ISSN: 0973-7510
E-ISSN: 2581-690X
Infections with P. aeruginosa are three times more common in people with diabetes than in non-diabetic individuals. Investigations disclosing the distinguishing traits of P. aeruginosa strains to cause respiratory and wound infection in diabetics is limited. Wound swab and sputum from infected diabetic patients were used for the isolation of P. aeruginosa. The confirmed isolates were evaluated for their virulence factor production, antibiotic susceptibility, and clonal relationship. The study confirmed the increased virulence of sputum isolates characterized by their multidrug resistant nature, strong biofilm formation at 72h [(p<0.05) =0.003)] and 96h [(p<0.05) =0.002)] and elaboration of proteolytic enzymes (40.0%). Albeit the fact that wound isolates were less virulent than the sputum isolates, there was an increased siderophore production (77.0%). Nearly 90.0% of the isolates including sputum and wound were resistant to colistin. Random Amplified Polymorphic DNA analysis showed no distinct lineages of wound and sputum isolates. The study disclosed the higher prevalence of virulent P. aeruginosa in causing infection in the diabetics. No distinct lineages of the wound and sputum isolates indicated their ability to adapt to different host environments. To the best of our knowledge, this is the first study to show the difference in virulence traits among the P. aeruginosa strains isolated from sputum and wound of diabetic patients. Our study distinctly reveals the significance of periodic examination of antibiotic resistance and virulence factors of P. aeruginosa in order to recognize the possible co-regulatory mechanism involved in their expression.
Pseudomonas aeruginosa, Diabetes, Antibiotic Resistance, Virulence Factors, Random Amplified Polymorphic DNA Analysis
Pseudomonas aeruginosa, an important nosocomial pathogen affecting convalescent patients in hospitals is a ubiquitous, Gram-negative, opportunistic bacteria that thrives in a variety of habitats. It depends on a wide array of virulence factors, including exoproteases, phenazines, exopolysaccharides, phospholipases, type III effectors, pili and flagella to survive and to establish an infection within the host.1 The production of many of these virulence factors is under the control of the quorum-sensing system, a communication that occurs via small chemical molecules.2 The emergence of Multidrug Resistant (MDR) P. aeruginosa is considered to be of medical importance for the presence of its advanced antibiotic resistance mechanisms. Treatment of infections caused by P. aeruginosa is more complicated due to its resistance, which leads to treatment failure or exposes patients to unexpected adverse outcomes. Infections with P. aeruginosa have been linked to a high risk of morbidity, with mortality rates ranging from 20% to 60% in hospital-acquired infections.3
The global incidence of diabetes mellitus has reached epidemic proportions and is considered to be the most prevalent metabolic disorder. According to the report of International Diabetes Federation, 366 million adults worldwide suffer with diabetes, accounting for more than 8% of the global population, and this number is anticipated to rise to over 552 million by 2030.4 India, a country with a fast developing economy is often called the ‘diabetes capital’ of the world with a considerable number of diabetic patients from the economic productive young population. People with diabetes are susceptible to infections due to their compromised defenses. Pneumonia, soft tissue infection, urinary tract infection, and periodontitis occur more frequently in people with diabetes than those without diabetes.5 It is believed that diabetic patients have a lifetime risk of developing foot ulcers. Diabetic foot ulcers by various Gram-positive and Gram-negative bacteria is the most common cause of diabetes-related hospital admissions.6 Diabetic foot infection is the most feared complication resulting in high morbidity that can end up with gangrene and amputation.7 In a majority of lower limb amputations, foot ulcers caused by biofilm forming bacteria precedes the event. Individuals with diabetes are at least ten times more likely to be hospitalized for soft tissue infections.8 In diabetic ulcers, P. aeruginosa is the most frequently detected Gram-negative pathogen and its presence in wounds is linked to a poor prognosis for healing.9 It has been proposed that uncontrolled hyperglycemia in diabetes raises the airway glucose levels, thus providing a congenial environment for the growth of microorganisms.10 As a consequence, diabetic patients with cystic fibrosis and chronic pulmonary obstruction are prone to ventilator associated pneumonia.11 P. aeruginosa is reported to be a common and troublesome multidrug resistant bacterium causing ventilator associated pneumonia.12
Studies have indicated distinct phenotypic and genotypic traits in P. aeruginosa isolated from different sites that facilitate its colonization,13,14 A better understanding of the virulence characteristics of P. aeruginosa would enable us to evaluate the heterogeneity between the strains. To the best of our ability, studies focusing on the diverse virulence attributes and clonal relationships of P. aeruginosa recovered from the wound and respiratory infection of diabetic patients is limited. This prompted us to undertake the current investigation which aimed at investigating the antibiotic susceptibility pattern, virulence factor production, and genetic relationship among P. aeruginosa strains isolated from wound and sputum samples of diabetic patients.
Study Setting
Clinical specimen (wound swab and sputum) from patients with lower respiratory tract infection and with a history of diabetes were collected from Justice K.S Hegde hospital, Mangaluru, and also from other primary health centers in and around Mangaluru city, Karnataka, India for a period of 12 months from August 2018-August 2019. Expectorated sputum was taken from the diabetic patients. The study protocol was approved by the Central Ethics Committee of the Nitte University (NU/CEC/2019/0229) and was conducted as per the ethical guidelines. The sputum samples were labeled as ADS (1-50) and wound specimen were labeled as ADW (1-50) and DWS (1-27).
Specimen Collection and Identification of P. aeruginosa
Sample collection was carried out only after the patient consent was taken and the names were anonymized. Wound swab was collected using a sterile swab from the site of infection. Sputum expectorated from diabetic patients were placed in a sterile container. All the samples collected were enriched in Pseudomonas asparagine broth (HiMedia Laboratories Pvt. Ltd., India) for a period of 8 h at 37°C. A loopful of the enriched culture was streaked onto cetrimide agar (HiMedia Laboratories Pvt. Ltd., India) and incubated at 37°C for 24 h. Typical colonies appearing on the selective plates were subjected to a battery of biochemical tests (catalase test, oxidation fermentation test, oxidase test, gelatin hydrolysis and citrate utilization test ) for the presumptive identification of P. aeruginosa.
