual plants in each treatment set) were taken and ANOVA (a = 0.05) was performed using the measured values of the plant traits.
Growth response of isolates on different concentrations of salt
Qualitative growth response of PGPR isolates to different concentrations of salt (NaCl in LB agar media: 0%, 1%, 2%, 3%, 4%, 5%, 6%, 7%, and 8%) was performed according to the method described by Swarupa and Kumar9. The plates were checked by observing the relative visible growth density and opacity against the light source of the bacterial culture spot on the culture plates, and the image was captured as a record.
Antibiotic sensitivity profiling of the isolates
An antibiotic (antimicrobial agents) sensitivity assay was performed on Mueller Hinton agar by the standard disc diffusion method45 using an antibiotic sensitivity teaching kit (HTM002-15PR; HiMedia Laboratories Pvt. Ltd, India). The bacterial isolates were grown overnight in LB broth at 37 °C with shaking at 160 rpm. Bacterial cultures were centrifuged (MIKRO 200R, Hettich ZENTRIFUGEN) at 10000xg for 2 min at room temperature. The pellet was washed and resuspended in sterile normal saline (0.85% NaCl). Bacterial suspensions in normal saline were spread on sterile Mueller-Hinton agar using sterile cotton swabs. The discs of the respective antibiotics, as given in Table 1, were placed on the bacterial lawn spread on Mueller-Hinton agar. The diameters of the inhibition zones were measured in millimeters after 24 h and 48 h of incubation at 37°C. The sensitivity and resistance of the bacteria to the respective antibiotics were inferred from the standard reference zone of inhibition (Table 1).
Table (1):
Reference zone-size of inhibition by antibiotics.
Antibiotics |
Concentration |
Resistant (mm or less) |
Intermediate (mm) |
Sensitive (mm or more) |
|---|---|---|---|---|
Chloramphenicol |
30 µg |
12 |
13-17 |
18 |
Gentamicin |
10 µg |
12 |
13-14 |
15 |
Kanamycin |
30 µg |
13 |
14-17 |
18 |
Tetracycline |
30 µg |
14 |
15-18 |
19 |
Vancomycin |
30 µg |
0 |
0 |
15 |
Penicillin-G |
10 units |
14 |
0 |
15 |
Ampicillin |
10 µg |
13 |
14-16 |
17 |
Source of reference value: Enterobacteriaceae, Staphlycoccus spp, Pseudomonas spp., Enterococcus spp., Neisseria meningitides, Streptococcus spp. (beta haemolytic group) (HiMedia Laboratories Pvt. Ltd, India).
Identification of isolates by 16S-rRNA
Phylogenetic analysis of bacterial isolates E-2, T-2, and T-1 was done using the 16S-rRNA gene sequence. Phylogenetic analysis services were procured from Xcelris Labs Ltd. (India). The genomic DNA of E-2, T-2, and T-1 isolates was subjected to 16S-rRNA PCR in a Veriti® 96 well Thermal Cycler (Model No. 9902) using specific universal primers 8F-(5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R-(5’-GGTTACCTTGTTACGACTT-3’). Samples of 16S-rRNA gene specific amplicons (~1.5 kb) from E-2, T-2, and T-1 were sequenced by Sanger sequencing on an ABI 3730xl Genetic Analyzer using the BDT v3.1 Cycle sequencing kit. The 16S-rDNA sequence was used to identify similarities in the GenBank database using the BLAST alignment search tool of the National Center for Biotechnology Information (NCBI). The first 15 sequences were selected based on the maximum identity score and aligned using the multiple alignment software program, ClustalW. The neighbor-joining method was used to establish the evolutionary history of the bacteria.46 The bootstrap consensus tree inferred from 1000 replicates was used to represent the evolutionary history of the taxa analyzed.47 Branches corresponding to partitions reproduced in less than 50% of bootstrap replicates were collapsed. Kimura 2 parameter method was used to infer evolutionary distance in the units of base substitution per site.48 Overall, 16 nucleotide sequences, including our query sequence, were used in this analysis, and evolutionary analyses were conducted in MEGA7.49
Effect of isolates on the growth of paddy plants
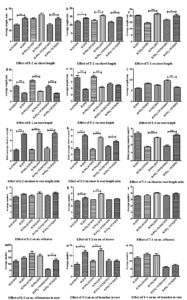
Paddy plants grown hydroponically under identical conditions with and without inoculation of isolates E-2, T-2, and T-1 were used to study the effect of bacteria on the growth of paddy plants. Phenotypic symptoms, such as leaf color, are indicators of the nutrient and health status of plants. Leaves become chlorotic under nitrogen/phosphorus deficiency and light green under nitrogen deficiency, whereas plant growth is stunted under phosphorus deficiency.50 Leaves of plants growing in nitrate-or phosphate-deficient media and without bacterial inoculum (Fig. 1: a-2, a-5, c-2, c-5, e-2, e-5) were yellowish-green and chlorotic, respectively, compared to plants grown in nitrate-or phosphate-deficient media but inoculated with bacteria (Fig. 1: a-1, a-6, c-1, c-6, e-1, e-6), where the leaves of plants were dark green.
Fig. 1. Photograph of plants under hydroponic study revealing morphological appearance of effect of PGPR isolates: letters a-b, c-d and e represent the influence of isolates E-2, T-2 and T-1 respectively; a, c and e represents standing paddy plants at day 21; b and d represents uprooted plants respectively from a and c at day 21; numerals 1-6 in boxes represents nutrient and inoculant condition in respective pots: 1- H-NO3¯(IN), 2- H-NO3¯(UNIN), 3- H(UNIN), 4- H(IN), 5- H-PO4¯+TCP(UNIN), 6- H-PO4¯+TCP(IN); where H- Hoagland, IN- Inoculated with bacterial isolate, UNIN- Uninoculated, TCP- Tricalcium phosphate, H-NO3– – nitrate deficient Hoagland medium, H-PO4 –– phosphate deficient Hoagland medium, +TCP- supplemented with TCP. Treatment condition “3” was omitted in setup “e” because the effect of inoculant had to be mainly compared with nutrient deficient condition.
