An acute respiratory disease (SARS-CoV-2, also recognized as COVID-19/2019-nCoV), caused by nCoV created a worldwide emergency. The World Health Organization declared the SARS-CoV-2 as epidemic of international concern on January 2020. After SARS-CoV in 2002 and MERS-CoV in 2012, the emergence of SARS-CoV-2 is marked as third highly pathogenic coronavirus of 21st century. Till now, various researches have been conducted, highlighting SARS-CoV-2 as β-coronavirus with high phylogenetic and genomic similarity with bat-CoV, indicating bats as natural reservoir of coronaviruses. It has also been confirmed that SARS-CoV-2 uses the same (ACE2) receptor for host cellular entry as of SARS-CoV, and primarily spread through respiratory pathway. Evidences shows continuous human-to-human viral transfer, with numerous worldwide exported cases. Currently, there is no specific approved drug available for the treatment of SARS-CoV-2, but various anti-parasitic and anti-viral drugs are being investigated. In this review, we have described several possible therapeutic modalities for SARS-CoV-2 infection based on (i) host protease inhibitors to block viral entry into the cell; (ii) gene silencing using siRNA-based RNAi and (iii) type I interferons (IFN1)-based therapeutics have been discussed in detail. Background knowledge on these strategies highlight them as potential therapeutic targets, which could be evaluated on urgent basis to combat COVID-19 epidemic.
SARS-CoV-2, COVID-19, Origin, Therapeutics, protease inhibitors, gene silencing (siRNA), interferons type I (IFN-1)
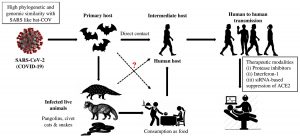
SARS-CoV-2 is an enveloped, positive-sense RNA virus with 120–160 nm in diameter, belongs to family Coronaviridae and genus Betacoronavirus. High genetic similarity between SARS-CoV and a bat-CoV (sub-genus Sarbecovirus) has been confirmed by the scientists1. They named as corona due to presence of prominent spikes or projections like petal in envelop. It is also their differential feature distinguished them from Orthomyxoviruses2. Among four-CoV genera (α, β, γ, δ), the α and β viruses are capable to infect animals, while δ and γ are found to infect avian population. Formerly, six coronavirus strains have been recognized as capable to infect humans. From which, four α-CoV spp. showed mild pathogenicity and respiratory symptoms while two β-CoV spp. (SARS-CoV and MERS-CoV) caused fatal respiratory infections3. Genome sequence analysis of SARS-CoV-2 was found to be highly identical (96.2%) with bat-CoV (RaTG13), whereas, the similarity was comparatively less (79.5%) with SARS-CoV. Based on evolutionary and viral genome sequence analyses, bats have been considered as primary carriers of SARS-CoV-2 and the infection might be transferred via intermediate animals (secondary hosts) to reach human population (Fig.1). It is found that ACE2 receptor is necessary for SARS-CoV-2 to infect humans, same as SARS-CoV4.
Fig.1. Source of infection and transmission of SARS-related coronaviruses to humans via intermediate hosts.
Transmission of SARS-CoV-2
The epidemic of SARS-CoV-2 was initially break out in China at the end of 2019, possibly due to seafood market. Various researches indicated that bats could be the natural reservoirs of SARS-related coronaviruses2,5 however, this fact still lacks potential evidences. Rather, bats serve as reservoir for variety of different CoVs including SARS-CoV and MERS-CoV6. Genome sequencing analysis showed that SARS-CoV-2 was 96.2% similar to bat-CoV5, which indicate that SARS-CoV-2 and bat-CoV might belong to the same ancestral specie, though bats are not sold in that marketplace7. Phylogenetic analysis and protein sequence alignment8 also showed different species (e.g. pangolin, civet cats, snakes and turtles) present similar residues of receptors, which put them also in the suspect category. Disease (COVID-19) transmission between human-to-human primarily occurs between friends and family who have close contact with infected persons or virus carrier person. Direct contact with infected animals or their utilization as foodstuff was assumed to be the major transmission pathway of SARS-CoV-2 infection. However, the transmission route(s) and the source(s) of infection of COVID-19 remains elusive (Fig.1).
