ISSN: 0973-7510
E-ISSN: 2581-690X
This study aims to evaluate the toxicity and genetic improvement of Bacillus thuringiensis isolates. Isolates were obtained from soil, insect and water samples from different regions of Assiut, Egypt for biological control of mosquito larvae. B. thuringiensis colonies were identified based on morphological and then by PCR which detect the Cry toxic genes in the isolates. Bioassays were performed to evaluate the toxicity of different strains of B. thuringiensis against mosquito larvae such as (Culex spp). In general, 36 B. thuringiensis isolates were obtained (31 from soil, 4 from insects, and 1 from water). And they were all toxic to mosquito larvae with different mortality percentages from 7 to 97% after 48 hours. Isolate Am2 recorded the highest mortality percentage 97% and Mn3 lowest mortality percentage 7%. PCR revealed that Am2 isolate which caused the highest mortality encodes three different types of Cry toxins, Cyt1AA, Cry1Ac and Cry2Aa. This isolate Am2 was examined by scanning electron microscopy to observe the shape of the Cry proteins. The results showed that the Am2 isolate contained of spherical and cuboidal toxic proteins. Then UV-mutagenesis was performed on the Am2 isolate to improve its toxicity. Out of 30 obtained UV-mutants, only one mutant showed improvement in the mortality of mosquito larvae since it caused a mortality rate of 100%. The results of the present study revealed the larvicidal efficacy of B. thuringiensis (Am2) isolate found in the soil of Assiut, could be used in biological control program of mosquito larvae.
B. thuringiensis, Cry Genes, Cyt Genes, UV- mutagenesis, Scanning Electron Microscope
Mosquitoes are among the most important insects that transmit diseases to humans, as they infect them with diseases due to their transmission of viruses and parasites.1,2 The mosquito transmits the virus that causes several diseases, including dengue fever, yellow fever, and chikungunya.3 Where they are found in places where humans and animals live and breed in places that contain ponds with stagnant water and swamps, as they multiply rapidly and the resulting eggs are resistant to drought and withstand harsh conditions until the appropriate conditions are available to complete the life cycle of the insect, so there is difficulty in controlling them.4
The current strategy for controlling insect vectors of different infectious diseases is based on the use of programs of chemical insecticides, the development of resistance is a threat to the program’s efficacy or by eliminating breeding sites.5,6 However, the frequent use of these chemicals has led to limiting the effectiveness, environmental pollution, toxicity in humans and animals and the development of resistance in mosquito populations.7-9 Therefore, it is necessary to search for alternative and more harmless control methods including biological methods such as the use of entomopathogenic bacteria, in which the bacterium B. thuringiensis represents a very promising alternative.10
B. thuringiensisis is a Gram-positive bacteria as well as facultative in that they are aerobic or anaerobic and produce spores in the form of protein crystals that are toxic against many orders of insects including B. thuringiensis subsp. israelensis (Diptera), B. thuringiensis subsp. tenebrionis (Coleoptera), B. thuringiensis subsp. kurstaki, B. thuringiensis subsp. aizawai (Lepidoptera) are examples of subspecies with specific bacterium against at variance groups of insects.11 These crystals are δ-endotoxins proteins, including Cry and Cyt proteins.12 These toxic proteins are encoded by the Cry and Cyt genes during the sporulation.1,13 More than 700 Cry genes have been sequenced and classified into at least 70 groups named Cry1, Cry2, Cry3 … Cry70, whose corresponding insect toxicity.13,14
The severity of toxicity to the insect also depends on the combinations of different proteins expressed by the Cry genes.15,16 Some strains can encode for more than one toxic protein, a well-known example is B. thuringiensis var. israelensis, which contains six proteins that are toxic to Diptera, this strain contains a plasmid called megaplasmid pBtoxis that encodes six different toxic proteins to mosquito larvae.17,18 It was observed in some strains that were isolated from different places that they contain different combinations of genes and have specialized toxicity to different insects order.19,20 These toxic proteins bind to receptors located in the lining membrane of the intestine of the mosquito larvae, where they feed on crystalline proteins, which leads to an imbalance of ions that leads to cell rupture and death of the larva.21-23
The main objective of this study was to collect different isolates of B. thuringiensis from different locations in Assiut governorate, Egypt that cause mortality to mosquito larvae, molecularly characterize its cry genes, and enhance the toxicity by UV- mutagenesis.
Isolation of B. thuringiensis
Samples were collected from different areas and different sources (soil, water and insects) of Assiut governorate, Egypt. Samples were collected at a depth of 10 cm below soil surface using cylindrical sampler and kept in polyethylene bags. All samples were immediately transported to the laboratory and stored in a refrigerator at 4°C until used for the isolation of B. thuringiensis.
B. thuringiensis was isolated according to previous studies with some modifications.19,24 One gram of soil sample was taken and mixed in 10 ml of sterilized distilled water. 100 μl of each sample was plated to a petri dish containing the nutrient agar supplemented with penicillin G (100 mg/L). Insect samples were cut into small pieces and mixed with 2 ml of sterilized distilled water then 100 μl were transferred to petri plates containing the selective media. And then, 100 μl of the water samples were transferred directly to petri plates. Then all the plates were placed in the incubation for 48 hours and incubated at a temperature of 28°C. The colonies showed round shapes with wavy edges and no pigment morphology were inoculated in a nutrient broth and selected for further studies.
