ISSN: 0973-7510
E-ISSN: 2581-690X
Antimicrobial agents are widely used for treatment of animal and human diseases. Heavy use of antimicrobial agents permits bacteria to develop resistance to these agents specifically when a dose of antibiotic is insufficient or course of treatment is incomplete. Antimicrobial resistance genes (ARGs) are usually associated with mobile genetic elements (MGEs) including Integron therefore; these genes can transmit among bacteria via horizontal transmission. The current study was conducted to assess the possible role of manure in dissemination of antimicrobial resistance. The presence, quantitate, and diversity of resistance genes associated with Integron class 1 have been assessed using conventional and quantitative polymerase chain reaction (PCR) combined with sequencing of gene cassette within Integron and analysis of sequenced data by blast tool. Thirty-eight samples were found a positive for Integron and concentration of Integron in positive sample ranged from from 106-1010 copies/g of manure. High frequencies were detected to genes that encoded to sulphonamide and ammonium compound resistance. These genes were detected in 25% and 23% respectively of the total manure samples. In general, the detected genes in manure functionally belong to five protein families including Efflux pump, DNA repair, heavy metal resistance, membrane protein, and antibiotic resistance. Manure might act as a hotspot from which ARGs emerge and transfer to the environment and then to the animal and human environments.
Antibiotic Rgene, Integrons, Manure and Mobile Genetic Elements
Resistance to antibiotics is a whole world thread to human and animal health. The global organization (OIE, FAO, and WHO) that deal with health and nutrition has reported that antibiotic resistance is one of three important risks that threaten human life.1 By 2050, antibiotic resistant infections could cause 10 million deaths a year with accumulative cost reach to 10 trillion dollars.2 Enteric Gram-negative bacteria are considered a major reservoir for antibiotic resistant genes. Transposons, plasmids, and Integrons are mobile genetic elements that mostly carry antibiotic resistance genes (ARGs) and are responsible for transferring these genes between bacterial species via a horizontal transmission (HT).3-5
Modification of cell components, an efflux pump, and enzymatic modification or degradation of antibiotic is such mechanisms that bacteria use to resist the harmful effect of antimicrobial substances. Some of these mechanisms specifically enzymatic ones rely on ARGs that are usually associated with mobile genetic elements; therefore, these genes are considered emerging contaminants and have a critical role in dissemination of antibiotic resistance.5-11
Using of manure as an organic fertilizer is involved in spreading of resistance genes to the human and animal environment.12,13
Based on WHO reports and several researches, further studies are needed to estimate the percentage of ARGs in the environment.14,15 Therefore, information about the discharge pattern and diversity of ARGs in animal farms is needed.16,17
The current study critically examines the role of manure in spreading of ARGs to the animal and human environment.
Sample collection
Four animal farms located within the AL-Qadisiyah governorate were selected and 30 manure samples were collected from each site in December 2021. Samples were collected from the deep layer of manure using sterile glass bottles.
Purification of DNA from manure
Soil DNA extraction kit from Biomedical Ana™ (South Korea) was used to purification of Metagnomic-DNA from manure. Purification of DNAs was performed according to the manufacturer’s instructions.
Primer, PCR and qPCR
Primers were designed to amplified class 1 Integron and 16s RNA (Table). Primers were synthesized by Integrated DNA Technologies (IDT). The presence of Integron and 16s rRNA genes in the tested sample were tested using Conventional PCR (Figure 1). The quantity of Integrons and reference gene in the manure were estimated using qPCR (Figure 2).