Therapeutic modalities for SARS-CoV-2 infection
Protease inhibitors to block viral entry into host cells
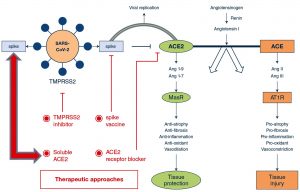
An attractive therapeutic strategy for SARS-CoV-2 infection, could be the blockage of viral entry into the cell by selectively targeting host factors (Fig.2). The viral spike (S)-glycoproteins are produced as inactive precursors which are converted into active form by host cell proteases (proteolytic cleavage – priming). The S-glycoprotein of SARS-CoV-2 attaches the angiotensin-converting enzyme 2 (ACE2), which allow the entry of coronavirus into the host cell9. Viral entrance also requires the cleavage of S-glycoproteins by host proteases, which consequences in a non-reversible conformational modification to S-glycoprotein, which allow the endocytosis of viral particle by the host cell9. Cleavage of S-glycoprotein (priming), can use host proteases i.e., cysteine proteases [cathepsin B/L (CatB/L)] and/or serine proteases [transmembrane protease (TMPRSS2)]10,11. Ziegler and coworkers described single-cell RNA-sequencing of non-human and human tissues and discovered the co-expression of three major cell types i.e., (i) enterocytes in terminal ileum; (ii) pneumocytes – type II, in lungs and (iii) goblet mucus-forming cells in nasal cavity12. A recent preprint also highlighted several other proteins which interact with 26/29 COVID-19 proteins13. Therefore, the respective proteolytic enzymes could be the potential targets for the development of broad-spectrum anti-viral therapeutics (Fig.2).
Fig. 2. Potential therapeutic targets to address ACE2-dependent SARS-CoV-2 infection. Both coronaviruses (SARS-CoV & SARS-CoV-2) use ACE2 for their transmission pathogenesis into the host cell. SARS-associate coronaviruses induce down-regulation of the ACE2 by selective binding of S-glycoprotein with ACE2 rather than ACE receptor. This causes viral transmission into host cell and its replication, as well as severe pulmonary infection. Therapeutics may comprise (i) S-glycoprotein-based vaccines; (ii) TMPRSS2 inhibitor to block the S-protein priming; (iii) ACE2 receptor blockage giving anti-ACE2 peptides or antibodies or (iv) slowdown viral entry by soluble form of ACE2, which competitively bind with SARS-CoV-2 and reduce viral spreading ultimately preventing pulmonary infection using exclusive enzyme function. Adapted from Zhang et al.4. Article distributed under creative common attribution (CC-BY) license.
The development of inhibitors for Cat-B, Cat-L, and other relevant proteases would be an amazing starting point towards the designing of broad-spectrum anti-viral therapeutics11,14. The designing of S-glycoprotein-based vaccines for SARS-CoV-2 may depend on the fact that the viral entry depends on ACE2 receptor binding. Cell-lines that enable viral multiplication in the existence of ACE2 may serve as the most effective in the development of large-scale vaccine4. The attachment sites between SARS-CoV and ACE2 has been recognized at atomic level. Hence, therapeutic approaches could also target this attachment site with smaller molecules or antibodies4. Recently, Hoffmann et al.11, described the priming of viral S-proteins by TMPRSS2, which is essential for viral entry into human cell types, and the spread of SARS-CoV-2 through binding with specific receptor (ACE2). The protease inhibitors such as ‘camostat mesylate’ has been studied to block TMPRSS2 activity, which has been approved in Japan14, which can be an interesting candidate for therapeutic purpose of SARS-CoV-2 infection.
