ISSN: 0973-7510
E-ISSN: 2581-690X
Lipase (Triacylglycerol acyl hydrolase), is a hydrolase enzyme that plays an important role in industries such as health, food, biotechnology, and energy. Lipase might be isolated from almost all living organism, either higher organisms or microbes. In this report, lipase was isolated from compost isolated microbe namely AL89 isolate. AL89 was previously identified closed to Pseudoxanthomonas taiwanensis. The crude extract of lipase was produced by incubating of the culture at 55°C for 19 hours. The crude extract was partially purified using acetone fractionation. Fractionation was carried out at concentrations of acetone at 0-20%, 20-40% and 40-60%. The specific activity was determined by hydrolytic activity of lipase with the substrate of para-nitrophenylpalmitate (pNPP). The result showed that the highest activity of the enzyme is 0.0971 U/mg protein from the fraction of 0-20%. Lipase from isolate AL89 showed optimum activity at 55°C and pH 9. In addition the enzyme prefers para-nitrophenyl laurate (pNPL) as substrate. Using zymography analysis showed that the active protein at the size of 70 kDa. All the data suggested the enzyme is thermo and alkali-tolerant lipase.
Biotechnology, lipase enzyme, isolate AL89, microorganisms, compost.
Lipase is one of the enzymes in the hydrolase group that catalyzes the breakdown and the formation of carboxyl ester bonds reaction. In water solvents, lipases catalyze the hydrolysis of carboxylic ester bonds to produce free fatty acids and organic alcohols. While in the environment of organic solvents, this enzyme has transesterification activities such as alcoholysis and acidolysis reaction1.
Lipase is a multipurpose biocatalyst group and has an important role in biotechnology and industrial processes, including additives in bio detergents, pulp and paper; leather and food processing, organic compound synthesis, biofuel, and biodegradable polymers. Lipase has also been exploited as an inexpensive and versatile catalyst to degrade lipids in more modern applications. High lipase activity is used as an alternative to produce fuel from vegetable oils2. Lipase is used as a biocatalyst that substitutes the traditional catalysts in biodiesel processing. This enzyme replaces inorganic catalysts in processes that require energy-intensive production of biodiesel3. By using lipase, the production process becomes more environmentally friendly and safe.
Lipases are produced in various sources in three domains of life including animals, plants, and microorganisms4. When compared with enzymes from plants or animals, enzymes from microbes have many advantages including production capacity in a short time, can be produced at any time and are not affected by conditions, easy for genetic engineering. Microbes can also grow quickly in inexpensive media. Enzymes from microbes also have relatively more stable activity5. For this reason, enzymes from microbes are preferred for economic reasons. In this case, lipases from microbes are preferred because they are more stable and cheaper.
Lipases are produced by microbes including bacteria, fungi, yeasts, and actinomyces. Various lipases obtained from these microbial sources have diverse characteristics. However, not all of these enzymes are suitable for commercial applications. Several factors such as pH range, tolerance to emulsifier or surfactant, and thermostability, are properties that must be considered in the selection and development of enzymes as commercial products6.
During the last five years, a lot of efforts carried out in order to produce active lipases using different producing microorganisms7. Various sources of microorganisms producing lipase were explored, such as from hot spring around West Java8,9, and domestic compost10,11. Thermostable lipases from several thermophilic microorganisms have been identified. Lipase from Thermus aquaticus, isolated from Indonesian hot spring, was obtained through homologous expression and purified by hydrophobic interactions-based chromatography12. Thermostable lipases from Pseudoxanthomonas sp., isolated from domestic compost was reported and showed tolerance towards methanol and various organic solvent13.
This report describes partial purification of lipase from domestic compost isolated bacteria. The enzyme was characterized as thermo and alkali tolerant lipase.