B. thuringiensis Bioassay Activity
To test the ability of different B. thuringiensis isolates to kill the mosquito larvae, all the B. thuringiensis isolates were incubated in nutrient broth for five days at 28°C and 180 rpm until the sporulation was observed. The concentration of each isolate was measured and adjusted to be equal at a concentration of OD600= 0.2. The sporulated isolate were added to 10 ml of sterile water that contains 10 mosquito larvae and incubated at of 28 ± 2°C. The number of deaths mosquito larvae was recorded after 48 hours of the treatment.25 The treatment was carried out in three replicates for each isolate. For comparison, an additional replicate containing the commercial bio-insecticide named Dacron 54% WP (SAFA TARIM. A. S. Turky) was used as a positive control.
Detection of the Presence of Toxic Genes within the Genome of the Isolates by PCR
Different Cry toxic proteins are produced by different B. thuringiensis strains.1,26-28 PCR technique was used to screen the presence of some toxic genes in collected B. thuringiensis isolates. Five primer pairs were newly designed to target 5 different Cry and Cyt genes (Cry11AA, Cyt1AA, Cry1Ac, Cry2Aa and Cry4AA) from B. thuringiensis (Table 1). All gene sequences were obtained from NCBI (www.ncbi.nlm.nih.gov). Sequences of DNA primers were manually selected by considering, sequence location, GC content, number of nucleotides and simulated with In-Silico PCR amplification website (http://insilico.ehu.es/PCR/). There is no probability of complementarity between one primer with other or bases in the same primer.29
DNA isolation was performed by the boiling method.24,30 PCR was performed in a final volume of 20 μl containing 10 μl of GoTaq blue master mix (Promega, Madison, WI, USA), 1 μl DNA sample, 0.2 μl of each primer and 8.5 μl of water, nuclease-free. The amplification reactions were carried out in a thermocycler (Sensoquest, Biomedical Electronics, Germany) under the following initial denaturation at 93°C for 5 min, followed by 30 cycles of, 30 sec of denaturation at 95°C, 1 min for annealing at 55 – 58°C and 2 min for the extension at 72°C, then followed by 5 min final extension at 72°C and a final hold at 4°C. Then 1% agarose gel and 1X TAE were used to perform the electrophoresis. The gel was stained with ethidium bromide, visualized under UV rays and photographed. The molecular sizes of DNA fragments were measured by comparison with a 100-bp molecular marker (Hyperladder 100 bp, Bioline, Meridian Biosience, UK).
Scanning Electron Microscope
To observe the crystalline proteins under the electron microscope, isolate Am2 was grown on nutrient agar medium for 5 days at 28°C until sporulation. The spores were washed three times by cacodylate buffer for 13 minutes and post fixed in 1 % osmium tetroxide for two hours. The sample was washed by cacodylate buffer for 13 minutes for three times and then dried by an ascending series of ethanol 30, 50, 70 and 90 % for two hours and 100% for two days and then transferred to amyl acetate for two days. The sample was dried by using liquid carbon dioxide. The sample was stuck on metallic blocks using silver paint. By using gold sputter coating apparatus, the sample was evenly gold’ coated in a thickness of 15 nm. The sample was examined in the electron microscope unit at Assiut University, Egypt by uSIng JEOL JSM 5400 LV scanning electron microscope 15- 25 kV at a voltage of 15 kV and magnification of 5000x.31,32
Induction of Mutagenesis with Ultraviolet Rays (UV)
The most mortal B. thuringiensis isolate to mosquito larvae was selected to improve its efficiency by UV-mutagenesis. 1 ml of an overnight culture was poured and spread on the petri plates that contain nutrient agar medium. The plates were exposed to UV source at different time intervals (0 sec as control), 20, 40, 80, 120, 160, 200, 240 and 280sec). The plates were covered in a black polyethylene bag and incubated at 28°C for 48h to get first generation mutants. The plates that were exposed to the time causing the death of half the colonies (LC50) were used to isolate the mutants.33 And all the selected mutants were screened for their mortality by the bioassay test which was carried out in three replicates.
Isolation of B. thuringiensis isolates from different Locations and Sources
Around 100 bacterial isolates with similar morphological characteristics to B. thuringiensis (Table 2) were obtained from 9 different locations (Assiut university farm, Menqabad, Al-Ghanaim, Posra, Arab almadabigh, Sedfa, Sahel Salim, Al-Fath, West country) and different sources (water, soil and insects). The 100 bacterial isolates were morphologically screened, and colonies showed round shapes with wavy edges and no pigment morphology were furthermore checked by inoculation on a selection media containing penicillin G (100 mg/L).
Soil is the main natural reservoir for B. thuringiensis bacteria and is currently the preferred substrate for the isolation of Bacillus species.34-36 However, B. thuringiensis was also found in insects and water samples.37-42 Our results showed that soil samples are wealthy with B. thuringiensis isolates since we could isolate from soil 31 different isolate.