It has been demonstrated that SARS-CoV down-regulates ACE2, by specifically binding with viral S-glycoprotein, contributing to severe pulmonary infection15. Following this, it could be suggested that excessive ACE2 may competitively conjugate with SARS-CoV-2 to counterbalance the viral particles, as well as to rescue ACE2 activity, which negatively regulates the RAS activity, and protect from pulmonary infection16. Indeed, decreased ACE2 availability and increased ACE activity contribute to pulmonary infection16,17. Treatment using soluble type of ACE2 may exert dual functionalities; (i) slowdown viral spreading into host cells18 and (ii) protecting from pulmonary infection17,19.
Suppression of ACE2 using siRNA
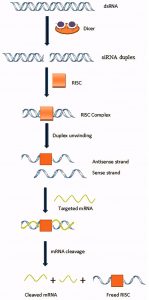
RNA-dependent gene silencing comprises post-transcriptional gene silencing in plants and RNA interference (RNAi) in mammalian/animal cells. Transfection of siRNA into animal cells causes degradation of target mRNA and consequently suppression of its expression20. siRNA-based RNAi may provide a new therapeutic approach for viral diseases (Fig.3). RNAi has been used against HCV and HIV to suppress the replication of viruses in cultured human cells21-24. SARS-CoV-2 is a positive-sense, enveloped RNA virus25, which predominantly requires the attachment of its S-glycoproteins with ACE2 receptor for its entry into host cells as described previously. That S-glycoprotein is almost identical in all SARS-associated coronaviruses, though bit differences in SARS-CoV-2 as compared with other related coronaviruses. Moreover, according to Diao et al.26, SARS-CoV-2 manage to clearly decrease the population of immune CD8+ and CD4+ cells causing enhanced viral spread inside the host’s body. According to Wang et al.27, SARS-CoV-2 also attaches with CD147 receptor.
Fig.3 Mechanism of gene silencing using siRNA-based RNAi. Adapted from Alam et al., [31]. Article distributed under creative common attribution (CC-BY) license.
Organs with high expression of ACE2 are highly affected by the SARS-CoV-2, as it binds with the receptor for its entry. Following the fact, reducing the availability of those receptors should reduce viral capability to enter into the cell, ultimately reducing the SARS-CoV-2 infection. Blood pressure (BP) regulating hormone (angiotensin II), which is a part of renin-angiotensin system (RAS) is mediated by ACE2 receptor28. Therefore, observed distribution of SARS-CoV-2 indicate a pattern of BP variability among population29. Also, diabetes, hypertension, viral pneumonia and cardiovascular disorders are the common comorbidities increasing the organism’s susceptibility to SARS-CoV-2, which are primarily related to BP regulatory mechanism of the body14. Hence, the medications used for the treatment of hypertension are being studied for their effectiveness in the treatment of SARS-CoV-2. However, their side effects and metabolic pathways limit their utilization as anti-hypertensive medications30.
As described previously, viral S-glycoprotein specifically attaches with ACE2 receptor. It has been shown that S-protein of SARS-CoV-2 is highly similar to that of SARS-CoV32, which caused epidemic in 2002/2003. Researches using mutagenic deletion of ACE2 suggested that the cytoplasmic tail of receptor is essential for shedding. Thus, siRNA-based suppression of ACE2 impaired viral capability to infect host cells33. Alam et al.31, reported various successful experiments using nanocarriers-mediated delivery of siRNA as a therapeutic strategy for hypertension. Tambe et al.34, described the gene silencing using surface engineered dendrimers with siRNA loading. The presented therapeutic techniques indicate the effectiveness of siRNA as a potential tool to reshape RAS-system. Following that, Merril35, proposed therapy based on temporarily silencing ACE2 expression by designing specific siRNA molecules, this would effectively block the viral entry by modulating immune system to efficiently prevent from viral infection.
There could be two possible carriers able for targeted delivery such as liposomes (stable nucleic acid lipid particles – SNALP) and the virus itself. It has been described that SNALP can efficiently deliver siRNA36. The use of inactivated virus as carrier of siRNA could be effective in targeted delivery to cells which are susceptible by live viral infection. The use of S-glycoproteins and viral coat that could deliver siRNA at targeted place could assist two purposes i.e., (i) targeted delivery at places which are more susceptible to SARS-CoV-2 infection and (ii) blocking the attachment sites for live viruses, ultimately reducing the disease severity. Hence, the use of viral envelope would efficiently perform targeted delivery of siRNA.