Statistical analyses of the plant growth responses to isolates E-2, T-2, and T-1 under different test conditions are shown in Fig. 2-3. The contribution of the bacterial inoculum was mainly focused on nutrient (nitrate/phosphate)-deficient treatment sets. With all the isolates, the length of shoots of plants growing in ‘treatment sets with bacterial inoculum’ were more (significant except E-2 under nitrate deficient condition) than shoot length of plants grown in ‘treatment sets without inoculum’ (Fig. 2a-c). With all the isolates, the length of roots of plants growing in ‘treatment sets with bacterial inoculum’ were shorter (significant except T-2 under phosphate deficient condition and T-1 under nitrate deficient condition) than the roots of plants grown in ‘treatment sets without inoculum’ (Fig. 2d-f). The ratios of the shoot-length to root-length (SL/RL) of plants in ‘treatment sets with bacterial inoculum’ were significantly higher than the ratios SL/RL of plants in ‘treatment sets without inoculum’ (Fig. 2g-i). Increases in the number of leaves were recorded under all test conditions when augmented by bacterial inoculum, although the increases were insignificant except for T-2 under nitrate-deficient conditions (Fig. 2j-l). The augmentation of media by bacterial inoculum resulted in a significant increase in the number of roots per plant under phosphate-deficient conditions in the case of E-2, the control (without nutrient deficiency in Hoagland medium), and nitrate-deficient conditions in the case of T-2 (Fig. 2m-o). In the other cases, the number of roots either increased or decreased insignificantly (Fig. 2m-o).
Fig. 2. Graph showing effect of isolates E-2, T-2 and T-1 on various parameters of the plants: shoot length (a-c), root length (d-f), shoot to root length ratio (g-i), number of leaves (j-l), number of branches in root (m-o); * corresponds to statistical significant difference as shown in ANOVA tables; attributes of axes ‘X’: H-NO3¯(IN), H-NO3¯(UNIN), H(UNIN), H(IN), H-PO4¯+TCP(UNIN), H-PO4¯+TCP(IN); where H- Hoagland, IN- Inoculated with bacterial isolate, UNIN- Uninoculated, TCP- Tricalcium phosphate, H-NO3– – nitrate deficient Hoagland medium, H-PO4– – phosphate deficient Hoagland medium, +TCP- supplemented with TCP. ns- not significant, *- P <0.05, **- P <0.01, ***- P <0.001
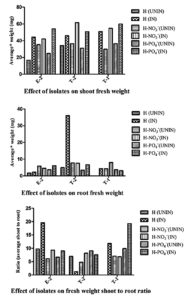
All isolates contributed to increased average fresh weight of shoots under all growth media conditions when inoculated with bacterial inocula E-2, T-2, or T-1 (Fig. 3). Although a definite trend was not seen, the observations on root biomass were represented in Fig. 3. Fresh biomass of roots increased in ‘control’ or ‘phosphate deficient media’ when inoculated with bacteria E-2 or T-2. Fresh biomass of roots was found to be either slightly decreased or comparable in nitrate-deficient media when inoculated with bacteria E-2 or T-2, respectively. The fresh biomass of roots was found to increase seven-fold when the control medium was inoculated with bacteria T-2. The fresh biomass of roots increased in nitrate-deficient media when inoculated with bacteria T-1, whereas it decreased in phosphate-deficient media when inoculated with the same bacteria. Similarly, the shoot biomass to root biomass ratio did not follow a definite trend, except when inoculated with E-2.
Fig. 3. Effect of isolates E-2, T-2 and T-1 on biomass of the plants: average fresh weight of shoot and root, fresh weight shoot to root ratio; average was calculated as (to avoid measurement errors): total fresh weight of shoots/roots of surviving plants in a treatment set divided by total number of surviving plants of respective treatment set pot; legend symbols of the graph: H-NO3¯(IN), H-NO3¯(UNIN), H(UNIN), H(IN), H-PO4¯+TCP(UNIN), H-PO4¯+TCP(IN); where H- Hoagland, IN- Inoculated with bacterial isolate, UNIN- Uninoculated, TCP- Tricalcium phosphate, H–NO3– nitrate deficient Hoagland medium, H-PO4– – phosphate deficient Hoagland medium, +TCP- supplemented with TCP.
Growth Response of Isolates to Salt and Antibiotics
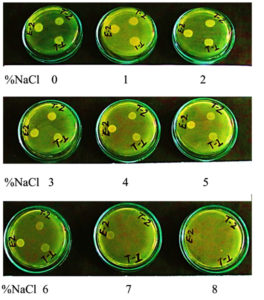
Bacterial isolates E-2, T-2, and T-1 grew in a wide range of salinity (0.0-7% NaCl), although growth was inhibited at higher salt concentrations (7% and beyond) of NaCl except E-2, which grew even at 7% NaCl (Fig. 4). The antibiotic sensitivity profile indicated that all three 3 isolates (E-2, T-2, and T-1) were resistant to ampicillin, penicillin, and vancomycin, but were intermediate or sensitive to kanamycin, tetracycline, gentamicin, and chloramphenicol (Table 2).