Strain
The thermostable bacterial culture was obtained from the Biochemistry Laboratory of Institut Teknologi Bandung, named AL89. This bacterium has been selected from bacterial cultures from domestic compost. This bacterium has hydrolytic activity which characterized by a clear zone around the colony in media containing rhodamine B. Cultivation was carried out with Luria Bertani media in an incubator shaker at 55°C, 150 rpm for 19 hours.
Crude Extract Production
Isolate AL89 was inoculated in 100 mL media (0.1% CaCl2, 0.5% yeast extract, 0.5% meat extract, 0.1% NaCl in 0.05 M phosphate buffer pH 8.0),then incubated at 55oC 150 rpm for 19 hours, as was done in a previous study13. Extracellular lipase was obtained from culture supernatants which had been centrifuged for 45 minutes at 12000 g.
Partial Purification
The crude lipase extract is purified using acetone fractionation at concentrations of 0-20%, 20-40%, and 40-60%. Supernatant added with acetone and mixed with a magnetic stirrer for 15 minutes. Afterward, the mixture was centrifuged for 45 minutes at 12000 g. The protein precipitate was then dissolved using a 0.05 M phosphate buffer pH 8.0.
SDS-PAGE Protein Lipase
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed with 12% polyacrylamide gel, as described by Smith (1984)14.
Zymography
The activity of lipase was analysis used zymographic method15. Protein samples were run in SDS-PAGE together with protein marker for 1.5 hours. The samples were incubated in 50 mL of phosphate buffer at0.05 M pH 8.0 containing 1 mM substrate a-naphtyl acetate and 3 mM fast blue. Gel was incubated at 50°C for 1 hour. The result of zymography was shown with brown band at gel.
Lipase Activity Assay
The lipolytic activity assay for this lipase was carried out by the spectrophotometric method as explained by Syihab (2017)13.
Characterization of The Enzyme
The purpose of this lipase activity assay is to find out the optimum activity of the enzyme on the substrate based on the length of the carbon chain. For the assay carried out with pNP ester substrate with short chains (C2 and C4) and long chains (C10, C12, C14, and C16).
Analysis of thermostability and pH was carried out using the best substrate above. The optimum temperature was carried out in the range of temperature from 35-85°C, with an interval of 10. Enzyme activity was tested at various pH ranges, to find out the optimum pH of the enzyme. This activity assayed by by carried out in the pH range from 6-12 at intervals of 1.
Production and Partial Purification of Lipase
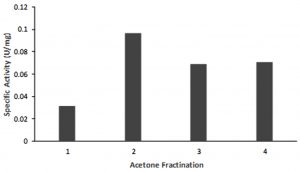
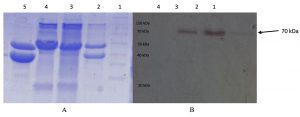
At 19 hours incubation at a temperature of 55 oC lipase with optimal activity was produced. In this condition, 1 L of culture produces an extracellular protein with an average protein level of 0.46 mg / mL. The enzyme activity assay shows the specific activity of the crude extract enzyme is 0.03 U / mg showed in Fig. 1. One unit of lipase is defined as the amounts of the enzyme-producing 1 mmol PNP per minute under the assay condition16. Analysis using SDS PAGE and zymograms of the crude extract enzyme reveals a thin band of about 70 kDa. This suggested that the crude extract of the enzyme has a hydrolase activity on protein with a size of about 70 kDa. Its showed in Fig.2.
Fig. 1. Activity of the partial purified enzyme. One unit of lipase is defined as the amount of enzyme producing 1 mmol PNP per minutes under the assay conditions. (1) crude of extract enzyme, (2) Fraction 0-20%, (3) Fraction 20-40%, (4) Fraction 40-60 %.
Fig. 2. SDS-PAGE Electrophoregram of partial purified lipase. (A) SDS-PAGE Lane 1, protein marker; lane 2, crude extract; lane 3, 0-20%; lane 4, 20-40%; lane 5, 40-60% acetone fraction (B) zymography of partial purified enzyme. Lane 1, crude extract; lane 2, 0-20 %; lane 3, 20-40 %; lane 4, 40-60% acetone fraction.