Genomic DNA Extraction and Molecular Confirmation
The method as described by Ausubel et al.15 was employed with minor modifications for the extraction of genomic DNA from cultures of P. aeruginosa. Isopropanol was used to precipitate the pure genomic DNA, which was then re-suspended in 100μl of 1× Tris-EDTA (TE, pH 8.0) buffer. The concentration and purity of the extracted DNA were measured using the spectrophotometer at 260 and 280nm (Bio Spectrophotometer, Eppendorf, Germany). Further, the purified DNA was confirmed by using the molecular method with the species-specific primer for outer membrane lipoprotein, OprL (5’ATTCTCTGCTCTGGCTCTGG3’; 5’CAGAGCTTCGTCAGCCTTG3’) designed using the Primer3 (v. 0.4.0) web tool (https://bioinfo.ut.ee/primer3-0.4.0/). The Polymerase Chain Reaction (PCR) was performed with a 25μl of reaction mixture containing 40 ng of genomic DNA, 40 pmol of oligonucleotide primer, 200 μM of deoxynucleotide triphosphates, 1.0 unit of Taq DNA polymerase, and 3μl of 1×buffer. The PCR programming constituted an initial 5min denaturation at 94°C, followed by 35 cycles of denaturation at 94°C for 30s, annealing at 55°C for 30s, and primer extension at 72°C for 1min (Bio-Rad T100, USA). Confirmed isolates were preserved as glycerol stocks at -80°C.
Amplification of Virulence Genes
The isolates were screened for different virulence associated genes using specific primers including Pili A, PhzH, LasB, AprI, PvdA, PslA, AlgD, and PelA. Details of the primers used in this study are provided in Table 1. The genes were amplified using PCR and the products were electrophoresed in an agarose gel (1.5%) and recorded using a gel documentation system.
Table (1):
List of virulence gene primer sequences used in this study.
Gene |
Sequence (5’ – 3’) |
Product length (bp) |
References |
|---|---|---|---|
Pill |
F:ATCATGGACCTCTGCGGTTT R :ACGCCATGAATGAAGGGTTG |
200 |
Alva et al., 2021a |
PhzH |
F: GACCATGAGCTGGTGGAGTA R : TGGCAGAAGTCGGATAAGGG |
151 |
Alva et al., 2021a |
LasB |
F: ACCATGTTCTATCCGCTGGT R :AGAACGCTTCGTTCATTCCG |
121 |
Alva et al., 2021a |
PvdA |
F: GTGGTGGATACCGACCTGAT R : CGTCGTAGGTCTCTACGCTT |
184 |
This study |
AprI |
F:CAGGATGGTTTGCTTGCTCT R : CCCAGGTCGTAGCCACTG |
183 |
This study |
pslA |
F: GTTCTGCCTGCTGTTGTTCA R : GGTTGCGTACCAGGTATTCG |
214 |
Alva et al., 2021a |
AlgD |
F: GGGCTATGTCGGTGCAGTAT R : AACGATACGTCGGAGTCCAG |
219 |
Alva et al., 2021b |
PelD |
F: AAGTCAGCGGCAACAACAC R : CGCTTCTCCCAGTACCTCAA |
208 |
Alva et al., 2021b |
Virulence Assays
Swarming motility assay: The assay was carried out according to the method of Murray et al.18 with slight modifications. 10μl of 0.6 OD culture was inoculated at the centre of a Petri plate containing 0.3% nutrient agar medium (HiMedia Laboratories Pvt. Ltd., India). The Petri plate was incubated at 37°C overnight and the swarm zone developed was measured in millimeters.
Pyocyanin Assay
The method of Essar et al.19 with a few modifications was employed. Pyocyanin was extracted from the supernatant fraction of P. aeruginosa isolates grown in Luria Bertani (LB) broth (HiMedia Laboratories Pvt. Ltd., India) for 48h and centrifuged. Five ml of the chloroform (Finar, India), was added to 5 ml of supernatant, and the lower organic layer was separated. To this layer, 1 ml of 0.2 N HCl (HiMedia Laboratories Pvt. Ltd., India), was added, vortexed for 15 min and the pyocyanin rich organic layer was separated. Pyocyanin produced in the extract was detected by measuring the absorbance at 520 nm using a spectrophotometer. The amount of pyocyanin produced was expressed as µg / ml of the culture supernatant and calculated by multiplying the optical density by the extinction coefficient 17.072.
Chrome Azurol S Assay for Siderophore Production
Preparation of Chrome Azurol S (CAS) Medium
60.5 mg of chrome azurol S (CAS) (Sigma-Aldrich) was dissolved in 50 ml of distilled water. It was then mixed with 10 ml iron (III) solution (1 mmol/L Fecl3 .6H20, 10 mmol/L HCL). This solution was slowly added to 72.7 mg of cetrimonium bromide dissolved in 40 ml water. The resultant dark mixture was diluted 20 fold and autoclaved at 121°C for 15 minutes. 2% of agar was added to prepare a solid medium.
Qualitative Method
Simple double layered chrome azurol S agar (SD-CASA) plate assay was used for the determination of siderophore production.20 10ml of chrome azurol S -blue (Sigma-Aldrich) agar was poured into Petri plates and allowed to solidify. Approximately, 6ml of nutrient agar was poured above the solidified chrome azurol S- blue agar. Test isolates were streaked onto the SD-CASA plates and incubated at 37°C for 24 h. Change in the colour from blue to yellowish orange around the colony indicated siderophore production.
Quantitative Method
The assay was performed as per the method of Arora et al. 21 with a minor modification. Ten microlitre of the 0.6 O.D bacterial culture was added 1 ml of LB broth, incubated for 24h at 37°C and then centrifuged for 10 min at 10,000 rpm. 100 µl of the culture supernatant was transferred to a 96 well microtiter plate and 100 µl of chrome azurol S reagent (Sigma-Aldrich), was added. The plate was incubated for 30 min. A reference consisting of 100µl of uninoculated broth with 100 µl of CAS reagent was also maintained. The absorbance was measured at 630 nm in a spectrophotometer. The amount of siderophore (%) produced was estimated as percent siderophore unit (psu) using the formula:
Siderophore production (psu) = (Ar-As) × 100/Ar
Where Ar = absorbance of reference (CAS reagent and uninoculated broth) and As = absorbance of the sample (CAS reagent and cell-free supernatant)
Biofilm Assay
The assay was accomplished according to the method of O’’Toole22 with minor modifications. 2μl of the overnight culture (1×106CFU) was mixed with 198μl of fresh LB broth taken in each well of a microtitre plate. The plates were incubated for 24h, 48h, 72h, and 120h at 37°C. After incubation, the plates were tilted to remove the medium. The adherent cells were thrice washed with 100µl of phosphate buffered saline (PBS) (HiMedia Laboratories Pvt. Ltd., India), of pH 7.4. The pellets were dried by keeping the plates upside down for half an hour. 125µl of freshly prepared 0.1% crystal violet (HiMedia Laboratories Pvt. Ltd., India), was added to the dried pellets, and incubated for 10 min. The stain was removed by washing the pellet thrice with PBS. 200µl of 30% acetic acid (HiMedia Laboratories Pvt. Ltd., India), was added to the stained and washed pellets and incubated for 15 min for stain solubilization. To a fresh plate 100µl from each well was transferred and the optical density was measured at 600 nm in an ELISA reader. The biofilm former’s were grouped as non-biofilm former’s (OD600 ≤ 0.071), weak biofilm former’s (OD600 0.071 – 0.142), moderate biofilm former’s (OD600 0.142 – 0.284) and high biofilm former’s (OD600 ≥ 0.284)23.