Interferons type 1 (IFN1)-based therapeutics
IFN-1 designate a group of cytokines including various subtypes such as α and β (further subdivided into various isoforms), ω, κ and ε-subtypes37. These are produced by numerous cells in response to a viral infection. However, upon viral component identification by pattern recognition receptor (PRR), plasmacytoid dendritic cells (pDCs) majorly contribute in the production of IFN-1 in the body38. Hence, they initiate the first responsive mechanism as a result of viral infection. IFN-1 are recognized by interferon-a/β receptor (IFNAR) existent at the cell membrane of various cells. Their conjugation on IFNAR causes the addition of phosphate group to the transcription factors (phosphorylation) e.g., STAT1 and its relocalization, ultimately switching interferon-stimulating genes (ISG), which are mostly involved in immunomodulation, signaling and inflammation39. They inhibit viral multiplication and initiate various responsive mechanisms e.g., reduction in cellular metabolic activity or discharge of cytokines which ultimately activates the host’s adaptive immunity. ISGs comprise PRRs, which enhance their sensitivity towards pathogens, preventing membrane fusion or viral egress, proteins which reduce membrane fluidity, and antivirals which particularly inhibit single stage of viral replication40,41. IFN-1, due to their immunomodulatory characteristics, has been used in the treatment of various disorders e.g., subcutaneous injection of IFNβ is being successfully used against multiple sclerosis42.
SARS-CoV and MERS-CoV exhibit closely related characteristics to COVID-19 coronaviruses, despite variances in their pathology, several proteins, and their epidemiology43. Treatment via IFN-1 against MERS-CoV and SARS-CoV has already been described by various in vivo and in vitro studies44, with and/or without lopinavir/ritonavir as a combinatorial medication45,46. Several other combinatorial drugs have been reported against coronavirus infection such as ribavirin47-49, chloroquine and remdesivir43,50. IFNα and β showed good therapeutic potential in various experimental animals 51 but no significant results were obtained in humans44. For instance, Sheahan et al.45, studied combinatorial effect of IFNβ with ritonavir/lopinavir against 2012 respiratory syndrome (MERS-CoV). The results indicated significant improvement in lung functionality. However, the effect was non-significant against viral replication or lung pathology severity. In a previous study, combination of ribavirin with IFNa2b showed amazing outcomes in monkeys52, while unconvincing in humans53. The insignificant results in various studies using IFN-1 therapy can be explained by several factors, i.e., (i) the inhibitory mechanism of IFN-signaling used by SARS-CoV and MERS-CoV; (ii) trouble to interpret agents behind the improvements (viz. IFN or combinatorial drug) and (iii) The limited number of study subjects. Moreover, results may also vary from study to study because of discrepancies in clinical conditions and experimental settings44. It has also been hypothesized that IFN-1 were effective in the population only if they lacked any co-morbid disorders47.
Following previous clinical investigations, IFNβ1a and IFNβ1b were the most potent types of interferons against MERS-CoV54, and SARS-CoV55. In addition, IFNβ-γ synergistically inhibited the replication of coronaviruses56. Cinatl et al.57, studied the effect of IFNs (both a and β) on SARS-CoV persuaded cytotoxicity in Vero cells while the drugs were added after 1 hour of adsorption period. IFNβ was active against SARS-CoV (FFM-1 strain), whereas, combination of IFNa-β was not. Therefore, IFNβ was the only agent used for the inhibition of coronavirus replication. Vero cells were infected with coronavirus (FFM-1) and treated with various concentrations of IFNs, and viral replication was studied using peroxidase staining with immune serum. In vivo clinical experiments suggest that the time of IFN-1 administration plays an important part. The reduce the time of IFN-1 administration after viral infection, the maximum the chances of the cure are and vice versa. In addition, later administration may inhibit viral replication cause severe complications58.