Table (2):
Antibiotic sensitivity profile of different bacterial isolates.
| ha | Isolates | Diameter (mm) of zone of inhibition on antibiotic sensitivity test platesb (I/R/S) | ||||||
|---|---|---|---|---|---|---|---|---|
| Ampicillin | Penicillin | Vancomycin | Kanamycin | Chloramphenicol | Gentamicin | Tetracycline | ||
| 24 | E-2 | 0(R) | 0(R) | 0(R) | 17(I) | 20(S) | 16(S) | 15(I) |
| T-2 | 9(R) | 0(R) | 0(R) | 19(S) | 22(S) | 16(S) | 16(I) | |
| T-1 | 8(R) | 0(R) | 0(R) | 19(S) | 20(S) | 16(S) | 15(I) | |
| 48 | E-2 | 0(R) | 0(R) | 0(R) | 17(I) | 21(S) | 17(S) | 16(I) |
| T-2 | 9(R) | 0(R) | 0(R) | 20(S) | 22(S) | 17(S) | 17(I) | |
| T-1 | 9(R) | 0(R) | 0(R) | 19(S) | 21(S) | 16(S) | 15(I) | |
aHours of incubation with respective antibiotics in test plate. bLetters I/R/S in parentheses stand for Intermediate/Resistant/Sensitive respectively, which were inferred from reference zone size in millimeter (mm) as per manual of HiMedia Laboratories Pvt. Ltd, India: Ampicillin (R<13, I- 14-16, S-≥17), Penicillin (R<14, I- 0, S- ≥15), Chloramphenicol (R<12, I-13-17, S->18), Vancomycin (R-0, I-0, S->15), Tetracycline (R<14, I-15-18, S->19), Kanamycin (R<13, I-14-17, S>18) and Gentamicin (R<12, I-13-14, S->15).
Fig. 4. Growth of isolates E-2, T-2 and T-1 on different concentration of salt (0-8% of NaCl) constituted in LB agar plates. White spots on the plates represent growing bacterial lawn at 24 h of incubation following spot inoculation of 5µL of respective bacterial suspension at ~1.0 optical density.9
Identification of Isolates by Similarity Search
A BLAST search and phylogenetic clustering of consensus nucleotide sequences of 16S-rDNA of E-2 (1447 bp), T-2 (1447 bp), and T-1 (1449 bp) resulted in 99% similarity and clustering of E-2, T-2, and T-1 isolates, respectively, with Klebsiella sp. strain BAB-6433, Citrobacter freundii strain R2A5, and Citrobacter sp. DY1981 (Fig. 5). Nucleotide sequences (partial sequence) of these isolates, E-2, T-2, and T-1, as identified above, have been submitted to GenBank NCBI under accession numbers MK424328, MT103108, and MT103105, respectively.
Fig. 5. 16S-rRNA gene based phylogenetic tree revealing identification of PGPR isolates E-2, T-2, T-1 and its evolutionary history. The phylogenetic tree was constructed with MEGA7 by Neighbour-Joining method with 1000 bootstrap. QC- Query coverage, MI- Maximum identity, EV- Expect value. Red triangular shape represented query sequences of isolates E-2, T-2 and T-1.
Plant growth-promoting bacteria comprise a heterogeneous mix of bacteria that can colonize plants or around the root system and also provide beneficial effects to the plants by many different mechanisms, including nitrogen fixation, phosphate solubilization, siderophore production, and IAA production among others.11,51,20,8,15 Three bacterial isolates were used in this study, which had been tested biochemically for their potential for nitrogen fixation, phosphate solubilization, siderophore production and IAA production.10
These isolates exhibited a positive influence on the growth parameters of the plants when inoculated under hydroponic conditions (Fig. 1-3). The three isolates, E-2, T-2, and T-1, were identified by 16S-rRNA gene sequencing as Klebsiella sp. strain BAB-6433, Citrobacter freundii strain R2A5, and Citrobacter sp. DY1981, respectively (Fig. 5). Other studies also reported similar findings, where Klebsiella pneumoniae type strain (KPY17657) and Citrobacter were found to be associated with plant roots of rice, wheat, and fruit plants, and also exhibited plant growth-promoting activities, such as nitrogen fixation, phosphate solubilization, siderophore production and IAA production.52,53 Klebsiella and Citrobacter freundii have also been associated with other plant species, including Zea mays,54 Triticum aestivum,55 Saccharum officinarum,56,57 and Glycine max.58 Citrobacter freundii and Klebsiella have been used for biocontrol, bioremediation, and plant growth promotion in tomato,59 maize,60,61 and sugarcane.62 However, in the present study, these bacteria were isolated from different plants such as eggplant (E-2) and tomato-plant (T-2 and T-1) but exhibited positive influence by enhancing the growth of the tested paddy plants. It also substantiated that these bacteria might not show a strict association with a particular plant, but can also be applied to improve the growth of other plants.