Isolates Toxicity Evaluation
After the morphological screening, all the isolates were examined for their mortality to mosquito larvae. Out of the 100 isolates, only 36 isolates showed a mortality effect on mosquito larvae (Table 2), so these isolates were selected for further investigations. The rest of the isolates (64 isolates) did not give a mortality rate (results are not shown), so they were excluded from further experiments.
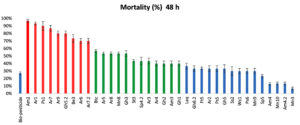
According to the mortality rate of mosquito larvae (Figure 1), the isolates were divided into 3 different levels (high, intermediate, and low). The potency of the tested isolates after 48 hours indicated that 9 isolates were highly toxic (Am2, Ar1, Ps1, Ar7, Gh5.2, Ar9, Be3, Ar6 and Ar7.2) with a mortality rate ranging from 70 to 97%; 12 intermediate isolates (Btc, Ar8, Mn8, Gh3, Ar5, St3, Sp4.2, Ar3, Ar4, Gh2, Am3 and Gh1) causing 40-57%; the rest of the isolates, 15 isolates, (Laq, Gh6.2, Ft5, Ar2, Ps5, Gh5, Ss2, Wc1, Ps6, Mn9, Sp5, Am4, Mn10, Am4.2 and Mn3), showed low mortality rate ranged from 7 to 37%.
Am2 isolate showed the highest mortality rate (97%) among all the isolates, while Mn3 isolate showed the lowest one (7%) after 48h of incubation.
The results illustrated in (Figure 1) showed that, the used bio-pesticide recorded a very low mortality rate (27%) compared to bacterial isolates especially the Am2 isolate which showed the highest mortality rate.
Previous studies indicated that it is possible to distinguish between different isolates with the mortality rate of mosquito larvae, which can be used to develop biological control tools to help control diseases caused by mosquitoes.14,43,44 The results showed that B. thuringiensis isolated from soil samples were high toxicity and more abundant than those from other sources (insects and water), these result were also found by Asokan and Puttaswamy45 who also isolated different B. thuringiensis isolates from different sources.
Detection of the Presence of B. thuringiensis Toxin Genes by PCR
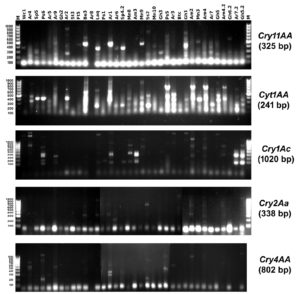
Polymerase chain reaction (PCR) was performed to determine the presence of different Cry genes (Cry11AA, Cyt1AA, Cry1Ac, Cry2Aa and Cry4AA) in the 36 selected isolates using specific primers illustrated in (Table 1). The results of the PCR screening were illustrated in (Figure 2) and (Table 3).
Table (1):
Primers used in the PCR to amplify Cry and Cyt genes of B. thuringiensis that cause toxic activity against mosquito, showing the primer sequences, the size of the target fragment, and the annealing temperature
| Gene | Accession No. | Nucleotide sequence (5⸌-3⸌) | Fragment size (bp) | Tm (◦C) | Activity |
|---|---|---|---|---|---|
| cry11AA | NC_010076 | CCAGGTTCTCAACCTGCTAC | 325 | 56 | Dip |
| CTCCCTGCTAGGATTCCGTC | |||||
| cyt1AA | NC_010076 | CCCCTCAATCAACAGCAAGG | 241 | 56 | Dip |
| CTCACTACAGCACCCATCGG | |||||
| cry1Ac | NC_020249 | GAGTGGGAAGCAGATCCTAC | 1020 | 55 | Lep |
| CCAAGAGAACATAGGAGCTC | |||||
| cry2Aa | NZ_CP013056 | GGTAGTGGACCACAGCAGAC | 338 | 58 | Lep/Dip |
| GTAGAGGTAGCAACGCCCTC | |||||
| cry4AA | NC_010076 | CCCAGGGGTAGCAGAACTGG | 802 | 58 | Dip |
| CACAGCGGACGTCTCACAAC |
Table (2):
Different isolates code, location, type of sample, number of bacterial isolates, and number of B. thuringiensis isolates.
| Isolates code | Sample’s location | Type of sample | No. of Bacterial isolates | No. of isolates caused Mortality |
|---|---|---|---|---|
| Am2, Am3, Am4, Am4.2 | Arab Almadabigh | Soil | 5 | 4 |
| Laq | Water | 10 | 1 | |
| Ar1, Ar7, Ar9, Ar6, Ar7.2, Ar5, Ar8, Ar3, Ar4, Ar2 | Assiut University Farm | Soil | 30 | 10 |
| Be3, Sp4.2, Sp5 | Insects | 8 | 3 | |
| Ps1, Ps5, Ps6 | Posra | Soil | 6 | 3 |
| Gh5.2, Gh3, Gh2, Gh1, Gh6.2, Gh5 | Al-Ghanaim | Soil | 8 | 7 |
| Mn8, Mn9, Mn10, Mn3 | Menqabad | Soil | 10 | 4 |
| Wc1 | West Country | Soil | 5 | 1 |
| St3 | Sedfa | Soil | 6 | 1 |
| Ft5 | Al-Fath | Soil | 7 | 1 |
| Ss2 | Sahel Salim | Soil | 4 | 1 |
| Btc | Egypt Microbial Culture Collection (EMCC) | Insects | 1 | 1 |
| Total | 100 | 36 |
Table (3):
Genes detected in the B. thuringiensis isolates.