Following previous knowledge, the utilization of IFN-1 as a therapeutic tool against SARS-CoV-2 infection demands high scientific attention. MERS-CoV and SARS-CoV are capable to interrupt the IFN-signaling mechanism. For instance, karyopherin transportation is disrupted by Orf6 protein of SARS-CoV, and hence, inhibiting the transport of transcriptional factors in the nucleus, ultimately triggering IFN’s responsive mechanism. Likewise, the phosphorylation of IRF3 is inhibited by Orf3b protein of SARS-CoV59 which causes switching on the process of IFN expression. However, Orf3b and Orf6 of novel coronavirus-2 are docked and may have lost their anti-IFN functionalities. This could address the fact that why novel SARS-CoV-2 showed high in vitro sensitivity to IFNa60. Though the replication was not completely stopped by interferon treatment, several folds decrease in viral titers was definitely a significant finding. SARS-CoV-2 showed much higher sensitivity to IFN-1, which indicates that IFN-1 based therapeutics should be as efficient for the previous than for the latter.
Following previous knowledge, it can be concluded that IFN-1 treatment might be safer and very effective against SARS-CoV-2 infection. The available data on SARS-CoV and MERS-CoV would be potential assets in this regard. For instance, studies suggest that IFN-1 should be administered at earlier stages of infection and IFNβ should be the most effective subtype to avoid complications58. Moreover, SARS-CoV-2 pathology, primarily consist in lung injury, exhibits closer properties to interferonopathies. This may indicate that COVID-19 causes excessive interferon-medicate response, ultimately resulting in tissue damage. Treatment via IFN-1 should be given at early stages of infection61, and inflammatory biomarkers in infected patients are linked with increased mortality5. At latter stages of infection, it is probable that anti-interferon medications should be prescribed to normalize the adverse events17.
The SARS-CoV-2 outbreak, which was started from city level has become a matter of global concern. Scientists are working to characterize the novel coronavirus strain (CoVID-19) and to explore potential therapeutic targets to overcome the epidemic. In this review, we have summarized the available knowledge related to the source of infection and its human-to-human transmission with possible therapeutic modalities i.e., host protease inhibition, ACE2 suppression using siRNA based RNAi and the IFN-1-based therapeutic, which presented excellent therapeutic potential in 2002 SARS-CoV and 2012 MERS-CoV infection. As, the SARS-CoV-2 shares the same ancestral organism, which high phylogenetic and genomic similarity to other coronaviruses these therapeutic modalities might prove to be the efficient targets to overcome COVID-19 pandemic. However, there is a need of more urgent researches to explore the in-depth knowledge of this novel virus to make clear evolutionary path and stop this fatal disease. Moreover, to explore molecular mechanisms of viral multiplication and host cellular entry, which provide the basics of future studies to develop targeted vaccines and drugs.
ACKNOWLEDGMENTS
The authors are grateful to their representative institute for providing literature facilities.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270-273.
Crossref - Tang X, Wu C, Li X, et al. On the origin and continuing evolution of SARS-CoV-2. Nat Sci Rev. 2020;7(6):1012-1023.
Crossref - Yin Y, Wunderink RG. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2018;23(2):130-137.
Crossref - Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46:1-5.
Crossref - Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):P1054-1062.
Crossref - Shi Z, Hu Z. A review of studies on animal reservoirs of the SARS coronavirus. Virus Res. 2008;133(1):74-87.
Crossref - Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265-269.
Crossref - Liu Z, Xiao X, Wei X, et al. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J Med Virol. 2020;92(6):595-601.
Crossref - Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444-1448.
Crossref - Simmons G, Zmora P, Gierer S, Heurich A, Pohlmann S. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antivir Res. 2013;100(3):605-614.
Crossref - Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Muller MA. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271-280.
Crossref - Ziegler C, Allon SJ, Nyquist SK, et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is enriched in specific cell subsets across tissues. Cell. 2020;181(5):1016-1035.
Crossref - Gordon DE, Jang GM, Bouhaddou M, et al. A SARS-CoV-2-human protein-protein interaction map reveals drug targets and potential drug-repurposing. BioRxiv. 2020.