Alkaline Protease Assay
Qualitative Method
A modified method of Laux et al.24 was used for the alkaline protease assay. 100µl of 0.6 OD culture of P. aeruginosa was inoculated to 5ml LB broth (HiMedia Laboratories Pvt. Ltd., India) and incubated for 24h. 1ml of the culture was centrifuged and 50µl of the culture supernatant was loaded into wells (6mm) punched on skim milk agar plates and incubated at 37°C for 24 h. The zone of clearance around the well was considered as a positive result. Clear zones were measured to assess the protease activity. A zone of less than 10mm was considered as a negative result.24
Quantitative Method
The assay was carried out according to the method of Murachi et al.25 with minor modifications. 100µl of 0.6 OD culture was inoculated to 5ml LB broth and incubated for 24h. 1ml of the overnight culture was centrifuged and the supernatant was collected. 0.5g of casein was dissolved in 100ml PBS (pH 7.5) and incubated at 37°C for 10min. To 800μl of pre-incubated casein, 200μl of the culture supernatant was mixed and incubated at room temperature for 20min. 10% (w/v) trichloroacetic acid (HiMedia Laboratories Pvt. Ltd., India), was added and incubated for 1h to stop the reaction. The reaction mixture was centrifuged for 5min at 10,000rpm, and the collected supernatant was analysed for the protein content by Bradford method using tyrosine (HiMedia Laboratories Pvt. Ltd., India) as the standard.
Las B Elastolytic Assay
The elastin Congo red (ECR) assay, as reported earlier by Pearson et al.26 was performed with minor changes. The cells were grown in LB broth for 24 hs at 37°C. The overnight cultures were centrifuged at 10,000 rpm for 10 min and 1ml of culture supernatant was mixed with the 1 ml of ECR buffer (HiMedia Laboratories Pvt. Ltd., India), containing 20 mg of elastin Congo red (Sigma-Aldrich, USA). After 14 h of incubation at 37°C, the mixture was centrifuged, and the released Congo red (200µl) was transferred to a 96 well microtiter plate, and released Congo red was measured at 495 nm in a spectrophotometer. Elastase activity was expressed as change in OD 495 / µg of protein.
Antibiotic Susceptibility Testing
P.aeruginosa isolates were analyzed for their antibiotic susceptibility by using the standard Kirby-Bauer disc diffusion method27 following the Clinical and Laboratory Standards Institute guidelines (CLSI 2018). 0.1 ml of 0.6 OD culture grown in Mueller Hinton broth was spread onto Mueller Hinton agar plates. The plates were incubated for 16-18h at 37°C after placing the antibiotic discs on the medium. Antipseudomonal drugs such as levofloxacin (5 µg), imipenem (10 µg), aztreonam (30 µg), netillin (30 µg), ciprofloxacin (5 µg) moxifloxacin (5 µg), procured from HiMedia Laboratories Pvt. Ltd., India were used for the assay. The measured zone of inhibition (mm) was compared with the interpretive chart of CLSI. Susceptibility to colistin was carried out by broth dilution method. Mueller-Hinton broth was used for broth MIC testing as described in the CLSI guidelines. Briefly, serial two-fold dilutions of colistin (HiMedia Laboratories Pvt. Ltd., India) were prepared in the medium (500μl) ranging from 0.125 to 256μg/ml. 100 μl of 105 CFU/ml of the culture was inoculated into the medium containing the antibiotic and incubated at 37°C for 24 hs. Based on the turbidity, the isolates were categorized into susceptible (≤2μg/ml) and resistant (≥4μg/ml) as per the current CLSI colistin breakpoints. The Multiple Antibiotic Resistance (MAR) index was calculated as described by Joseph et al.28 in which the number of antibiotics to which an isolate is resistant was divided by the total number of antibiotics used in the study.
Random Amplified Polymorphic DNA Analysis (RAPD)
The PCR was performed with a 25 µl of reaction mixture containing 40 ng of genomic DNA, 40 pmol of oligonucleotide [272: 5’-AGCGGGCCAA-3’]29, 1.0 unit of Taq polymerase, 200 μM of deoxynucleotide triphosphates, 3μl of 1× buffer). The PCR amplification was carried out in a thermocycler, with an initial denaturation for 5 min at 94°C, followed by 30 cycles of denaturation for 1 min at 94°C, annealing for 1 min at 36°C, extension for 2 min at 72°C, and final delay for 10 min at 72°C. The PCR amplified product was electrophoresed on a 1.5% agarose gel and documented using a gel documentation system. Bionumerics software package version 7.6 was employed to analyse the RAPD pattern of the isolates.
Statistical Analysis
The experimental results were expressed as mean ± standard deviation (SD) of triplicate measurements. One-way analysis of variance (ANOVA), z-test and student t-test were used to check for the significance in the test results. Data was considered significant at p≤ 0.05.
A total of 127 samples (77 wound swabs and 50 sputum) were collected from diabetic patients. Based on the biochemical test results (oxidase positive, catalase positive, oxidative positive, gelatinase positive and citrate positive) and molecular identification method using species specific primer OprL, 77 isolates (53 wound and 24 sputum) were confirmed to be as P. aeruginosa.
Detection of Virulence Genes
The screening of the confirmed isolates for the presence of virulence genes encoding for swarming (PiliA), pyocyanin (PhzH), siderophore (PvdA), protease (LasB, AprI), and biofilm (PslA, AlgD, and PelA) resulted in all the isolates being positive for the tested genes (gel image provided in supplementary file).
Virulence Assays
Swarming Motility Assay
A characteristic dendritic pattern was observed in all the 77 P. aeruginosa isolates along with control PAO1. There was a difference in the swarm diameter between the isolates from the two sources and also a difference between the isolates and the control. The average swarm diameter produced by wound and sputum isolates were 39 mm and 33 mm respectively. A large number of wound isolates (43.5%) showed a swarm zone between 20-30mm, (Table 2). Although, majority of the isolates did not produce a swarm zone greater than the control (63mm), five wound isolates gave rise to a swarm zone more than the control.
Table (2):
Difference in the virulence activity of the wound and sputum isolates.