Paddy plants grown under nitrate-or phosphate-deficient media were short, had yellowish-green leaves, were narrow, and had fewer leaves. Application of bacterial isolates resulted in improved growth of plants with better plant height, green leaves, and an increased number of leaves, revealing that these isolates are fit to be called, as stated by Vessey,21 PGPR as a biofertilizer. Paddy growth was significantly enhanced following inoculation with E-2, T-2, or T-1, although these isolates manifested improved growth in different ways (Fig. 1-3). Isolates E-2, T-2, and T-1 increased the shoot length of plants compared to un-inoculated sets, indicating that these isolates promoted plant growth, as reported by Souza et al., (2013).30 On the other hand, root length decreased following inoculation with E-2, T-2, or T-1, which is corroborated by the interpretation of Fageria and Moreira (2011)63 that “when there is deficiency of a determined nutrient, roots try to grow longer to take nutrients from lower soil depths”. Therefore, under nutrient deficit conditions, an increase in root length should not be regarded as a growth-promoting parameter, as is the case with IAA production, which increased root length under optimum nutrient condition.64-67 Increased root length under nitrogen-or phosphorus-deficient uninoculated plants indicated that the plants are experiencing nutrient deficiency, and hence, roots grow faster to acquire more nutrients. In contrast, roots were short under conditions of nitrogen or phosphorus deficient media but were inoculated with isolates E-2, T-2, or T-1, which substantiated that these isolates supplemented deficiency of nitrogen or phosphorus by nitrogen fixation or phosphate solubilization, respectively. Similarly, fresh shoot biomass increased when plants were inoculated with isolates E-2, T-2, or T-1. The increase in shoot length to root-length ratio under inoculated nitrogen-or phosphorus-deficient media compared to its respective uninoculated sets further evidenced the growth-promoting characteristics of isolates E-2, T-2, and T-1. These findings are consistent with those of earlier reports.64-67 Thus, isolates E-2, T-2, and T-1 led to phenotypic and statistically improved plant growth. These isolates also led to increased leaf number, as reported by Sharma et al25 On the other hand, increased shoot-length to root-length ratios under N-or P-deficient conditions (Fig. 1-3) evidenced that seeds/plants were more likely to be intolerant to N and P, which has also been reported by others.68-71 Growth-promoting effects on paddy plants were also observed when isolates E-2 and T-2 were inoculated into plants growing in Hoagland media, where both ‘N’ and ‘P’ are available. The present setup seems insufficient to explain this, but nutrient enrichment by isolates may be one of the reasons especially, during the late stages when plants start experiencing nutrient deficiency.
Due to irregular rainfall patterns, human activities, extreme climatic changes, improper drainage, and inadequate leaching of mineral salts, soil salinity has increased at an alarming rate.72,73 Increased levels of soil salinity lead to physiological, molecular, and biochemical changes and reduce crop productivity and yield.74,75 The ability of these isolates E-2, T-2, and T-1 to grow at a wide range of salt concentrations (E-2: up to 7%, T-2 and T-1: up to 6% of NaCl) is of additional significance for their application to alleviate the abiotic stresses experienced by most agricultural plants, which are sensitive to high salt concentrations in agricultural fields. E-2, T-2, and T-1 showed moderately halo-tolerant character.76 Therefore, these isolates, as PGPR in saline soils, will be effective in helping glycophyte plants (such as rice) ameliorate or cope with abiotic stress and enhance crop productivity by adopting one or the other mechanisms.77-81
The isolates were identified as Klebsiella sp. strain BAB-6433, Citrobacter freundii strain R2A5, and Citrobacter sp. DY1981 annotated the risk status of the isolates E-2, T-2 and T-1, respectively. Studies have revealed the pathogenic status of Citrobacter as a nosocomial opportunistic pathogen, which may cause human diseases of insignificant clinical concern to serious complications.82-84 However, they are associated with secondary infections, either in immunocompromised patients or in patients with underlying serious medical conditions such as diabetes, burns, and use of internal medical devices, rendering the patients weak.82-84 In view of the safety aspects of the field application of isolates T-2 and T-1, the above-mentioned studies have clearly mentioned that these opportunistic pathogens are commensal to the human gut and are not the primary cause of diseases. Therefore, the pathogenesis of these bacteria is not due to the inherent virulence of Citrobacter. Despite the emergence of antibiotic resistance and the risk factor blaTEM-1 resistance gene of these bacteria, there are still reliable antimicrobial agents such as cephalosporins, amikacin, and quinolones available for controlling these resistant strains.84 Klebsiella spp. belong to two types of habitats: environment (soil, sewage, water, and plants) and mammals (humans, horses, and swine).85-90 Klebsiella infection is generally nosocomial in nature and generally affects individuals who are either immunocompromised or have underlying diseases such as type 2 diabetes, cancer, and pulmonary infections.88,89 In this study, Klebsiella spp. were isolated from rhizospheric soil; hence, its probability of being pathogenic to humans may be very low. In fact, some strains of Klebsiella pneumonia and Klebsiella oxytoca have been used as effective bioinoculants in some studies,91-93 which themselves belong to one of the most pathogenic species of Klebsiella genera.89 Therefore, based on the above facts, isolates E-2, T-2 and T-1 may be assigned Risk Group 2 (with moderate individual risk and limited community risk, includes opportunistic pathogens).94 Besides susceptibility to some tested antibiotics, E-2, T-2, and T-1 showed resistance to ampicillin, penicillin, and vancomycin, indicating the emergence of drug resistance among a number of culturable PGPR or soil microbes, which is also supported by previous finding.95 It is possible that the indiscriminate disposal of unused prescribed antibiotics in garbage and subsequent use of this garbage in agricultural fields as compost fertilizers or other unknown circumstances might have created selective pressure that consequently led to the development of antibiotic-resistant microbes in the soil.95-97 Ferjani et al., (2018)98 also found a low rate of antibiotic resistance among isolated PGPR isolates, which could be used for the development of microbial inoculums to enhance plant growth while considering human health. The sensitivity of isolates E-2, T-2, and T-1 to broad-spectrum antibiotics such as chloramphenicol, kanamycin, and gentamicin makes them suitable for safe (readily available antibiotics may help immediate prevention of infection or disease outbreak) industrial production and formulation of biofertilizers for field application. Although antibiotic resistance of PGPR has been sporadically investigated by others,98 the risk status of PGPR has also been explored in this study by literature search and 16S-rDNA based identification.