Table :
Oligonucleotides used in PCR, RT-PCR and sequencing of integrons 1
| Primer name | Target gene | Sequence | Reference | |
|---|---|---|---|---|
| 1- | Int1 F | Integron integrase class 1 | GCCTTGATGTTACCCGAGAG | (25) |
| 2- | Int1 R | GATCGGTCGAATGCGTGT | ||
| 3- | GC F | Gene Cassette Integron 1 | GGCATCCAAGCAGCAAG | |
| 4- | GC R | AAGCAGACTTGACCTGA |
Genetic sequencing of Integron 1 gene cassette
PCR products that were positive to the presence of resistance genes within a gene cassette were further analyzed by sequencing. Sequencing of Gene cassette within Integrons performed by Macrogen (Seoul, South Korea). An Integron database (http://integrall.bio.ua.pt/?search#) and Blast tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi) is an online tool used to analyse the sequence and diversity of resistance gene within cassette.
Data analysis
The statistic program, prism 5 was used to analyse data. Differences between groups were statistically performed using one-way ANOVA, when p value was less than 0.5 the difference was considered significant. The number of intregron1 integrase and 16S rRNA copies was calculated to estimate the amount of Integron to the total number of bacteria.
Ethical approval
The primary studies under which the samples were collected received ethical clearance from the Medical Ethics Committee of Al-Qadisiyah University (approval number 262/2022).
Integron identification
Pair of primers from the conservative area of interon1, Int1-F (GCCTTGATGTTACCCGAGAG) and Int1-R (GATCGGTCGAATGCGTGT) was used to detect the presence of the target gene in farm animal manure. As shown in Figure 1, 70-80% of farm animal manure was positive for integron1. Integrons were found in 100 out of 120 (83%) of total samples. Integron 1 was found in 43 of 50 (86%) sheep manure samples, 38 of 45 (84%) of the Cow manure samples, and 19 of 25 of the Goat samples (76%) (Figure 3).
Figure 1. PCR Gel electrophoresis image (agarose 1%).Example of Positive result of integron 1 from DNA animal manure. line 1 represent DNA marker (1 KB) (NEB BioLabs®)
Figure 2. Examples of qPCR amplification plot of Positive result of integrons 1 from DNA animal manure
The abundance of integrons1 in the farm animal manure
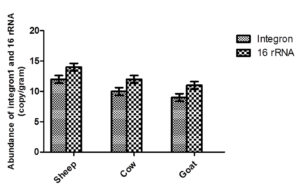
Quantitate of Integron 1 and 16S rRNA in manure was estimated using qPCR. As the result show, the concentrate of Integron – ranged from 106 – 1010 copies per gram of manure (Figure 4). The highest (but not significant difference p<0.05) of Integron was recorded in sheep manure as compared with other tested species manure, followed by cow manure, then goat. While the concentration of 16S rRNA in manure samples ranged from 108 – 1012 copies per gram of sample (Figure 4). The current study suggests that manure might act as a hotspot from which ARGs emerge and transfer to the natural environment.
Figure 4. Amount of class one Intgron1 and 16s rRNA in some farm animal manure. The value represents log 10 of genes copy number
High-throughput analysis of class 1 Integron GCs
The positive sample for Integron and their GCs were further analysed using High-throughput sequencing. 50% of Integron have no resistance gene within gene cassette. Also, 50% gene sequence within cassettes shows no hint in the NCBI gene bank.
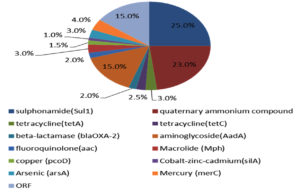
Sequencing and blast analysis show that sulphonamide resistance gene (Sul1) and & quaternary ammonium compound were detected in 25% and 23% respectively of the tested sample (Figure 5). Gene encoding resistant protein to Arsenic was recorded in 15% of the tested sample. As shown in Figure 5 the detected genes in manure at a percentage less than 5% included tetracycline genes (tetA and tetC), beta-lactamase (blaOXA-2), aminoglycoside (AadA), fluoroquinolone (aac) and Macrolide (Mph). Also, we found resist genes to heavy metals which include copper (pcoD), Cobalt-zinc-cadmium (silA), and Mercury (merC).