A
| Swarming zone (mm) | No. of isolates (%) | |
|---|---|---|
| W | S | |
| 0-29 | 43 | 42 |
| 30-49 | 32 | 50 |
| 50-70 | 25 | 8 |
B
| Alkaline protease production (µg/ml) | No. of isolates (%) | |
|---|---|---|
| W | S | |
| 0 to 5 | 77 | 100 |
| 0 to 5 | 77 | 100 |
C
| Elastase production (µg/ml) | No. of isolates (%) | |
|---|---|---|
| W | S | |
| 0 – 5 | 93 | 96 |
| 5 – 10 | 3 | 0 |
| 10 – 15 | 1 | 0 |
| more than 15 | 3 | 4 |
D
| Pyocyanin production (µg/ml) | No. of isolates (%) | |
|---|---|---|
| W | S | |
| 0 to 5 | 45 | 75 |
| 5 to 10 | 12 | 7 |
| 10 to 15 | 17 | 7 |
| More than 15 | 26 | 11 |
Pyocyanin Assay
The average pyocyanin produced by the wound and sputum isolates was 10.86 µg/ml and 4.6 µg/ml respectively. The control strain, PAO1 elaborated 13.7µg/ml of pyocyanin. Pyocyanin production of greater than 13.7µg/ml was seen in 19 wound isolates and 3 sputum isolates. One isolate of the wound (ADW26) sample showed the pyocyanin production of about 25µg/ml which was significantly higher than the other isolates and also the control. The No. of isolates producing pyocyanin of different concentrations is represented in Table 2. Student t-test was performed to check the level of significance between the amount of pyocyanin produced in wound and sputum isolates. There was a significant difference in the pyocyanin production between the wound and sputum isolates at a concentration of 0 to 5µg/ml (p<0.05) = 0.012) and more than 15µg/ml (p<0.05) = 0.02).
Chrome Azurol S Assay
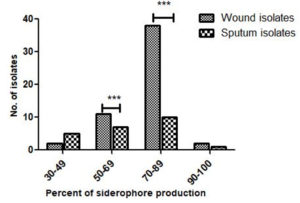
Qualitative assay indicated the ability of all the 77 isolates of P. aeruginosa to produce siderophore based on the color change from dark blue to orange around the colonies. Siderophore production of more than 50.8% was observed in a large number of isolates (Figure 1). The average siderophore produced by the wound isolates (77.0%) was more than the sputum isolates (66.8%). Siderophore output by PAO1 (48.0%) was far less than the isolates from both the sources. Student t-test was performed to check the difference in the siderophore production by the sputum and wound isolates. A significant difference in the siderophore production between the wound and sputum isolates at 50-69% and 70-89% siderophore concentration was observed [(p<0.05) = 0.0001)].
Figure 1. Percent siderophore production in wound and sputum isolates of P. aeruginosa (*P ≤ 0.05; ** P ≤ 0.01; ***P ≤ 0.001)
Alkaline Protease Assay
Of the 77 isolates of P. aeruginosa, only 62 isolates were found to be elaborating alkaline protease when qualitatively evaluated. The amount of casein degraded by the isolate was estimated using tyrosine as the standard. The No. of isolates producing alkaline protease production is represented in Table 2. Sputum isolates produced an average of 4.7 µg/ml of alkaline protease which was greater than the wound (2.9µg/ml) isolates and the control (3.6µg/ml). Although the average alkaline protease production by the wound isolates was less than sputum isolates, few isolates secreted more than 7 µg/ml of alkaline protease.
Las B Elastolytic Assay
The amount of elastase produced by the isolates was estimated using elastin congo red. The No. of isolates producing elastase shown in Table 2. The control strain PAO1 produced 6.4µg/ml of elastolytic activity which is lesser than the wound and sputum isolates. Elastolytic activity in the range of 1 to 5µg/ml was found to be more in sputum isolates (96.0%), whereas secretion of 5 to 10µg/ml was found to be higher in the wound isolates. Four sputum isolates were found to elaborate more than 15 µg/ml of elastase. Three wound isolates and one sputum isolate showed an increased production of elastase B with greater than 17 µg/ml.
Biofilm Assay
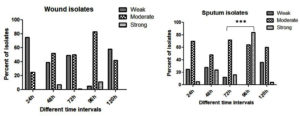
At all-time intervals the number of sputum isolates forming a strong biofilm exceeded the number of wound isolates. Strong biofilm formation was found as a trait in a number of sputum isolates. For a given time period, there was a significant difference in biofilm forming ability between sputum and wound isolates (Figure 2). Sputum isolates at 72h [(p<0.05) =0.003)] and 96h [(p<0.05) =0.002)] showed a significant difference in the ability to produce biofilm as compared to the wound isolates. Majority of the wound isolates were moderate biofilm former at all the time intervals where as sputum isolates produced strong biofilm formation.
Figure 2. Percent wound and sputum isolates of P.aeruginosa showing biofilm formation at different time intervals ***P ≤ 0.001
Antibiotic Susceptibility Testing
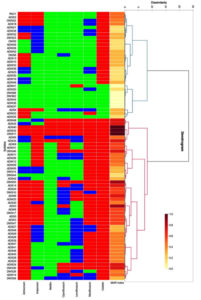
Antibiotic susceptibility test for all the 77 isolates was conducted along with the control, P. aeruginosa ATCC 27853. The number of sputum isolates exhibiting resistance to the antibiotics used was more than the wound isolates. 70.0% of the sputum isolates were found to be resistant to more than 3 class of antibiotics indicating their multidrug resistant nature. While only 45.0% of the wound isolates were multidrug resistant. Surprisingly, 90.0% of both wound and sputum isolates were resistant to colistin. Around 80.0% resistance was observed in sputum isolates for aztreonam. In wound isolates, resistance of 40% was observed with imipenem. The majority of the wound and sputum isolates were susceptible to netillin. Two wound (ADW22 and DWS11) and one sputum isolate (ADS40) was resistant to all the tested antibiotics with a MAR index of 1.0. Of the 77 isolates tested, 71 isolates (92.22%) presented a MAR index between 0.2 and 1.0. Only a few isolates were with a MAR index of less than 0.2 (Figure 3).