| Code of Isolate | Gene | Mortality (%) | |||||
|---|---|---|---|---|---|---|---|
| Cry11AA | Cyt1AA | Cry1Ac | Cry2Aa | Cry4AA | 48 h | Total | |
| Am2 | – | + | + | + | – | 97% | 3 |
| Ar1 | – | – | – | + | – | 93% | 1 |
| Ps1 | – | – | – | – | – | 90% | 0 |
| Ar7 | – | + | – | – | – | 87% | 1 |
| Ar9 | – | – | – | – | – | 80% | 0 |
| Gh5.2 | – | + | – | – | – | 80% | 1 |
| Be3 | – | – | – | + | – | 73% | 1 |
| Ar6 | + | – | – | – | – | 70% | 1 |
| Ar7.2 | – | – | – | – | – | 70% | 0 |
| Btc | – | – | – | – | – | 57% | 0 |
| Ar5 | – | – | – | – | – | 53% | 0 |
| Ar8 | – | – | – | – | – | 53% | 0 |
| Mn8 | – | – | – | – | – | 53% | 0 |
| Gh3 | – | – | – | – | – | 53% | 0 |
| St3 | – | – | – | – | – | 43% | 0 |
| Sp4.2 | + | + | – | – | – | 43% | 2 |
| Ar3 | – | – | – | – | – | 43% | 0 |
| Ar4 | – | – | – | – | – | 40% | 0 |
| Gh2 | – | – | – | – | – | 40% | 0 |
| Am3 | – | – | – | – | – | 40% | 0 |
| Gh1 | – | – | – | – | – | 40% | 0 |
| Laq | + | + | – | + | – | 37% | 3 |
| Gh6.2 | – | – | – | – | – | 33% | 0 |
| Ft5 | – | – | – | – | – | 33% | 0 |
| Ar2 | – | – | – | – | – | 33% | 0 |
| Ps5 | – | – | – | – | – | 33% | 0 |
| Gh5 | – | – | – | – | – | 33% | 0 |
| Ss2 | – | + | – | – | – | 30% | 1 |
| Wc1 | – | – | – | – | – | 30% | 0 |
| Ps6 | – | – | – | – | + | 30% | 1 |
| Mn9 | – | – | + | – | – | 30% | 1 |
| Sp5 | – | – | – | – | – | 23% | 0 |
| Am4 | – | – | + | – | – | 13% | 1 |
| Mn10 | – | – | – | – | – | 13% | 0 |
| Am4.2 | – | – | + | – | – | 13% | 1 |
| Mn3 | – | – | – | – | – | 7% | 0 |
| total | 3 | 6 | 4 | 4 | 1 | ||
+: amplified the gene; -: did not amplify the gene.
The presence of Cry11AA gene was only detected in three isolates (Ar6, Sp4.2 and Laq isolate). While Cyt1AA gene was found in 6 isolates (Am2, Ar7, Gh5.2, Sp4.2, Laq, and Ss2 isolate). Results also showed that the Cry1Ac gene are present in four isolates (Am2, Mn9, Am4 and Am4.2 isolate). Four tested isolates (Am2, Ar1, Be3, and Laq isolate) produced Cry2Aa gene. Cry4AA gene was detected in only one isolate (Ps6).
Based on the results of the PCR amplification listed in Table 3, Am2 isolate amplified three different types of toxic genes (Cyt1AA, Cry1Ac and Cry2Aa) and caused the highest mortality (97%). On the other hand, Laq isolate also showed the presence of three different types of toxic genes (Cry11AA, Cyt1AA and Cry2Aa) but caused low mortality (37%). Both Am2 and Laq isolates contained 3 toxic genes, but they showed different mortality rates this could be due the differences in the detected genes.
The Cyt1AA gene was more frequent among all the 5 tested genes and was present in six different isolates. Cyt1AA was found to cause high mortality since Ar7 and Gh5.2 isolates showed only the presence of Cyt1AA gene and caused a mortality percent of 87% and 80%, respectively. On the other hand, lepidopteran-specific Cry1Ac gene causes the least mortality rate, since it was detected individually in three isolates (Mn9, Am4 and Am4.2) and the maximum mortality rate was 30%, thus it can be concluded that the Cry1Ac gene has no major effect on the mortality. The results showed the lowest frequent amplified gene was Cry4AA since it was detected in only one isolate (Ps6) which showed a mortality rate with 30%.
In this study, 23 isolates did not show any presence of any tested toxic genes, but these isolates still have toxicity on mosquito larvae. This indicates the presence of other toxic genes that were not included in this study. Among those isolates, Ps1 isolate showed the highest mortality percentage 90%. On the other hand, Mn3 isolate showed the least mortality percentage 7%.
The absence of the PCR product in some isolates was detected was also obtained by46,47 that found 89 out of 215 field-collected strains of B. thuringiensis did not produce PCR products with universal primers.