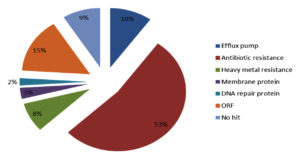
According to their function the detected genes belong to five protein families including Efflux pump, DNA repair, heavy metal resistance, membrane protein, and antibiotic resistance (Figure 6).
Figure 6. The relative abundance of protein family. Five protein family including heavy metal resistance, efflux pump, antibiotic resistance, DNA repair protein and membrane protein were show in functionally analysis of Integron gene cassette
Our study suggests that manure represents a reservoir for ARGs and MGEs and one of the important source for emerge and spread of the genes to the animal and human environment.
A strong relationship between using manure as soil fertilizer and spreading of antibiotic resistance gene and bacteria have been reported in many studies.16,17 Manure may represent a hotspot for dissemination of antibiotic resistance and as a big storage for ARGs and antibiotic resistant bacteria (ARB). Therefore manure has a critical role in role in emerging and disseminating ARGs & ARB in the natural environments.18,19
Integron is one of the mobile genetic elements that can capture, integrate, and express resistance and virulence genes. It is associated with plasmid and transposon and plays a critical role in the transfer of antibiotic resistance genes from resistance to sensitive bacteria. Furthermore, Integron represents a biological indicator for the assessment of ARGs and ARB in the environment than in animal and human.15
In the current study, PCR, qPCR, DNA sequencing, and the blast tool were used to assess the presence and abundance of Integron associated resistance genes in farm animal manure.
Results of the current study suggest that class one Integron is a dominant mobile genetic element in farm animal manure and gene cassette within Integrons carrying a wide range of antibiotic and metal resistance genes.
According to some studies, a strong relationship between the types of ARGs found in manure of certain farms and antimicrobial agents used in the same farm.20-22
Horizontal gene transfer (HGT) is the main mechanism in bacteria that facilitate transfer of ARGs from manure to soil microbes which can infect human or animal directly or by transfer to human or animal pathogen.15,23 The evidence from this study indicates that manure is one of the important sources of Integrons and resistance genes.
Recently, ARGs classified as micro-contaminants that might hurt animal and human health.24,25 Further researches are urgently needed to prevent the dissemination of ARGs to the environment. Find efficient methods for inactivation of ARGs in animal wastes, manure, and hospital wastes to reduce of emergence and dissemination of ARGS in the natural environments.23,25
Antibiotic resistance genes which are associated with mobile genetic elements including Integrons represent a potent risk factor for animal and human health. Manure has a high density of Integron and ARGs within the gene cassette of the Integron. Action is needed to prevent the spreading of these genes to various environments.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
Both authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
ETHICS STATEMENT
This study was approved by the Medical Ethics Committee of Al-Qadisiyah University, Iraq, with reference number 262/2022.
- Bolon I, Mason J, O’Keeffe P, et al. One Health education in Kakuma refugee camp (Kenya): From a MOOC to projects on real world challenges. One Health. 2020;10:100158.
Crossref - Sugden R, Kelly R, Davies S. Combatting antimicrobial resistance globally. Nat Microbiol. 2016;1(10):16187.
Crossref - Bahl MI, Sorensen SJ, Hansen LH, Licht TR. Effect of tetracycline on transfer and establishment of the tetracycline-inducible conjugative transposon Tn 916 in the guts of gnotobiotic rats. Appl Environ Microbiol. 2004;70(2):758-764.
Crossref - Goren MG, Carmeli Y, Schwaber MJ, Chmelnitsky I, Schechner V, Navon-Venezia S. Transfer of carbapenem-resistant plasmid from Klebsiella pneumoniae ST258 to Escherichia coli in patient. Emerg Infect Dis. 2010;16(6):1014-1017.
- Van Boeckel TP, Brower C, Gilbert M, et al. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci. 2015;112(18):5649-5654.
Crossref - Pruden A, Pei R, Storteboom H, Carlson KH. Antibiotic resistance genes as emerging contaminants: studies in northern Colorado. Environ Sci Technol. 2006;40(23):7445-7450.