Figure 3. Hierarchical clustering analysis of the antibiotic resistance combinations and antibiotic resistance (MAR) index of Pseudomonas aeruginosa obtained from wound and sputum samples of diabetic cases. The cells are coded in tricolour with green for sensitive, blue for intermediate and red for resistance to antibiotics. ATM: Aztreonam; IPM: Imipenem; NET: Netillin; CIP: Ciprofloxacin; LE: Levofloxacin; MO: Moxifloxacin; C: Colistin
Based on the resistance pattern, the isolates were grouped into two clusters. Both the clusters comprised of isolates from sputum and wound. Isolates with a greater resistance were grouped in cluster 2 and isolates with a greater susceptibility were grouped in cluster 1. 20 of the 24 sputum isolates were found in cluster 2. Of the two clusters, cluster 2 was the biggest with 47 strains and cluster 1 comprised of 30 strains. Colistin was the only antibiotic to which the majority of the isolates belonging to both the clusters were resistant. In addition to colistin, a number of strains of cluster 2 were resistant to moxifloxacin. As compared to strains of cluster 2, resistance in strains of cluster 1 was minimal. DWS11, ADW22, and ADS40 resistant to all the tested antibiotics were grouped together. While ADW37, ADW36, ADW44, DWS62, and DWS68 which showed the least resistance, were grouped together in cluster 1.
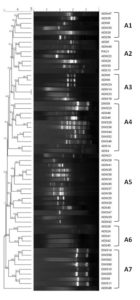
RAPD Analysis
There was an amplification of 2-11 fragments with a band size ranging from 0.2 to 2kb. Polymorphic bands were observed only in 58 isolates of P. aeruginosa and no bands were found in the rest of the isolates. The molecular typing showed 7 well-defined clusters and 37 major groups at 80% similarity. The dendrogram classified all strains into 7 clusters (A1–A7) with an average similarity score of 80%, revealing diversity in their profiles. Among strains, 14 (24.1%) belong to A5, six (10.3%) each to A1 and A3, nine (15.5%) belong to A7, 11 (18.9%) to A4, seven (12%) to A2, five (8.6%) to A6. As seen in the Figure 4, the clusters A1, A2, A4, A6, have both wound and sputum isolates clubbed together along with the control PAO1, whereas clusters A3, A5, A7 have solely wound isolates that are clubbed together.
In the present study, attempts were made to isolate and identify P. aeruginosa from sputum and wound specimens collected from people with diabetes. The recovered isolates from the two different sources were compared for their virulence properties, antibiotic sensitivities, and clonal relationship.
Samples collected from patients with a history of diabetes were confirmed using the species-specific primer oprL, considered to be specific and sensitive for detecting P. aeruginosa30. In the present study, ~61.0% of isolates were recovered from wound swab and sputum samples collected from diabetic patients. The number of isolates retrieved from wound and sputum was found to be 68.0% and 48.0%, respectively. Earlier studies conducted have reported a lower rate of isolation from wound samples. Investigations by Sivanmaliappan et al.31 and Qayoom et al.32 have revealed 37.5%, 14.30% and 9.1% respectively. Similarly, the recovery from sputum samples was also less as shown in a study by Boyer et al. 33 who reported an isolation rate of 6.2%. These studies have employed the method of direct inoculation of the sample on cetrimide agar, a selective medium for P. aeruginosa. Whereas, our study has adopted the enrichment method of the sample in asparagine broth followed by subculturing on cetrimide agar. This could be the probable reason for the increased recovery rate of P. aeruginosa in the current investigation.
The characteristic dendritic pattern as observed on the semisolid medium indicated swarming in P. aeruginosa strains. Studies conducted by Murray et al.18 and Deligianni et al. 34 reported 63.0% and 52.0% of swarming in isolates of P. aeruginosa recovered from diverse sites and sputum, respectively. In the current investigation around 92.0% of the strains isolated from sputum and wound possessed swarming motility which indicates their increased virulence with which they can move to different sites in a coordinated manner leading to an increased morbidity and mortality. The difference in the swarm motilities as seen in the isolates is due to the differences in the strain and also the site of collection.
It has been reported that pyocyanin production is greatly enhanced in the clinical isolates.35 Although the amount of pyocyanin produced in wound and sputum isolates varied significantly, all the isolates produced the phenazine pigment. The results from our study correlates with a study which has shown varied pyocyanin production in the clinical isolates retrieved from different sites such as urine, pus, sputum, wound swab, and ear swab.35 The likely reason for the difference in the pyocyanin production among the isolates lies with their genetic variability.
Siderophores produced by P. aeruginosa enables them to survive under iron-restricted conditions found in the host cells. The present study indicated the elaboration of siderophore by all the 77 isolates of wound and sputum. A study by Nezhad et al. 36 demonstrated that 31.0% of the strains were siderophore producers. A number of investigations carried out on P. aeruginosa strains isolated from burn wounds indicated a lower number of isolates with siderophore production.37,36 The rate of occurrence of siderophore producers in our study is significantly higher than any of the other studies conducted. A positive association between siderophore production and multidrug resistance has been indicated.38 The results from the present investigation concur with the earlier finding as a large number of isolates recovered were multi-drug resistant. More number of siderophore producers in diabetic wound as revealed in this study gives them an advantage over other bacteria in causing chronic infection.
In the current investigation, strong biofilm formation was observed in the sputum isolates. In addition, these isolates were resistant to more than 3 drugs indicating their multidrug-resistant nature. A similar result was observed in a study by Elmaraghy et al.39 where sputum isolates showed strong biofilm formation compared to strains from other specimens. Infection with such multidrug resistant isolates with the ability to form a thick biofilm could lead to a chronic condition resulting in an increased use of antimicrobials and extended hospital stay. It is well established that bacteria with biofilm forming ability is more resistant to antibiotics than their counterparts because of the structure and physiology of biofilms.40 The study of Murali et al.41 showed that only 8.0% of the strains of P. aeruginosa isolated from diabetic wounds were with the ability to form high amounts of biofilm. Whereas, in the present study, 19.5% of the recovered isolates formed significantly high biofilm formation indicating the chronic nature of the diabetic wound. In addition, two wound isolates were completely resistant to all the antibiotics used in the study. Albeit the fact that wound isolates were moderate biofilm formers, the persistent nature of infection and the ability of cells to tolerate antibiotics can lead to severe tissue damage.