- Zhou Y, Vedantham P, Lu K, et al. Protease inhibitors targeting coronavirus and filovirus entry. Antivir Res. 2015;116:76-84.
Crossref - Kuba K, Imai Y, Rao S, et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005;11(8):875-879.
Crossref - Yu L, Yuan K, Phuong HTA, Park BM, Kim SH. Angiotensin-(1-5), an active mediator of renin-angiotensin system, stimulates ANP secretion via Mas receptor. Peptides. 2016;86:33-41.
Crossref - Zhang R, Pan Y, Fanelli V, eta l. Mechanical stress and the induction of lung fibrosis via the midkine signaling pathway. Am J Respir Crit Care Med. 2015;192(3):315-323.
Crossref - Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864-1868.
Crossref - Wosten-van Asperen RM, Lutter R, Specht PA, et al. Acute respiratory distress syndrome leads to reduced ratio of ACE/ACE2 activities and is prevented by angiotensin-(1-7) or an angiotensin II receptor antagonist. J Pathol. 2011;225(4):618-627.
Crossref - Hannon GJ. RNA interference. Nature. 2002;418(6894):244-251.
Crossref - Wilson JA, Jayasena S, Khvorova A, et al. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc Nat Acad Sci. 2003;100(5):2783-2788.
Crossref - Leung RK, Whittaker PA. RNA interference: from gene silencing to gene-specific therapeutics. Pharmacol Therapeut. 2005;107(2):222-239.
Crossref - Ter Brake O, Konstantinova P, Ceylan M, Berkhout B. Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol Ther. 2006;14(6):883-892.
Crossref - Scarborough RJ, Gatignol A. RNA interference therapies for an HIV-1 functional cure. Viruses. 2018;10(1):8.
Crossref - Chen D, Xu W, Lei Z, et al. Recurrence of positive SARS-CoV-2 RNA in COVID-19: A case report. Int J Infect Dis. 2020.
Crossref - Diao B, Wang C, Tan Y, et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19). Medrxiv. 2020.
Crossref - Wang K, Chen W, Zhou YS, et al. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. BioRxiv. 2020.
Crossref - Xiang Z, Bendt K, Griffiths R, Giani JF, Ortiz Melo DI, Gurley SB. Renal Proximal Tubule-derived Angiotensin Converting Enzyme 2 (ACE2) in Blood Pressure Regulation. Hypertension. 2017;70(suppl_1):A010-A010.
- Young BE, Ong SWX, Kalimuddin S, et al. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. Jama. 2020.
Crossref - Tignanelli CJ, Ingraham NE, Sparks MA, et al. Antihypertensive drugs and risk of COVID-19?. Lancet Resp Med. 2020.
Crossref - Alam T, Khan S, Gaba B, Haider MF, Baboota S, Ali J. Nanocarriers as treatment modalities for hypertension. Drug Deliv. 2017;24(1):358-369.
Crossref - Ou X, Liu Y, Lei X, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11(1):1-12.
Crossref - Wu S, Miao L, Zhou Q, et al. Suppression of Androgen Receptor (AR)-ACE2/TMPRSS2 Axis by AR Antagonists May Be Therapeutically Beneficial for Male COVID Patients. SSRN. 2020.
Crossref - Tambe V, Thakkar S, Raval N, Sharma D, Kalia K, Tekade RK. Surface engineered dendrimers in siRNA delivery and gene silencing. Curr Pharm Des. 2017;23(20):2952-2975.
Crossref - Merril J. Proposal for supportive/preventive treatment of COVID19 using siRNA to suppress expression of ACE2 receptors and vaccines based phage-display technology developed by epitopeRX. 2020. Available at: https://bitlattice.org
- Nolte A, Schneider M, Walker T, Wendel HP. Gene-silencing for treatment of cardiovascular diseases. In Regenerative Medicine and Tissue Engineering-Cells and Biomaterials. Intech Open. 2011.
Crossref - Samuel CE. Antiviral actions of interferons. Clin Microbiol. 2001;14(4):778-809.