PCR was used as a basic parameter to detect the different types of toxic proteins of the tested isolates and their activity against mosquito larvae.13,48 The genes/toxins Cry11AA, Cyt1AA, Cry2Aa and Cry4AA are associated with poisoning against mosquito larvae.1,26-28 The Cry1Ac gene is associated with toxicity against Lepidoptera larvae.49
Results in Figure 2 showed that, most of the isolates did not produce a positive single band with the specific primers, and an extra band/bands were amplified. Similar findings were reported by50,51 who suggested that, isolates containing novel Cry genes may give PCR products different in size relative to the standard or may completely lack PCR products. This could be because two or three Cry genes might be positioned next to each other, forming an operon.52
Scanning Electronic Microscopy (SEM)
Am2 isolate which caused the highest mortality 97% were preliminarily screened with phase contrast microscopy 100X magnification to observe the presence of shape of the Cry crystals. Scanning electron microscopy showed crystal architecture of cuboidal shapes for the toxic proteins and confirmed the presence of rod-shaped bacterial cells, the morphological characteristics of B. thuringiensis (Figure 3).
Figure 3. Scanning electronic microscopy (SEM) of B. thuringiensis isolates, showing the characterized parasporal inclusion shapes. Sp: spore and crystal cuboidal (CC)
A previous study,53-55 reported different shapes of crystals protein cuboidal, spherical, irregular, bipyramidal, balloon shaped and flat square all these crystal structures were found in different isolates of B. thuringiensis and isolated from different regions. In the present study, Am2 isolate amplified three different types of toxic genes Cyt1AA, Cry1Ac and Cry2Aa and the scanning electron microscopy showed crystal structure of cuboidal shape for the toxic proteins. Cry proteins related to some crystal structures.46,56,57 In this study, Am2 isolate demonstrated cuboidal crystals related to amplify Cry2Aa protein.56,58
Genetic Improvement with Mutagenesis by Ultraviolet rays (UV)
The radiation of sunlight is crucial in the decrease of biological activity of pesticides of B. thuringiensis due to ultraviolet rays that break down spores and their internal toxins.59-61 Previous studies were obtained in B. thuringiensis mutants that were produced by successive rounds of ultraviolet rays, the mutants were more UV resistant and some of them were lost encoding their toxin Cry genes due to mutations.33,62,63
UV-mutagenesis was performed to improve the toxicity of the best killing BT isolate (Am2). Am2 isolate was exposed to UV irradiation for different time intervals (0 sec (as control), 20, 40, 80, 120, 160, 200, 240 and 280sec). The mutants were selected from plates that showed a survivability rate of 50% after exposure (280 sec of UV irradiation).
Thirty mutants were obtained from the mutagenesis of the wild Am2 isolate. All the mutants were screened for their mortality. Out of 30 mutants, only one mutant Mu25 showed better mortality than the wild type Am2 isolate. The improved Mu25 mutant showed toxicity to mosquito larvae with a mortality rate of 100% after 48 hours (Figure 4).
Results in Figure (4) showed that, the toxicity of the isolates was decreased in all the isolates after the UV-mutagenesis. These results were also found by64,65 since they found that, insecticidal activity of B. thuringiensis were negatively affected by UV rays, which leads to a decrease in toxicity and a decrease in stability. Only one mutant Mu25 was improved and caused a high mortality for mosquito larvae after UV-mutagenesis.
In this study, the genetic variability was obvious among different isolates and caused variation in their toxicity against mosquito larvae. The isolate Am2 amplified the highest number of different types of genes (Cyt1AA, Cry1Ac and Cry2Aa) and caused high mortality rates against mosquito larvae. The Cyt1AA gene toxin has predominant efficiency against dipterous according to.19,66 The Cry2Aa protein demonstrated form cuboidal crystal in Am2 isolate under scanning electron microscopy according to.58,67,68 UV-mutagenesis showed high toxicity and stability in mutant Mu25 against mosquito larvae. Further studies will be performed to investigate the effect of UV mutagenesis on different cry gene sequences. Moreover, the gene expression levels of different cry gene will be determined by qRT-PCR and SDS-PAGE.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
AEH and AEE conceptualized the study. AR conducted the experiment. AEH and AR wrote original draft. AAA and GAM revised original draft. AEH wrote, reviewed and edited the manuscript. AEH, AEE, AAA and GAM performed supervision. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
- Ben-Dov E. Bacillus thuringiensis subsp. israelensis and its dipteran-specific toxins. Toxins. 2014;6(4):1222-1243.
Crossref - Shaw WR, Catteruccia F. Vector biology meets disease control: using basic research to fight vector-borne diseases. Nat Microbiol. 2019;4(1):20-34.
Crossref - Merle H, Donnio A, Jean-Charles A, et al. Ocular manifestations of emerging arboviruses: Dengue fever, Chikungunya, Zika virus, West Nile virus, and yellow fever. J Fr Ophtalmol. 2018;41(6):e235-e243.
Crossref - Williams DD. The ecology of temporary waters. Springer Science & Business Media; 2012.