Proteolytic enzymes play a significant role during different stages of infection. In the current study, nearly 80.0% of the isolates were found be producing alkaline protease. A study conducted in Iraq has revealed a lesser number of isolates (30.0%) with the ability to produce alkaline protease.42 The number of isolates secreting the protease enzyme was significantly higher in the current study. Of the isolates from the two different sites, sputum isolates produced significant amounts of alkaline protease which can be correlated with the damage that it causes during pulmonary infection. Corroborating with our findings, implication of this enzyme in the hydrolysis of biologically important proteins in cystic fibrosis and other respiratory infection has been indicated.43 Elastolytic activity has been found in most of the clinical and environmental isolates.44 The presence of elastolytic activity in both sputum and wound isolates, as observed in the current investigation, validates the statement. However, numerous studies have indicated the amount of elastase produced by strains isolated from lower respiratory clinical specimens including sputum, tracheal aspirate, and bronchoalveolar lavage to be significantly higher than isolates from other sites.45,46 In our study, the amount of elastase produced by wound and sputum isolates were very much identical. The amounts of alkaline protease and elastase produced by the strains varied immensely which signifies isolate-dependence. Irrespective of the isolation sites, the proteases produced by strains of P. aeruginosa recovered in the present study indicates their role in causing tissue damage, in breaking down the human innate immune system, and modifying the adaptive immune system, thus favoring a more localized chronic infection.47 The results from our study is not in agreement with a study by Delden et al.48 who has reported that isolates recovered from chronic infection produce reduced extracellular proteases. Contrary to the report some of the strains isolated in the present study, produced significantly higher amounts of proteases regardless of the isolation site.
Research carried out on P. aeruginosa during the recent years have confirmed an increase in its MDR traits. An earlier report by Mirzaei et al.49 observed 16.50% of wound isolates of P.aeruginosa being multidrug resistant. Whereas our findings observed an increase in the number of wound isolates being multidrug resistant (45.0%). Similarly, the percent of MDR sputum isolates of P. aeruginosa was also elevated (70.0%) in contrast to other studies which have shown the percent of MDR isolates as 30.0% and 39.44% (Perez et al.50; Samad et al.51). Although, three isolates were found to be resistant to all the drugs used in the present study, it is difficult to judge Pan Drug Resistant (PDR) strains as the isolates are not frequently assessed against all possible antibiotics. A shocking result as noticed by us is the resistance of a majority of isolates (~90.0%) towards colistin, the drug of last resort for treating multidrug-resistant P. aeruginosa infection. Except for one sputum isolate and nine wound isolates, all other isolates displayed resistance towards colistin. Studies conducted in the past have indicated least resistance of P. aeruginosa strains towards colistin.52-54 Contrary to this, majority of the isolates in the current study were resistant to colistin which clearly indicates the increase in resistance in P. aeruginosa to colistin. This gives a clear forewarning that susceptibility testing towards colistin should be performed before its clinical use. The likely cause of more number of isolates resistant to colistin could be attributed to the increased use of colistin in the hospitals as it has been noted that prolonged exposure of isolates to the antibiotic can trigger the development of resistance. Carbapenems are believed to be effective antibiotics for the treatment of P. aeruginosa infections and studies have disclosed more than 90% sensitivity of strains towards this antibiotic.51 A contradictory result was seen in our study where 33.0% and 40.0% sputum and wound isolates respectively were resistant to imipenem. Sputum and wound isolates were with an intermediate resistance of 54.0% and 22.0%, respectively. This clearly indicates the gradual increase in resistance in isolates of P. aeruginosa over the years and in the next few years, strains with intermediate resistance would become completely resistant to imipenem. In accordance with the results from our study, netillin could be considered as the best drug for treating MDR P. aeruginosa infections as ~90% of the isolates were susceptible. A MAR index of greater than 0.2 indicates a high risk source of contamination due to the frequent use of antibiotics.55 Based on our findings, majority of the isolates had MAR indices of more than 0.2, confirming that the isolates are from a place where there is extensive use of antibiotics and are with a high selective pressure to acquire resistance.
RAPD primer 272 was used in the current investigation as it is dependable because of its higher discriminatory power and reproducible profiles.56 In the current study, of the 77 isolates, only 58 gave polymorphic bands which is not concordant with a study from Iran where all the 54 isolates showed the presence of polymorphic bands.57 The probable reason for the 23 isolates not producing any band might be due to a mismatch between the template and the primer used. Based on the Dice similarity coefficient of 80%, a study by Vaez et al.57 reported the presence of 39 groups in P. aeruginosa isolates recovered from five different specimens. Whereas our study could identify 37 groups in the isolates retrieved from only two samples. This indicates a vast genetic diversity in the strains isolated in the present study. The probable reason for greater genetic diversity in the isolates as seen in this study, could be attributed to the fact that the samples were collected from diabetic patients residing in different localities of Mangaluru and neighbouring places. The greater genetic diversity among the isolates provides a selective advantage to acclimatize to different environments. Each group consisted of isolates from both sputum and wound that indicated their genetic relatedness and their potential to cause infection at different sites. Report by Nanvazadeh et al.58 identified 9 groups out of 50 isolates. The possible explanation for low diversity among the isolates as observed in the previous study might be due to the limited source of recovery of the isolates. The RAPD and the antibiotic resistance profiles of the isolates revealed a weak correlation which is in accordance with other studies.59,57 Although the isolates belonged to the same cluster, there was variation in their resistance pattern. Different antibiotic resistance mechanisms could be the predicted as the cause for this diversity.60
Collectively, the results from the current investigation have clearly indicated the prevalence of P. aeruginosa as one of the most significant nosocomial pathogens causing infection in people with diabetes. The study highlights the increased virulence of sputum isolates as recognized by their resistance pattern, strong biofilm formation, and elaboration of proteolytic enzymes that can result in both acute and chronic lung infection. Although the wound isolates were less virulent than the sputum isolates, the production of siderophores and moderate biofilm formation gives them an advantage in causing chronic diabetic foot infection. RAPD analysis showed no distinct lineages of wound and sputum isolates indicating their ability to adapt to different environments in the host causing a wide range of infections. Furthermore, the presence of MDR strains in both wound and sputum raises an alarm to develop novel therapeutic strategies to combat the infection with P. aeruginosa. The outcome of our study distinctly reveals the significance of periodic examination of antibiotic resistance and virulence factors of P. aeruginosa in order to recognize the possible co-regulatory mechanism involved in their expression.
Additional file: Additional supplementary file.
ACKNOWLEDGMENTS
The authors would like to thank Nitte (Deemed to be University), Mangaluru, Karnataka, India for all the facilities provided.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
RP conceptualized the study, received funds and supervised. SS performed the experiments and analysed the data. KSP performed formal software analysis. SS drafted the manuscript. BKK and RP reviewed and edited the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
The work was supported by the Nitte University Research Grant with the grant number NUFR2/2018/10/27.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript and/or the supplementary files.
ETHICS STATEMENT
The study protocol was approved by the Central Ethics Committee of the Nitte University, Mangaluru, Karnataka, India (NU/CEC/2019/0229).
- Ballok AE, O’Toole GA, Pouring salt on a wound: Pseudomonas aeruginosa virulence factors alter Na+ and Cl− flux in the lung. J. Bacteriol. 2013;195(18):4013-4019.