The cited studies from the literature and the antibiotic sensitivity study of this study are suggestive of the safe application of E-2, T-2, and T-1. However, safety parameters must be evaluated experimentally in detail or caution should be taken prior to the release of antibiotic-resistant bacteria, especially if such resistance genes are borne on mobile genetic elements. Further field studies using these isolates may be required to evaluate their response under different prevailing environmental conditions, including the influence of other biotic communities that influence the growth of PGPR microbes.99,9,10
The isolates E-2, T-2, and T-1 were identified as Klebsiella sp. strain BAB-6433, Citrobacter freundii strain R2A5 and Citrobacter sp, DY1981 respectively, which revealed their positive impact on plant progression parameters in different manners under hydroponic study, by improving plant height, maintaining healthy leaves, and increasing the number of leaves. Their moderate halo-tolerant character make them fit under the abiotic stress of saline soil. Being sensitive to broad-spectrum antibiotics and belonging to a low-risk group of organisms, these isolates are safe and reliable for biofertilizer formulation for field application.
Additional file: Additional Tables, Table 1-12
ACKNOWLEDGMENTS
The authors would like to thank Dr. PN Jha, Ph.D., Associate Professor, Department of Biological Science, Birla Institute of Technology & Science, Vidya Vihar, Pilani 333031, Rajasthan, India for Pre-Submission Review of the manuscript.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
MK, PS and AK designed the experiments. MK and PS performed the experiments. MK, PS and AK analyzed the data and wrote the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
This study was partially supported by grants CSIR (09/1126(0001)/2014-EMR-1) and partly by DBT-Builder program [BT/PR9028/INF/22/193/2013] from the DBT-GoI and Central University of Jharkhand.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript and in the supplementary files. Nucleotide sequences of E-2, T-2 and T-1 are available on the https://www.ncbi.nlm.nih.gov/ server under the accession numbers MK424328, MT103108, MT103105 respectively.
- Gupta PK. Pesticide exposure- Indian scene. Toxicology. 2004;198(1-3):83-90.
Crossref - Aktar MW, Sengupta D, Chowdhury A. Impact of pesticides use in agriculture: their benefits and hazards. Interdiscip Toxicol. 2009;2(1):1-12.
Crossref - Lucy M, Reed E, Glick BR. Applications of free living plant growth-promoting rhizobacteria. Antonie Leeuwenhoek. 2004;86(1):1-25.
Crossref - Adesemoye AO, Torbert HA, Kloepper JW. Plant growth-promoting rhizobacteria allow reduced application rates of chemical fertilizers. Microb Ecol. 2009;58(4):921-929.
Crossref - Hungria M, Nogueira MA, Araujo RS. Co-inoculation of soybeans and common beans with rhizobia and azospirilla: strategies to improve sustainability. Biol Fertil Soils. 2013;49(7):791-801.
Crossref - Altaf MM, Khan MSA, Ahmad I. Functional Diversity of Plant Growth-Promoting Rhizobacteria: Recent Progress and Future Prospects. In Singh D, Gupta V, Prabha R (eds.), Microbial Interventions in Agriculture and Environment. Springer, Singapore. 2019:229-253.
Crossref - Ahmad F, Husain FM, Ahmad I. Rhizosphere and Root Colonization by Bacterial Inoculants and Their Monitoring Methods: A Critical Area in PGPR Research. In: Ahmad I, Ahmad F, Pichtel J (eds.), Microbes and Microbial Technology. Springer, New York. 2011:363-391.
Crossref - Sivasakthi S, Usharani G, Saranraj P. Biocontrol potentiality of plant growth promoting bacteria (PGPR) – Pseudomonas fluorescence and Bacillus subtilis: A review. Afr J Agri Res. 2014;9(16):1265-1277.
- Swarupa P, Kumar A. Impact of Chlorpyrifos on Plant Growth Promoting Rhizobacteria Isolated from Abelmoschus esculentus. J Pure Appl Microbiol. 2018;12(4):2149-2157.
Crossref - Kumar A, Kumari M, Swarupa P, Shireen. Characterization of pH Dependent Growth Response of Agriculturally Important Microbes for Development of Plant Growth Promoting Bacterial Consortium. J Pure Appl Microbiol. 2019;13(2):1053-1061.
Crossref - Kloepper JW, Leong J, Teintze M, Schroth MN. Enhanced plant growth by siderophores produced by plant growth promoting rhizobacteria. Nature. 1980;286(5776):885-886.
Crossref - Dimkpa C, Weinand T, Asch F. Plant-rhizobacteria interactions alleviate abiotic stress conditions. Plant, Cell Environ. 2009;32(12):1682-1694.
Crossref - Grover M, Ali SZ, Sandhya V, Rasul A, Venkateswarlu B. Role of microorganisms in adaptation of agriculture crops to abiotic stresses. World J Microbiol Biotechnol. 2011;27(5):1231-1240.
Crossref - Glick BR. Plant growth-promoting bacteria: mechanisms and applications. Scientifica. 2012;2012:963401.
Crossref - Souza RD, Ambrosini A, Passaglia LM. Plant growth-promoting bacteria as inoculants in agricultural soils. Genet Mol Biol. 2015;38(4):401-419.
Crossref - Whipps JM. The Rhizosphere Carbon utilization. In: Lynch JM, ed. UK, Wiley-Interscience, Chichester; 1990:59-97
- Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol. 2006;57(1):233-266.
Crossref - Badri DV, Vivanco JM. Regulation and function of root exudates. Plant Cell Environ. 2009;32(6):666-681.
Crossref - Badri DV, Weir TL, van der Lelie D, Vivanco JM. Rhizosphere chemical dialogues: plant-microbe interactions. Curr Opin Biotechnol. 2009;20(6):642-650.