The crude extract of lipase was partially purified using acetone fractination. The highest activity was shown at the 0-20% acetone fraction, with the activity more than 3 times higher (0.97 U/mg)compared to that the crude extract (Fig 1).SDS PAGE and zymographic analysis of acetone fraction 0-20%, showed that there was a clear band around 70 kDa (Fig. 2). This suggested that a protein with a size of 70 kDa is a lipase enzyme produced by AL89 isolates
Characterization of the Enzyme
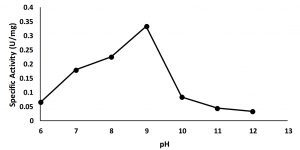
Characterization of partially purified enzyme aims to study the properties of the enzyme in the form of optimum conditions for it activity including optimum pH and temperature.The activity of enzyme in various condition of pH were assayed in the range of pH 6.0 to 12.00. The result showed that the maximum activity at pH 9 (Fig. 3), suggested that the enzymewas an alkali tolerant lipase10,13. The high activity of the isolated at alkaline condition suggests that the lipase shows high potential application on many industries such as detergent, biopolymers, pharmacy, cosmetics and flavor17.
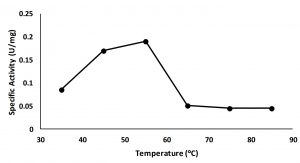
The optimum temperature of the enzyme was assayed by measuring the lipolytic activity at various temperatures from 35 to 85°C. The result showed that the highest lipolytic activity at 55°C (Fig. 4). This is suggested that enzyme belonged as a thermostablelipase9,10.
Fig. 4. Activity of 0-20 % fraction in wide different temperature, maximum activity is showed on the temperature 55°C.
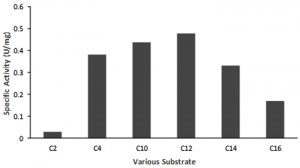
Other lipase properties are shown by the results of tests on various substrates based on chain length C. The test results show that lipase has activity on substrates with C 4 to C 16 chain lengths. The highest lipase activity was obtained at C 12 (Fig. 5). This suggests that the lipase produced by AL89 isolates has broad substrate specificity in both esterase and lipase activity.
Fig. 5. Activity of 0-20 % fraction toward different length of carbon range. (1) p-NP acetate (C2); (2) p-NP butyrate (C4); (3) p-NP caprate (C10); (4) p-NP laurate (C12); (5) p-NP myristate (C14); (6) p-NP palmitate. Maximum activity was showed on p-NP laurate (C12).
From all of the data suggested that the enzyme is alkaline and thermostablelipase.
Bacterial isolate from domestic compost, AL89, is a thermophilic bacterium that has hydrolytic activity. This bacterium produces lipases with a size of about 70 kDa. The lipase produced by this bacterium has optimum activity at 55°C and pH 9. This lipase has broad substrate specificity in both esterase and lipase activity, with highest activity at C12. All data obtained suggested that this lipase is grouped in alkaline and thermostable lipases.
ACKNOWLEDGMENTS
The authors fully acknowledge the Institute Teknologi Bandung, which made this study viable and effective.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work and approved it for publication.
FUNDING
This work was supported by a grant from P3MI Research Grant, Institut Teknologi Bandung, Ministry of Research, Technology and Higher Education, Republic of Indonesia.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
- Borelli, G. M. And Trono D. Recombinant lipases and phospolipases and their use as biocatalysts for industrial application. International Journal of Molecular Sciences, 2015; 16: 20774-20840.
Crossref - Gupta R, Gupta N, Rathi P. “Bacterial lipases: an overview of production, purification and biochemical properties”. Appl. Microbiol. Biotechnol., 2004; 64(6): 763–81.