Crossref - Hossain MA, Sattenapally N, Parikh HI, Li W, Rumbaugh KP, German NA. Design, synthesis, and evaluation of compounds capable of reducing Pseudomonas aeruginosa virulence. Eur J Med Chem. 2020;1(1)85:111800.
Crossref - Gholami S, Tabatabaei M, Sohrabi N. Comparison of biofilm formation and antibiotic resistance pattern of Pseudomonas aeruginosa in human and environmental isolates. Microb Pathog. 2017;109:94-98.
Crossref - Yaribeygi H, Bo S, Ruscica M, Sahebkar A. Ceramides and diabetes mellitus: An update on the potential molecular relationships. Diabet Med. 2020;37(1):11-19.
Crossref - Toniolo A, Cassani G, Puggioni A, Rossi A, Colombo A, Onodera T, Ferrannini E. The diabetes pandemic and associated infections: Suggestions for clinical microbiology. Rev Med Microbiol. 2019;30(1):1-17.
Crossref - Jneid J, Lavigne JP, La Scola B, Cassir N. The diabetic foot microbiota: A review. Hum Microbiome J. 2017;5-6:1-6.
Crossref - Premanath R, Suresh S, Alva PP, Akash SK. Biofilm forming abilities of microorganisms associated with diabetic wound infection: A study from a tertiary Care Hospital. Biomed Pharmacol J. 2019;12(2):669-676.
Crossref - Shanmugam P, MJ, Susan SL. The bacteriology of diabetic foot ulcers, with a special reference to multidrug resistant strains. J Clin Diagnostic Res. 2013;7(3):441-445.
Crossref - Goldufsky J, Wood SJ, Jayaraman V, et al. Pseudomonas aeruginosa uses T3SS to inhibit diabetic wound healing. Wound Repair Regen. 2015;23(4):557-564.
Crossref - Gill SK, Hui K, Farne H, et al. Increased airway glucose increases airway bacterial load in hyperglycaemia. Sci Rep. 2016;6(1):1-10.
Crossref - Huber P, Basso P, Reboud E, Attree I. Pseudomonas aeruginosa renews its virulence factors. Environ Microbiol Rep. 2016;8(5):564-571.
Crossref - Yang X, Xing B, Liang C, Ye Z, Zhang Y. Prevalence and fluoroquinolone resistance of Pseudomonas aeruginosa in a hospital of South China. Int J Clin Exp Med. 2015;8(1):1386-1390.
- Koohler T, Curty,LK, Barja F, Van Delden C, Pechere JC. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J Bacteriol. 2000;182(21):5990-5996.
Crossref - Hickey C, Schaible B, Nguyen S, et al. Increased virulence of bloodstream over peripheral isolates of Pseudomonas aeruginosa identified through post-transcriptional regulation of virulence factors. Front Cell Infect Microbiol. 2018;8:357.
Crossref - Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology, 3rd ed.; John Wiley & Sons Inc. Media: Hoboken, NJ, USA. 1989.
Crossref - Alva PP, Suresh S, Nanjappa DP, et al. Isolation and identification of quorum sensing antagonist from Cinnamomum verum leaves against Pseudomonas aeruginosa. Life Sci. 2021;267:118878.
Crossref - Alva PP, Sundar S, D’Souza C, Premanath R. Increased expression of genes involved in biofilm formation in a multidrug-resistant environmental Pseudomonas aeruginosa isolate. J Datta Meghe Inst Med Sci Univ. 2021;16:(2):357.
Crossref - Murray TS, Ledizet M, Kazmierczak BI. Swarming motility, secretion of type 3 effectors and biofilm formation phenotypes exhibited within a large cohort of Pseudomonas aeruginosa clinical isolates. J Med Microbiol. 2010;59(5):511-520.
Crossref - Essar DW, Eberly LE, E, Hadero A, Crawford IP. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol. 1990;172(2):884-900.
Crossref - Hu QP, Xu JG. A simple double-layered chrome azurol S agar (SD-CASA) plate assay to optimize the production of siderophores by a potential biocontrol agent Bacillus. Afr J Microbiol Res. 2011;5(25):4321-4327.
Crossref - Arora NK, Verma M. Modified microplate method for rapid and efficient estimation of siderophore produced by bacteria. 3 Biotech. 2017;7(6):381.
Crossref - O”Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: A genetic analysis. Mol Microbiol. 1998; 28(3):449-461.
Crossref - Percival SL. Wounds and infection. Biofilms in infection prevention and control. 2014;127-139.
Crossref - Laux DC, Corson JM, Givskov M, et al. Lysophosphatidic acid inhibition of the accumulation of Pseudomonas aeruginosa PAO1 alginate, pyoverdin, elastase and LasA. Microbiology. 2002;148(6):1709-1723.
Crossref - Murachi T. Bromelain enzymes. Methods in enzymology. 1976;45:475-485.
Crossref - Pearson JP, Pesci EC, Iglewski BH. Roles of Pseudomonas aeruginosa lLas and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol. 1997;179(18):5756-5767.
Crossref - Bauer AW. Antibiotic susceptibility testing by a standardized single disc method. Am J Clin Pathol. 1966;45:149-158.
Crossref - Joseph AA. Odimayo MS, Olokoba LB, Olokoba AB, Popoola GO. Multiple antibiotic resistance iIndex of Escherichia coli isolates in a tertiary hospital in south-west Nigeria. Med J Zamb. 2017:44(4):225-232.
- Mahenthiralingam E, Campbell ME, Foster J, Lam JS, Speert DP. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J Clin Microbiol. 1996;34(5):1129-1135.
Crossref - Lavenir R, Jocktane D, Laurent F, Nazaret S, Cournoyer B. Improved reliability of Pseudomonas aeruginosa PCR detection by the use of the species-specific ecfX gene target. J Microbiol Methods. 2007;70(1):20-29.
Crossref - Sivanmaliappan TS, Sevanan M. Antimicrobial Susceptibility Patterns of Pseudomonas aeruginosa from Diabetes Patients with Foot Ulcers. Int J Microbiol. 2011;2011:605195.
Crossref - Qayoom S, Rashid A, Kohli A, Masoodi T, Amin M. Prevalence and antibiotic sensitivity pattern of Pseudomonas aeruginosa isolates from respiratory samples, pus samples and body fluids in a tertiary care hospital, Kashmir. Indian J Microbiol Res. 2019;6(4):345-349.
Crossref - Boyer A, Doussau A, Thiebault R, et al. Pseudomonas aeruginosa acquisition on an intensive care unit: Relationship between antibiotic selective pressure and patients” environment. Crit Care. 2011;15(1):1-10.