Crossref - Vacheron J, Desbrosses G, Bouffaud ML, et al. Plant growth-promoting rhizobacteria and root system functioning. Front Plant Sci. 2013;4(356):1-19.
Crossref - Vessey JK. Plant growth promoting rhizobacteria as biofertilizers. Plant Soil. 2003;255(2):571-586.
Crossref - Khan AA, Jilani G, Akhtar MS, Naqvi SMS, Rasheed M. Phosphorus solubilizing bacteria: occurrence, mechanisms and their role in crop production. J Agric Biol Sci. 2009a;1(1):48-58.
- Zaidi A, Khan MS, Ahemad M, Oves M. Plant growth promotion by phosphate solubilizing bacteria. Acta Microbiol Immunol Hung. 2009;56(3):263-284.
Crossref - Gaby DH, Buckley JC. A comprehensive evaluation of PCR primers to amplify the nifH gene of nitrogenise. PLoS One. 2012;7(7):1-12
Crossref - Sharma SB, Sayyed RZ, Trivedi MH, Gobi TA. Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. Springer Plus. 2013;2(1):1-14.
Crossref - Vansuyt G, Robin A, Briat JF, Curie C, Lemanceau P. Iron acquisition from Fe-pyoverdine by Arabidopsis thaliana. Mol Plant Microbe Interact. 2007;20(4):441-447.
Crossref - Saharan B, Nehra V. Plant Growth Promoting Rhizobacteria: A Critical Review. Life Sci Med Res. 2011;21(1):1-30.
- Majeed A, Abbasi MK, Hameed S, Imran A, Rahim, N. Isolation and characterization of plant growth-promoting rhizobacteria from wheat rhizosphere and their effect on plant growth promotion. Front Microbiol. 2015;6:1-10.
Crossref - Islam S, Akanda AM, Prova A, Islam MT, Hossain MM. Isolation and identification of plant growth promoting rhizobacteria from cucumber rhizosphere and their effect on plant growth promotion and disease suppression. Front Microbiol. 2016;6(1360):1-12.
Crossref - Souza R, Beneduzi A, Ambrosini A, et al. The effect of plant growth-promoting rhizobacteria on the growth of rice (Oryza sativa L.) cropped in southern Brazilian fields. Plant soil. 2013;366(1):585-603.
Crossref - Khush G. Productivity improvements in rice. Nutr Rev. 2003;61(Suppl 6):114-S116.
Crossref - Shoebitz M, Ribaudo CM, Pardo MA, Cantore ML, Ciampi L, Cura JA. Plant growth promoting properties of a strain of Enterobacter ludwigii isolated from Lolium perenne rhizosphere. Soil Biol Biochem. 2009;41(9):1768-1774.
Crossref - Yanni YG, Dazzo FB. Enhancement of rice production using endophytic strains of Rhizobium leguminosarum bv. trifolii in extensive field inoculation trials within the Egypt Nile delta. Plant Soil. 2010;36(1):129-142.
Crossref - Xiao AW, Li WC, Ye ZH. The effect of plant growth-promoting rhizobacteria (PGPR) on arsenic accumulation and the growth of rice plants (Oryza sativa L.). Chemosphere. 2020;242:125136.
Crossref - Nasution RA, Tangapo AM, Taufik I, Aditiawati P. Comparison of plant growth promoting rhizobacteria (PGPR) diversity and dynamics during growth of Cilembu sweet potato (Ipomoea batatas L var. Rancing) in Cilembu and Jatinangor Site, Indonesia. J Pure Appl Microbiol. 2017;11(2):837-845.
Crossref - Govindarajan M, Balandreau J, Muthukumarasamy R, Revathi G, Lakshminarasimhan C. Improved yield of micropropagated sugarcane following inoculation by endophytic Burkholderia vietnamiensis. Plant Soil. 2006;280(1):239-252.
Crossref - Ahemad M, Kibret M. Mechanisms and applications of plant growth promoting rhizobacteria: Current perspective. J King Saud Univ – Sci. 2014;26(1):1-20.
Crossref - Kumar A, Maurya BR, Raghuwanshi R. Isolation and characterization of PGPR and their effect on growth, yield and nutrient content in wheat (Triticum aestivum L.). Biocatal Agric Biotechnol. 2014;3(4):121-128.
Crossref - Dinesh R, Anandaraj M, Kumar A, Bini YK, Subila KP, Aravind R. Isolation, characterization, and evaluation of multi-trait plant growth promoting rhizobacteria for their growth promoting and disease suppressing effects on ginger. Microbiol Res. 2015;173:34-43.
Crossref - Meena M, Swapnil P, Divyanshu K, et al. PGPR-mediated induction of systemic resistance and physiochemical alterations in plants against the pathogens: Current perspectives. J Basic Microbiol. 2020;60(10):828-861.
Crossref - Basu A, Prasad P, Das SN, et al. Plant Growth Promoting Rhizobacteria (PGPR) as Green Bioinoculants: Recent Developments, Constraints, and Prospects. Sustainability. 2021;13(3):1140-1159.
Crossref - Miche L, Balandreau J. Effects of rice seed surface sterilization with hypochlorite on inoculated Burkholderia vietnamiensis. Appl Environ Microbiol. 2001;67(7):3046-3052.
Crossref - Oyebanji OB, Nweke O, Odebunmi O, et al. Simple, effective and economical explant-surface sterilization protocol for cowpea, rice and sorghum seeds. Afr J Biotechnol. 2009;8(20):5395-5399.
Crossref - Albdaiwi RN, Khaymi-Horani H, Ayad JY, Alananbeh KM, Kholoud M, Al-Sayaydeh, R. Isolation and Characterization of Halotolerant Plant Growth Promoting Rhizobacteria from Durum Wheat (Triticum turgidum subsp. durum) Cultivated in Saline Areas of the Dead Sea Region. Front Microbiol. 2019;10:1-16.