- Macoris MdLdG, Andrighetti MTM, Otrera VCG, Carvalho LRd, Caldas Junior AL, Brogdon WG. Association of insecticide use and alteration on Aedes aegypti susceptibility status. Mem Inst Oswaldo Cruz. 2007;102(8):895-900.
Crossref - Rather IA, Parray HA, Lone JB, et al. Prevention and Control Strategies to Counter Dengue Virus Infection. Front Cell Infect Microbiol. 2017;7:336.
Crossref - Grisales N, Poupardin R, Gomez S, Fonseca-Gonzalez I, Ranson H, Lenhart A. Temephos resistance in Aedes aegypti in Colombia compromises dengue vector control. PLoS Neglected Tropical Diseases. 2013;7(9):e2438.
Crossref - Moyes CL, Vontas J, Martins AJ, et al. Contemporary status of insecticide resistance in the major Aedes vectors of arboviruses infecting humans. PLoS Negl Trop Disses. 2017;11(7):e0005625.
Crossref - Aponte A, Penilla RP, Rodriguez AD, Ocampo CB. Mechanisms of pyrethroid resistance in Aedes (Stegomyia) aegypti from Colombia. Acta tropica. 2019;191:146-154.
Crossref - Ingabire CM, Hakizimana E, Rulisa A, et al. Community-based biological control of malaria mosquitoes using Bacillus thuringiensis var. israelensis (Bti) in Rwanda: community awareness, acceptance and participation. Malar J. 2017;16(1):399.
Crossref - Seifinejad A, Jouzani GS, Hosseinzadeh A, Abdmishani C. Characterization of Lepidoptera-active cry and vip genes in Iranian Bacillus thuringiensis strain collection. Biological Control. 2008;44(2):216-226.
Crossref - Santos F, Lopes J, Vilas-Boas G, Zequi JAC. Characterization of Bacillus thuringiensis isolates with potential for control of Aedes aegypti (Linnaeus, 1762)(Diptera: Culicidae). Acta tropica. 2012;122(1):64-70.
Crossref - Bravo A, Gomez I, Porta H, et al. Evolution of Bacillus thuringiensis Cry toxins insecticidal activity. Microbial Biotechnol. 2013;6(1):17-26.
Crossref - Soares-da-Silva J, QueirOs SG, de Aguiar JS, et al. Molecular characterization of the gene profile of Bacillus thuringiensis Berliner isolated from Brazilian ecosystems and showing pathogenic activity against mosquito larvae of medical importance. Acta tropica. 2017;176:197-205.
Crossref - Federici BA, Park H-W, Bideshi DK. Overview of the basic biology of Bacillus thuringiensis with emphasis on genetic engineering of bacterial larvicides for mosquito control. Open Toxinology Journal. 2010;3(1):83-100.
- Guo Z, Kang S, Sun D, et al. MAPK-dependent hormonal signaling plasticity contributes to overcoming Bacillus thuringiensis toxin action in an insect host. Nat Commun. 2020;11(1):303.
Crossref - Berry C, O’Neil S, Ben-Dov E, et al. Complete sequence and organization of pBtoxis, the toxin-coding plasmid of Bacillus thuringiensis subsp. israelensis. Appl Environ Microbiol. 2002;68(10):5082-5095.
Crossref - Gillis A, Fayad N, Makart L, et al. Role of plasmid plasticity and mobile genetic elements in the entomopathogen Bacillus thuringiensis serovar israelensis. FEMS Microbiol Rev. 2018;42(6):829-856.
Crossref - Lobo KdS, Soares-da-Silva J, Silva MCd, Tadei WP, Polanczyk RA, Pinheiro VCS. Isolation and molecular characterization of Bacillus thuringiensis found in soils of the Cerrado region of Brazil, and their toxicity to Aedes aegypti larvae. Rev Bras Entomol. 2018;62:5-12.
Crossref - Aynalem B, Muleta D, Venegas J, Assefa F. Isolation, molecular characterization and pathogenicity of native Bacillus thuringiensis, from Ethiopia, against the tomato leafminer, Tuta absoluta: Detection of a new high lethal phylogenetic group. Microbiol Res. 2021;250:126802.
Crossref - Boucias DG, Pendland JC. Principles of insect pathology. Springer Science & Business Media; 2012.
- Zhang Q, Hua G, Adang MJ. Effects and mechanisms of Bacillus thuringiensis crystal toxins for mosquito larvae. Insect Sci. 2017;24(5):714-729.
Crossref - Soberon M, Monnerat R, Bravo A. Mode of action of cry toxins from Bacillus thuringiensis and resistance mechanisms. Microbial Toxins. 2018;1.
Crossref - Guaycurus T. Controle biologico de mosquitos com Bacillus thuringiensis subsp. israelensis, modelo alemao e pesquisa de genes cry em linhagens auto-aglutinantes. UnB; 1999.
- Dulmage HT, Correa JA, Gallegos-Morales G. Potential for improved formulations of Bacillus thuringiensis israelensis through standardization and fermentation development. Bacterial Control of Mosquitoes & Black Flies. 1990:110-133.
Crossref - Bravo A, Gill SS, Soberon M. Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control. Toxicon. 2007;49(4):423-435.