Crossref - Deligianni E, Pattison S, Berrar D, et al. Pseudomonas aeruginosa cystic fibrosis isolates of similar RAPD genotype exhibit diversity in biofilm forming ability in vitro. BMC Microbiol. 2010;10(1):1-1338.
Crossref - Alva PP, Prasad R, Venkatesh T, Suresh PS, Premanath R. Increased virulence in Pseudomonas aeruginosa at pathological glucose levels. Infect Dis. 2019;51(2):153-156.
Crossref - Nezhad MS, Pordeli H, Ghasemi N, Ahani A. Evaluation of multidrug resistance patterns in siderophore-producing Pseudomonas aeruginosa from clinical and environmental samples in Gorgan, Iran. New Microbes New Infect. 2018;24:38-41.
Crossref - Mirsalehian A, Nakhjavani F, Bahador A, Bigverdi R, Goli H. Prevalence of MBL-producing Pseudomonas aeruginosa isolated from burn patients. Tehran Univ Med J. 2011;68(10):563-569.
- Finlayson EA, Brown PD. Comparison of antibiotic resistance and virulence factors in pigmented and non-pigmented Pseudomonas aeruginosa. West Indian Med. 2011;60(1):24-32.
- Elmaraghy N, Abbadi S, Elhadidi G, Hashem A, Yousef A. Virulence genes in Pseudomonas aeruginosa strains isolated at Suez Canal University Hospitals with respect to the site of infection and antimicrobial resistance. Int J Clin Microbiol Biochem Technol. 2019;2:8-19.
Crossref - James GA, Swogger E, Wolcott R, et al. Biofilms in chronic wounds. Wound Repair Regen. 2008;16(1): 37-44.
Crossref - Murali TS, Kavitha S, Spoorthi J, et al. Characteristics of microbial drug resistance and its correlates in chronic diabetic foot ulcer infections. J Med Microbiol. 2014;63(10):1377-1385.
Crossref - Saleem AJ. Relationship Study between the Alkaline protease production and the growth phases of Pseudomonas aeruginosa isolated from patients. Adv Appl Microbiol.2012;2(03):354.
Crossref - Butterworth MB, Zhang L, Heidrich EM, Myerburg MM, Thibodeau PH. Activation of the epithelial sodium channel (ENaC) by the alkaline protease from Pseudomonas aeruginosa. J Biol Chem. 2012;287(39):32556-32565.
Crossref - Thibodeau PH, Butterworth MB. Proteases, cystic fibrosis and the epithelial sodium channel (ENaC). Cell Tissue Res. 2013;351(2):309-323.
Crossref - Tingpej P, Smith L, Rose B, et al. Phenotypic characterization of clonal and non clonal Pseudomonas aeruginosa strains isolated from lungs of adults with cystic fibrosis. J Clin Microbiol. 2007;45(6):1697-1704.
Crossref - Karatuna O, Yagci A. Analysis of quorum sensing-dependent virulence factor production and its relationship with antimicrobial susceptibility in Pseudomonas aeruginosa respiratory isolates. Clin Microbiol Infect. 2010;16(12):1770-1775.
Crossref - Khalil MA, Sonbol FI, Mohamed AF, Ali SS. Comparative study of virulence factors among ESβL-producing and nonproducing Pseudomonas aeruginosa clinical isolates. Turk J Med Sci. 2015;45(1):60-69.
Crossref - Van Delden C, Iglewski BH. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg Infect Dis. 1998;4(4):551-560.
Crossref - Mirzaei B, Bazgir ZN, Goli HR, et al. Prevalence of multi-drug resistant (MDR) and extensively drug-resistant (XDR) phenotypes of Pseudomonas aeruginosa and Acinetobacter baumannii isolated in clinical samples from Northeast of Iran. BMC Res Notes. 2020;13:380.
Crossref - Perez A, Gato E, Perez-Llarena J, et al. High incidence of MDR and XDR Pseudomonas aeruginosa isolates obtained from patients with ventilator-associated pneumonia in Greece, Italy and Spain as part of the MagicBullet clinical trial. J Antimicrob Chemother. 2019;74(5):1244-1252.
Crossref - Samad A, Ahmed T, Rahim A, Khalil A, Ali I. Antimicrobial susceptibility patterns of clinical isolates of Pseudomonas aeruginosa isolated from patients of respiratory tract infections in a Tertiary Care Hospital, Peshawar. Pak J Med Sci.2017; 33(3):670-674.
Crossref - Yayan J, Ghebremedhin B, Rasche K. Antibiotic Resistance of Pseudomonas aeruginosa in Pneumonia at a Single University Hospital Center in Germany over a 10-Year Period. PLoS One. 2015;2:10.
Crossref - Goli HR, Nahaei MR, Rezaee MA, Hasani A, Samadi Kafil H, Aghazadeh M. Emergence of colistin resistant Pseudomonas aeruginosa at Tabriz hospitals, Iran. Iran J Microbiol. 2016;8(1):62-69. http://www.ncbi.nlm.nih.gov/pmc/articles/pmc4833742/
- Azimi L, Lari AR. Colistin-resistant Pseudomonas aeruginosa clinical strains with defective biofilm formation. GMS Hyg Infect Control. 2019;14:Doc12.
Crossref - Davis R, Brown PD. Multiple antibiotic resistance index, fitness and virulence potential in respiratory Pseudomonas aeruginosa from Jamaica. J Med Microbiol.2016;65(4):261-271.
Crossref - Saitou K, Furuhata, K, Fukuyama, M. Genotyping of Pseudomonas aeruginosa isolated from cockroaches and human urine. J Infect Chemother. 2010;16(5):317-321.
Crossref - Vaez H, Faghri J, Esfahani BN, et al. Antibiotic resistance patterns and genetic diversity in clinical isolates of Pseudomonas aeruginosa isolated from patients of a referral hospital, Isfahan, Iran. Jundishapur J Microbiol. 2015;8(8:e20130).
Crossref - Nanvazadeh F, Khosravi AD, Zolfaghari MR, Parhizgari N. Genotyping of Pseudomonas aeruginosa strains isolated from burn patients by RAPD-PCR. Burns. 2013;39(7):1409-1413.
Crossref - Nazik H, Ongen B, Erturan Z, Salcioglu M. Genotype and antibiotic susceptibility patterns of Pseudomonas aeruginosa and Stenotrophomonas maltophilia isolated from cystic fibrosis patients. Jpn J Infect Dis. 2007;60(2):82-86. PMID: 17515637
- Hirsch EB. Tam VH. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev Pharmacoecon Outcomes Res. 2010;10(4):441-451.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.