Crossref - Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol.1966;45:493-496.
Crossref - Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406-425.
Crossref - Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39(4):783-791.
Crossref - Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111-120.
Crossref - Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731-2739.
Crossref - Chen L, Lin L, Cai G, Sun Y, Huang T, Wang K, Deng J. Identification of nitrogen, phosphorus, and potassium deficiencies in rice based on static scanning technology and hierarchical identification method. PLoS ONE. 2014;9(11):1-17.
Crossref - Barea JM, Pozo MJ, Azcon R, Azcon-Aguilar C. Microbial co-operation in the rhizosphere. J Exp Bot. 2005;56(417):1761-1778.
Crossref - Govindarajan M, Kwon SW, Weon HY. Isolation, molecular characterization and growth-promoting activities of endophytic sugarcane diazotroph Klebsiella sp. GR9. World J Microbiol Biotechnol. 2007;23(7):997-1006.
Crossref - Farina R, Beneduzi A, Ambrosini A, et al. Diversity of plant growth-promoting rhizobacteria communities associated with the stages of canola growth. Appl Soil Ecol. 2012;55:44-52.
Crossref - Hinton DM, Bacon CW. Enterobacter cloacae is an endophytic symbiont of corn. Mycopathologia. 1995;129(2):117-125.
Crossref - Joshi P, Bhatt AB. Diversity and function of plant growth promoting rhizobacteria associated with wheat rhizosphere in North Himalayan region. Int J Environ Sci. 2011;1(6):1135-1143.
- Magnani GS, Didonet CM, Cruz LM, Picheth CF, Pedrosa FO, Souza EM. Diversity of endophytic bacteria in Brazilian sugarcane. Genet Mol Res. 2010;9(1):250-258.
Crossref - de Santi Ferrara FI, Oliveira ZM, Gonzales HH, Floh EI, Barbosa HR. Endophytic and rhizospheric enterobacteria isolated from sugar cane have different potentials for producing plant growth-promoting substances. Plant Soil. 2012;353(1):409-417.
Crossref - Kuklinsky-Sobral J, Araujo WL, Mendes R, Pizzirani-Kleiner AA, Azevedo JL. Isolation and characterization of endophytic bacteria from soybean (Glycine max) grown in soil treated with glyphosate herbicide. Plant Soil. 2005;273(1):91-99.
Crossref - Caballero-Mellado J, Onofre-Lemus J, Estrada-de los Santos P, Martinez-Aguilar L. The tomato rhizosphere, an environment rich in nitrogen-fixing Burkholderia species with capabilities of interest of agriculuture and bioremediation. Appl Environ Microbiol. 2007;73(16):5308-5319.
Crossref - Di Cello F, Bevivino A, Chiarini L, et al. Biodiversity of a Burkholderia cepacia population isolated from the maize rhizosphere at different plant growth stages. Appl Environ Microbiol. 1997;63(11):4485-4493.
Crossref - Estrada P, Mavingui P, Cournoyer B, Fontaine F, Balandreau J, Caballero-Mellado J. A N2 -fixing endophytic Burkholderia sp. associated with maize plants cultivated in Mexico. Can J Microbiol. 2002;48(4):285-294.
Crossref - Bramer CO, Vandamme P, da Silva LF, Gomez JG, Steinbuchel A. Polyhydroxyalkanoate-accumulating bacterium isolated from soil of a sugar-cane plantation in Brazil. Int J Syst Evol Microbiol. 2001;51(5):1709-1713.
Crossref - Fageria NK, Moreira A. The role of mineral nutrition on root growth of crop plants. Advances in Agronomy. 2011;110:251-331.
Crossref - Pilet PE, Saugy M. Effect on root growth of endogenous and applied IAA and ABA. Plant Physiol. 1987;83(1):33-38.
Crossref - Patten CL, Glick BR. Role of Pseudomonas putida indole acetic acid in development of the host plant root system. Appl Environ Microbiol. 2002;68(8):3795-3801.
Crossref - Perrig D, Boiero ML, Masciarelli OA, et al. Plant-growth-promoting compounds produced by two agronomically important strains of Azospirillum brasilense, and implications for inoculant formulation. Appl Microbiol Biotechnol. 2007;75(5):1143-1150.
Crossref - Spaepen S, Vanderleyden J, Remans R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol Rev. 2007;31(4):425-448.
Crossref - Mollier A, Pellerin S. Maize root system growth and development as influenced by phosphorus deficiency. J Exp Bot. 1999;50(333):487-497.
Crossref - Wissuwa M, Ae N. Genotypic variation for tolerance to phosphorus deficiency in rice and the potential for its exploitation in rice improvement. Plant Breed. 2001;120(1):43-48.
Crossref - Wissuwa M. Combining a modelling with a genetic approach in establishing associations between genetic and physiological effects in relation to phosphorus uptake. Plant Soil. 2005;269(1):57-68.
Crossref - Wissuwa M, Gamat G, Ismail AM. Is root growth under phosphorus deficiency affected by source or sink limitations? J Exp Bot. 2005;56(417):1943-1950.
Crossref - Zhu F, Qu L, Hong X, Sun X. Isolation and characterization of a phosphate-solubilizing halophilic bacterium Kushneria sp. YCWA18 from Daqiao Saltern on the coast of Yellow Sea of China. Evid Based Complement Alternat Med. 2011;2011:1-6.
Crossref - Mohan V, Devi KS, Anushya A, Revathy G, Kuzhalvaimozhi GV, Vijayalakshmi KS. Screening of salt tolerant and growth promotion efficacy of phosphate solubilizing bacteria. J Acad Ind Res. 2017;5(12):168-172.