Crossref - Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7(2):131-137.
Crossref - Wang W, Xu L, Su J, Peppelenbosch MP, Pan Q. Transcriptional regulation of antiviral interferon-stimulated genes. TRENDS Microbiol. 2017;25(7):573-584.
Crossref - Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513-545.
Crossref - Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14(1):36-49.
Crossref - Patti F, Amato MP, Bastianello S, et al. Effects of immunomodulatory treatment with subcutaneous interferon beta-1a oncognitive decline in mildly disabled patients with relapsing-remitting multiple sclerosis. Mult Scler J. 2010;16(1):68-77.
Crossref - Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;105924.
Crossref - Stockman LJ, Bellamy R, Garner P. SARS: systematic review of treatment effects. PLoS Med. 2006;3(9).
Crossref - Sheahan TP, Sims AC, Leist SR, et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat Commun. 2020;11(1):1-14.
Crossref - Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). 2020.
Crossref - Al-Tawfiq JA, Momattin H, Dib J, Memish ZA. Ribavirin and interferon therapy in patients infected with the Middle East respiratory syndrome coronavirus: an observational study. Int J Infect Dis. 2014;20:42-46.
Crossref - Omrani AS, Saad MM, Baig K, et al. Ribavirin and interferon alfa-2a for severe Middle East respiratory syndrome coronavirus infection: a retrospective cohort study. Lancet Infect Dis. 2014;14(11):1090-1095.
Crossref - Kim UJ, Won EJ, Kee SJ, Jung SI, Jang HC. Case report Combination therapy with lopinavir/ritonavir, ribavirin and interferon-α for Middle East respiratory syndrome. Antiviral therapy. 2016;21:455-459.
Crossref - Wang M, Cao R, Zhang L, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269-271.
Crossref - Chan JFW, Yao Y, Yeung ML, et al. Treatment with lopinavir/ritonavir or interferon-β1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J Infect Dis. 2015;212(12):1904-1913.
Crossref - Falzarano D, De Wit E, Rasmussen AL, et al. Treatment with interferon-α2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat Med. 2013;19(10):1313-1317.
Crossref - Arabi YM, Shalhoub S, Mandourah Y, et al. Ribavirin and interferon therapy for critically ill patients with middle east respiratory syndrome: a multicenter observational study. Clin Infect Dis. 2020;70(9):1837-1844.
Crossref - Shalhoub S, Farahat F, Al-Jiffri A, et al. IFN-a2a or IFN-β1a in combination with ribavirin to treat Middle East respiratory syndrome coronavirus pneumonia: a retrospective study. J Antimicrob Chemother. 2015;70(7):2129-2132.
Crossref - Hensley LE, Fritz EA, Jahrling PB, Karp C, Huggins JW, Geisbert TW. Interferon-β 1a and SARS coronavirus replication. Emerging Infect Dis. 2004;10(2):317.
Crossref - Sainz Jr B, Mossel EC, Peters CJ, Garry RF. Interferon-beta and interferon-gamma synergistically inhibit the replication of severe acute respiratory syndrome-associated coronavirus (SARS-CoV). Virology. 2004;329(1):11-17.
Crossref - Cinatl J, Morgenstern B, Bauer G, Chandra P, Rabenau H, Doerr HW. Treatment of SARS with human interferons. Lancet. 2003;362(9380):293-294.
Crossref - Channappanavar R, Fehr AR, Zheng J, et al. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J Clin Invest. 2019;129(9). Crossref
- Kopecky-Bromberg SA, Martinez-Sobrido L, Frieman M, Baric RA, Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. Virology. 2007;81(2):548-557.
Crossref - Lokugamage KG, Hage A, Schindewolf C, Rajsbaum R, Menachery VD. SARS-CoV-2 sensitive to type I interferon pretreatment. BioRxiv. 2020.
- Siddiqi HK, Mehra MR. COVID-19 illness in native and immunosuppressed states: a clinical-therapeutic staging proposal. J Heart Lung Transplant. 2020.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.