Crossref - Soberon M, Gill S, Bravo A. Signaling versus punching hole: How do Bacillus thuringiensis toxins kill insect midgut cells? Cell Mol Life Sci. 2009;66(8):1337-1349.
Crossref - Bideshi DK, Waldrop G, Fernandez-Luna MT, et al. Intermolecular interaction between Cry2Aa and Cyt1Aa and its effect on larvicidal activity against Culex quinquefasciatus. J Microbiol Biotechnol. 2013;23(8):1107-1115.
Crossref - Wahyuni FD, Saraswati H, Dewi KS. In-Silico Analysis for cryI gene amplification from Bacillus thuringiensis. BIOEDUKASI. 2020:8-14.
Crossref - Hesham AE-L. New safety and rapid method for extraction of genomic DNA from bacteria and yeast strains suitable for PCR amplifications. J Pure Appl Microbiol. 2014;8(1):383-388.
- Bozzola J, Russell L. Electron microscopy principles and techniques for biologists. Jones and Bartlitt publ. Inc; 1991.
- Kati H, Sezen K, Nalcacioglu R, Demirbag Z. A highly pathogenic strain of Bacillus thuringiensis serovar kurstaki in lepidopteran pests. J Microbiol. 2007;45(6):553-557.
- Sansinenea E, Salazar F, Ramirez M, Ortiz A. An ultra-violet tolerant wild-type strain of melanin-producing Bacillus thuringiensis. Jundishapur J Microbiol. 2015;8(7):e20910.
Crossref - Silva MC, Siqueira HA, Marques EJ, et al. Bacillus thuringiensis isolates from northeastern Brazil and their activities against Plutella xylostella (Lepidoptera: Plutellidae) and Spodoptera frugiperda (Lepidoptera: Noctuidae). Biocontrol Science and Technology. 2012;22(5):583-599.
Crossref - Soares-da-Silva J, Pinheiro VCS, Litaiff-Abreu E, Polanczyk RA, Tadei WP. Isolation of Bacillus thuringiensis from the state of Amazonas, in Brazil, and screening against Aedes aegypti (Diptera, Culicidae). Rev Bras Entomol. 2015;59:01-06.
Crossref - El-Kersh TA, Ahmed AM, Al-Sheikh YA, Tripet F, Ibrahim MS, Metwalli AAM. Isolation and characterization of native Bacillus thuringiensis strains from Saudi Arabia with enhanced larvicidal toxicity against the mosquito vector Anopheles gambiae (sl). Parasit Vectors. 2016;9(1):647.
Crossref - Chilcott C, Wigley P. Isolation and toxicity of Bacillus thuringiensis from soil and insect habitats in New Zealand. J Invertebr Pathol. 1993;61(3):244-247.
Crossref - Beron CM, Salerno GL. Characterization of Bacillus thuringiensis isolates from Argentina that are potentially useful in insect pest control. BioControl. 2006;51(6):779-794.
Crossref - Valicente FH, de Toledo Picoli EA, de Vasconcelos MJV, et al. Molecular characterization and distribution of Bacillus thuringiensis cry1 genes from Brazilian strains effective against the fall armyworm, Spodoptera frugiperda. Biological Control. 2010;53(3):360-366.
Crossref - Iriarte J, Porcar M, Lecadet M-M, Caballero P. Isolation and characterization of Bacillus thuringiensis strains from aquatic environments in Spain. Curr Microbiol. 2000;40(6):402-408.
Crossref - Konecka E, Baranek J, Hrycak A, Kaznowski A. Insecticidal activity of Bacillus thuringiensis strains isolated from soil and water. Scientific World Journal. 2012;2012:10501.
Crossref - Hernandez-Rodriguez C, Boets A, Van Rie J, Ferre J. Screening and identification of vip genes in Bacillus thuringiensis strains. J Appl Microbiol. 2009;107(1):219-225.
Crossref - Gobatto V, Giani S, Camassola M, Dillon A, Specht A, Barros N. Bacillus thuringiensis isolates entomopathogenic for Culex quinquefasciatus (Diptera: Culicidae) and Anticarsia gemmatalis (Lepidoptera: Noctuidae). Braz J Biol. 2010;70:1039-1046.
Crossref - Reyaz A, Gunapriya L, Indra Arulselvi P. Molecular characterization of indigenous Bacillus thuringiensis strains isolated from Kashmir valley. 3 Biotech. 2017;7(2):1-11.
Crossref - Asokan R. Isolation and characterization of Bacillus thuringiensis Berliner from soil, leaf, seed dust and insect cadaver. J Biol Control. 2007;21(1):83-90.
- Ben-Dov E, Zaritsky A, Dahan E, et al. Extended screening by PCR for seven cry-group genes from field-collected strains of Bacillus thuringiensis. Appl Environ Microbiol. 1997;63(12):4883-4890.
Crossref - Ben-Dov E, Wang Q, Zaritsky A, et al. Multiplex PCR screening to detect cry9 genes in Bacillus thuringiensis strains. Appl Environ Microbiol. 1999;65(8):3714-3716.