Crossref - Harding, KG, Dennis, JS, von Blottnitz, H, Harrison, STL. “A life-cycle comparison between inorganic and biological catalysis for the production of biodiesel”. Journal of Cleaner Production, 2008; 16(13): 1368–78.
Crossref - Olusesan, A.T., Azura, L.K., Abubakar, F., Hamid, N.S.A., Radu, S. and Saari, N. Phenotypic and molecular identification of novel thermophilic Anoxybacillus species: a lipase producing bacterium isolated from Malaysian hotspring. World J. Microbiol. Biotechnol., 2009; 25: 1981–1988.
Crossref - Wiseman, A. Introduction to Principles. In: Handbook of Enzyme Biotechnology, Wiseman, A. (Ed.). 3rd Edn., Ellis Horwood Ltd., T.J. Press Ltd., Padstow, Cornwall, UK, 1995; 3-8.
- Sharma, R., Y. Chistib and U.C. Banerjeea. Research review paper: Production, purification, characterization and applications of lipases. Biotechnol. Adv., 2001; 19: 627-662.
Crossref - Abdelmoez W and Mustafa A. Oleochemical industry future through biotechnology. J. Oleo Sci., 2014; 63(6): 545-554.
Crossref - Madayanti F., B.V.E. Viera, M.P Whidiastuty, and Akhmaloka. Characterization and Identification of Thermophilic Lipase Producing Bacteria from Thermogenic Compost. J Pure Appl Microbiol, 2008; 2(2): 325-332.
- Widhiastuty, M.P., Febriani, Yohandini, H., Maelita R. Moeis, Madayanti, F., Akhmaloka. Characterization and Identification of Thermostable Alkaline Lipase Producing Bacteria from Hot Spring around West Java. J Pure Appl Microbiol, 2009; 3(1): 27-40.
- Syihab SF, Madayanti F, Akhmaloka. Isolation, characterization, and identification of lipolytic thermophiles with methanol tolerance from domestic compost. J Pure Appl Microbiol, 2015; 9(2): 385-390.
- Nurhasanah, Nurbaiti S, Warganegara, FM, Akhmaloka. Diversity of gene en coding thermostable lipase from compost based on metagenome analysis. International Journal of Integrative Biology, 2015; 16(1): 7-12.
- Febriani, Ihsanawati, Hertadi R, MadayantiF, Akhmaloka. Thermostable alkaline lipase isolated from Thermus aquaticus. International Journal of Integrative Biology, 2013; 14(2): 104-112.
- Syihab SF, Madayanti F, Akhmaloka A, Widhiastuty MP. Purification and characterization of thermostable and alcohol tolerant lipase from Pseudoxanthomonas sp. African Journal of Biotechnology, 2017; 16(31): 1670-1677.
Crossref - Smith B.J. SDS Polyacrylamide Gel Electrophoresis of Proteins. In: Walker J.M. (eds) Proteins. Methods in Molecular Biology™, 1984; 1. Humana Press.
- Soliman, N., Knoll, M., Yasser, R., Fattah, A., Scmid, R.D., Lange, S.: Molecular cloning and characterization of thermostable esterase and lipase from Geobacillus thermoleovoran YN isolated from desert soil in Egypt, Process Biochemistry, 2007; 42: 1090-1100.
Crossref - Lee, D., Koh, Y., Kim, K., Kim, B., Choi, H., Kim, D., Suhartono, M. T., dan Pyun, Y.: Isolation and characterization of a thermophilic lipase from Bacillus thermoleovorans ID-1, FEMS Microbiology Letters, 1999; 179: 393-400.
Crossref - Abdelkafi S, Fouquet B, Barouh N, Durner S, Pina M, Scheirlinckx F, Villeneuve P, Carriere F. In vitro comparisons between Carica papaya and pancreatic lipases during test meal lipolysis: potential use of CPL in enzyme replacement therapy. Food Chem., 2009; 115: 488-494.
Crossref
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.