Crossref - Wang A, Yang Y, Lu Q, et al. Presence of qnr gene in Escherichia coli and Klebsiella pneumoniae resistant to ciprofloxacin isolated from pediatric patients in China. BMC Infect Dis. 2008;8:68.
Crossref - Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268-281.
Crossref - Xiong W, Wang Y, Sun Y, et al. Antibiotic-mediated changes in the fecal microbiome of broiler chickens define the incidence of antibiotic resistance genes. Microbiome. 2018;6(1):34.
Crossref - Ma Z, Wu H, Zhang K, Xu X, Wang C, Zhu W, Wu W. Long-term low dissolved oxygen accelerates the removal of antibiotics and antibiotic resistance genes in swine wastewater treatment. Chem Eng J. 2018;334:630-637.
Crossref - Agga GE, Cook KL, Netthisinghe AM, Gilfillen RA, Woosley PB, Sistani KR. Persistence of antibiotic resistance genes in beef cattle backgrounding environment over two years aftercessation of operation. PLoS One. 2019;14(2):e0212510.
Crossref - Chee Sanford JC, Mackie RI, Koike S, et al. Fate and transport of antibiotic residues and antibiotic resistance genes following land application of manure waste. J Environ Qual. 2009;38(3):1086-1108.
Crossref - Johnson TA, Stedtfeld RD, Wang Q, et al. Clusters of antibiotic resistance genes enriched together stay together in swine agriculture. MBio. 2016;7(2):e02214-15.
Crossref - Chen B, Hao L, Guo X, Wang N, Ye B. Prevalence of antibiotic resistance genes of wastewater and surface water in livestock farms of Jiangsu Province, China. Environ Sci Pollut Res. 2015;22(18):13950-13959.
Crossref - Yang F, Han B, Gu Y, Zhang K. Swine liquid manure: A hotspot of mobile genetic elements and antibiotic resistance genes. Sci Rep. 2020;10(1):15037.
Crossref - Berendonk TU, Manaia CM, Merlin C, et al. Tackling antibiotic resistance: the environmental framework. Nat Rev Microbiol. 2015;13(5):310-317.
Crossref - World Health Organization. Antimicrobial resistance: global report on surveillance. World Health Organization. 2014.
- Lima T, Domingues S, Da Silva GJ. Manure as a potential hotspot for antibiotic resistance dissemination by horizontal gene transfer events. Vet Sci. 2020;7(3):110.
Crossref - Alsultan A, Alsallami D. Efflux-Mediated bile Resistance in Gram-Positive Pathogens. J Pure Appl Microbiol. 2022;16(1):10-17.
Crossref - An XL, Chen QL, Zhu D, Zhu YG, Gillings MR, Su JQ. Impact of wastewater treatment on the prevalence of integrons and the genetic diversity of integron gene cassettes. Appl Environ Microbiol. 2018;84(9):e02766-17.
Crossref - Kivits T, Broers HP, Beeltje H, van Vliet M, Griffioen J. Presence and fate of veterinary antibiotics in age-dated groundwater in areas with intensive livestock farming. Environ Pollut. 2018;241:988-998.
Crossref - Waseem H. Evaluation of Antimicrobial Resistance Dissemination in the Environment. (Doctoral dissertation). Islamabad:Quaid-i-Azam University;2019. http://prr.hec.gov.pk/jspui/handle/123456789/10934
- Rosser SJ, Young HK. Identification and characterization of class 1 integrons in bacteria from an aquatic environment. J Antimicrob Chemother. 1999;44(1):11-18.
Crossref - Mahmood FRR, Ahmed IM. Molecular detection of ESBL/AmpC b-Lactamase Escherichia coli isolated from sheep in Mosul city. Iraqi J Vet Sci. 2022;36(2):387-392.
Crossref - Alkhafaje WK, Olama ZA, Ali SM. Molecular characterization and microbial resistance of different bacterial isolates in some dairy products. Iraqi J Vet Sci. 2022;36(3):333-339.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.