Crossref - Andrade-Ochoa S, Correa-Basurto J, Rodriguez-Valdez L, Sanchez-Torres L, Nogueda-Torres B, Nevarez-Moorillon G. In vitro and in silico studies of terpenes, terpenoids and related compounds with larvicidal and pupaecidal activity against Culex quinquefasciatus Say (Diptera: Culicidae). Chem Cent J. 2018;12(1):53.
Crossref - Sivasupramaniam S, Moar W, Ruschke L, et al. Toxicity and characterization of cotton expressing Bacillus thuringiensis Cry1Ac and Cry2Ab2 proteins for control of lepidopteran pests. J Econ Entomol. 2008;101(2):546-554.
Crossref - Carozzi NB, Kramer VC, Warren GW, Evola S, Koziel MG. Prediction of insecticidal activity of Bacillus thuringiensis strains by polymerase chain reaction product profiles. Appl Environ Microbiol. 1991;57(11):3057-3061.
Crossref - Kuo W-S, Chak K-F. Identification of novel cry-type genes from Bacillus thuringiensis strains on the basis of restriction fragment length polymorphism of the PCR-amplified DNA. Appl Environ Microbiol. 1996;62(4):1369-1377.
Crossref - Brown KL, Whiteley H. Molecular characterization of two novel crystal protein genes from Bacillus thuringiensis subsp. thompsoni. J Bacteriol. 1992;174(2):549-557.
Crossref - Noguera PA, Ibarra JE. Detection of new cry genes of Bacillus thuringiensis by use of a novel PCR primer system. Appl Environ Microbiol. 2010;76(18):6150-6155.
Crossref - Lone SA, Malik A, Padaria JC. Selection and characterization of Bacillus thuringiensis strains from northwestern Himalayas toxic against Helicoverpa armigera. Microbiologyopen. 2017;6(6):e00484.
Crossref - Nair K, Al-Thani R, Al-Thani D, Al-Yafei F, Ahmed T, Jaoua S. Diversity of Bacillus thuringiensis strains from Qatar as shown by crystal morphology, δ-endotoxins and cry gene content. Front Microbiol. 2018;9:708.
Crossref - Hofte H, Whiteley H. Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol Rev. 1989;53(2):242-255.
Crossref - Rosas-Garcia NM, Mireles-Martinez M, Hernandez-Mendoza JL, Ibarra J. Screening of cry gene contents of Bacillus thuringiensis strains isolated from avocado orchards in Mexico, and their insecticidal activity towards Argyrotaenia sp.(Lepidoptera: Tortricidae) larvae. J Appl Microbiol. 2008;104(1):224-230.
- Lopez-Meza JE, Ibarra JE. Characterization of a novel strain of Bacillus thuringiensis. Appl Environ Microbiol. 1996;62(4):1306-1310.
Crossref - Liu Y-T, Sui M-J, Ji D-D, Wu I-H, Chou C-C, Chen C-C. Protection from ultraviolet irradiation by melanin of mosquitocidal activity of Bacillus thuringiensis var. israelensis. J Invertebr Pathol. 1993;62(2):131-136.
Crossref - Pozsgay M, Fast P, Kaplan H, Carey P. The effect of sunlight on the protein crystals from Bacillus thuringiensis var. kurstaki HD1 and NRD12: a Raman spectroscopic study. J Invertebr Pathol. 1987;50(3):246-253.
Crossref - Pusztai M, Fast P, Gringorten L, Kaplan H, Lessard T, Carey PR. The mechanism of sunlight-mediated inactivation of Bacillus thuringiensis crystals. Biochem J. 1991;273(1):43-47.
Crossref - Patel K, Wyman J, Patel K, Burden B. A Mutant of Bacillus thuringiensis Producing a Dark-Brown Pigment with Increased UV Resistance and Insecticidal Activity. J Invertebr Pathol. 1996;67(2):120-124.
Crossref - Saxena D, Ben-Dov E, Manasherob R, Barak Ze, Boussiba S, Zaritsky A. A UV tolerant mutant of Bacillus thuringiensis subsp. kurstaki producing melanin. Curr Microbiol. 2002;44(1):25-30.
Crossref - Siegwart M, Graillot B, Blachere Lopez C, et al. Resistance to bio-insecticides or how to enhance their sustainability: a review. Front Plant Sci. 2015;6:381.
Crossref - Zhang L, Zhang X, Zhang Y, et al. A new formulation of Bacillus thuringiensis: UV protection and sustained release mosquito larvae studies. Sci Rep. 2016;6(1):1-8.
Crossref - Soberon M, Lopez-Diaz JA, Bravo A. Cyt toxins produced by Bacillus thuringiensis: a protein fold conserved in several pathogenic microorganisms. Peptides. 2013;41:87-93.
Crossref - Ibarra JE, Federici BA. Parasporal bodies of Bacillus thuringiensis subsp. morrisoni (PG-14) and Bacillus thuringiensis subsp. israelensis are similar in protein composition and toxicity. FEMS Microbiol Lett. 1986;34(1):79-84.
Crossref - Crickmore N, Zeigler D, Feitelson J, et al. Revision of the nomenclature for the Bacillus thuringiensis pesticidal crystal proteins. Microbiol Mol Biol Rev. 1998;62(3):807-813.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.