- Sairam RK, Tyagi A. Physiology and molecular biology of salinity stress tolerance in plants. Curr Sci. 2004;86(3):407-421.
Crossref - Acquaah G. Principles of plant genetics and breeding, 2nd ed. John Wiley & Sons, UK. 2009.
- Ollivier B, Caumette P, Garcia JL, Mah, RA. Anaerobic bacteria from hypersaline environments. Microbiol Rev. 1994;58(1):27-38.
Crossref - Munns R, Tester M. Mechanisms of salinity tolerance. Ann Rev Plant Biol. 2008;59:651-681.
Crossref - Munns R, Gilliham M. Salinity tolerance of crops-what is the cost? New phytol. 2015;208(3):668-673.
Crossref - Vannier N, Mony C, Bittebiere AK, Vandenkoornhuyse P. Epigenetic mechanisms and microbiota as a toolbox for plant phenotypic adjustment to environment. Front Plant Sci. 2015;6:1-8.
Crossref - Etesami H, Beattie GA. Mining Halophytes for Plant Growth-Promoting Halotolerant Bacteria to Enhance the Salinity Tolerance of Non-halophytic Crops. Front Microbiol. 2018;9:1-20.
Crossref - Alexander A, Mishra A, Jha B. Halotolerant Rhizobacteria: A Promising Probiotic for Saline Soil-Based Agriculture. In: Kumar M, Etesami H, Kumar V, eds. Saline Soil-based Agriculture by Halotolerant Microorganisms. Singapore, Springer; 2019:53-73.
Crossref - Lipsky BA, Hook III EW, Smith AA, Plorde JJ. Citrobacter infections in humans: experience at the Seattle Veterans Administration Medical Center and a review of the literature. Rev Infect Dis. 1980;2(5):746-760.
Crossref - Kus JV, Burrows LL. Infections due to Citrobacter and Enterobacter. xPharm: The Comprehensive Pharmacology Reference. Elsevier; 2007:1-12.
Crossref - Liu LH, Wang NY, Wu AY, Lin CC, Lee CM, Liu CP. Citrobacter freundii bacteremia: Risk factors of mortality and prevalence of resistance genes. J Microbiol Immunol Infect. 2018;51(4):565-572.
Crossref - Brown C, Seidler RJ. Potential pathogens in the environment: Klebsiella pneumoniae, a taxonomic and ecological enigma. Appl Microbiol. 1973;25(6):900-904.
Crossref - Matsen JM, Spindler JA, Blosser RO. Characterization of Klebsiella isolates from natural receiving waters and comparison with human isolates. Appl Microbiol. 1974;28(4):672-678.
Crossref - Seidler RJ, Knittel MD, Brown C. Potential pathogens in the environment: cultural reactions and nucleic acid studies on Klebsiella pneumoniae from clinical and environmental sources. Appl Microbiol. 1975;29(6):819-825.
Crossref - Bagley ST. Habitat association of Klebsiella species. Infect Control Hosp Epidemiol. 1985;6(2):52-58.
Crossref - Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11(4):589-603.
Crossref - Chen M, Lin L, Zhang Y, Sun L, An Q. Genome Sequence of Klebsiella oxytoca SA2, an Endophytic Nitrogen-Fixing Bacterium Isolated from the Pioneer Grass Psammochloa villosa. Genome Announc. 2013;1(4):1-2.
Crossref - Liu Y, Shi Z, Yao L, Yue H, Li H, Li CJ. Effect of IAA produced by Klebsiella oxytoca Rs-5 on cotton growth under salt stress. J Gen Appl Microbiol. 2013;59(1):59-65.
Crossref - Bhardwaj G, Shah R, Joshi B, Patel P. Klebsiella pneumoniae VRE36 as a PGPR isolated from Saccharum officinarum cultivar Co99004. J Appl Biol& Biotechnol. 2017;5(1):47-52.
Crossref - Liu D, Chen L, Zhu X, et al. Klebsiella pneumoniae SnebYK Mediates Resistance against Heterodera glycines and Promotes Soybean Growth. Front Microbiol. 2018;9:1-13.
Crossref - HS076. Classification of Microorganisms by Risk Group, UNSW, Australia: 2020. https://safety.unsw.edu.au/sites/default/files/HS076_Classification_of_
infective_microorganisms.pdf. Accessed July 29, 2020. - Smolinski MS, Hamburg MA, Lederberg J editors. Microbial Threats to Health: Emergence, Detection, and Response. 3, Factors in Emergence. Institute of Medicine (US) Committee on Emerging Microbial Threats to Health in the 21st Century; Washington (DC): National Academies Press (US). 2003. Available from: https://www.ncbi.nlm.nih.gov/books/NBK221497/
- Martinez JL. The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc Biol Sci. 2009;276(1667):2521- 2530.
Crossref - Chang Q, Wang W, Regev-Yochay G, Lipsitch M, Hanage WP. Antibiotics in agriculture and the risk to human health: how worried should we be? Evol Appl. 2015;8(3):240-247.
Crossref - Ferjani R, Gharsa H, Estepa-Perez V, et al. Plant growth- promoting Rhizopseudomonas: expanded biotechnological purposes and antimicrobial resistance concern. Ann Microbiol. 2019;69(1):51-59.
Crossref - Malusa E, Pinzari F, Canfora L. Efficacy of Biofertilizers: Challenges to Improve Crop Production. In: Singh DP, Singh HB, Prabha R, eds. Microbial Inoculants in Sustainable Agricultural Productivity. India, Springer; 2016:17-40.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.