Since the first detection of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the virus remains a public health concern. Several public health measures have been implemented in an effort to curb the infections. However, the effectiveness of these strategies was threatened with the emergence of numerous SARS-CoV-2 variants in all parts of the globe, due to the persistent mutations as part of the viral evolution. Mutations that usually occur in its spike glycoprotein, allow SARS-CoV-2 to possess advantageous characteristics for its survivability and persistence. This has led to poor performance of diagnostic kits which have caused non-specific and insensitive detection of these variants, resulting in undetermined infection. The variants also have caused the increased severity of COVID-19, involving hospitalisation rates, ICU admissions, and deaths. Many have reported the vaccine-breakthrough infections and reduced effectiveness of vaccination, which is supposed to provide an effective degree of protection against COVID-19 infections. Due to these issues, this review summarises the impacts related to SARS-CoV-2 variants emergence towards the performance of diagnostic kits, transmissibility of the virus, severity of disease, and effectiveness of COVID-19 vaccines.
SARS-CoV-2, Variants, Impacts, Detection, Vaccines
The detection and transmission of a novel coronavirus, SARS-CoV-2 responsible for the COVID pandemic came into existence at the end of 2019 in Wuhan City, China following which the World Health Organization (WHO) declared it as a global pandemic on March 11, 2020. The most common clinical presentations for COVID-19 include fever, fatigue, cough, anosmia, and myalgia, with some reports recently on gastrointestinal symptoms such as anorexia, nausea, vomiting, and diarrhoea.1 As of March 4, 2022, there have been more than 440 million reported cases accounting for more than 5.9 million fatalities worldwide.2
SARS-CoV-2 which is classified under genera Betacoronavirus of the Coronavirus family is a positive-sense single-stranded RNA virus.3 Morphologically, SARS-CoV-2 has four structural proteins, which are spike (S), membrane (M), envelope (E), and nucleocapsid (N), with the S protein being one of the most vital since it is directly involved in the first step of SARS-CoV-2 pathogenesis. S glycoprotein of SARS-CoV-2 recognizes and binds to the specific angiotensin-converting enzyme 2 (ACE2) and mediates the fusion of viral and host cell membrane. S protein is divided into two main subunits which are S1 and S2. The S1 subunit of the viral spike proteins will initiate the binding towards ACE2 receptors present on the host cells.4 Fusion of viral and host cellular membrane, leading to the entry of virus into the susceptible cells is primed by S2 subunit of spike proteins.5 Genetic materials of the virus will be released into the infected cell post-fusion, resulting in the replication of the viral RNA.6
Translation of viral RNA will lead to the assembly and release of the virions from the infected cells, which is facilitated by another three structural proteins, namely membrane, nucleocapsid, and envelope proteins.7 As the replication of SARS-CoV-2 continues, patients will transmit the virus to others and start to develop symptoms and illness, which can be further classified into several phases – asymptomatic, mild, moderate, severe and critical.8
There are three main approaches in the detection of SARS-CoV-2, which include the molecular, antigen, and serology tests. Molecular tests focus on the detection of specific viral genomes from the collected nasopharyngeal sample of suspected infected individuals. Reverse transcriptase-polymerase chain reaction (RT-PCR), clustered regularly interspaced short palindromic repeats (CRISPR), microarray assays, whole genome sequencing (WGS) and loop-mediated isothermal amplification (LAMP) are the instances of the techniques used for molecular testing.9 Another option for SARS-CoV-2 detection is antigen-based detection, which requires the application of lateral flow immunoassays, such as the rapid antigen detection (RAD) or rapid diagnostic tests (RDT). RDTs are known for being less sensitive and specific compared to RT-PCR, due to the varying types of samples which can be used and possible errors since it is operator-dependent.10 However, the SARS-CoV-2 RDT are also used globally as one of the major detection tools, as it offers an easier, faster, and cost-effective approach. This method requires neither laboratory skills nor specific facilities. Application of antibody-antigen reaction in serological assays is also beneficial in the determination of antibody response towards the infection.11
Due to the importance of spike glycoproteins towards SARS-CoV-2 pathogenesis, it has been widely applied as the main component for the potential vaccine candidates.12 Readily available COVID-19 vaccines, such as BNT162b2 are proven to be effective in the prevention of severe disease, hospitalisation, symptomatic COVID-19, and even deaths, with higher efficacy in a person who has completed two doses of vaccination.13 Two weeks post complete vaccination is required for the body to produce protection against the virus. The infections with SARS-CoV-2 can still occurs between the time of vaccination and the development of a sufficient level of protection against the virus.14
Regular mutations are an essential part of the viral evolution that allow SARS-CoV-2 variants to emerge, which either persist or disappear randomly. The mutation causes genetic variations, resulting in neutral, detrimental, or beneficial impacts for the virus. Most of the mutations lead to neutral or mildly deleterious effects, and only a small proportion of them results in beneficial properties for the virus.15 However, the rare mutations promoting fitness advantages for the SARS-CoV-2 can still occur, which is further enhanced with the encounter of varying hosts with highly diversified genetic backgrounds.16,17 The most beneficial mutation for the survival of an organism, if present, will then be the most dominant genome portfolio within a population.18
Variants are detected through numerous gene sequences deposited to the database, namely GISAID, and publications describing the phenotypic variations. These lead to the classification of SARS-CoV-2 variants into four major groups, by WHO. Some variants may result in concerning impacts, involving the increase in transmissibility of the virus, poor performance of diagnostic kits, increase in severity of COVID-19 as well as reduction of vaccines effectiveness.
Emergence of Variants
Variants refer to the virus with one or more gene mutations determined through whole-genome sequencing (WGS), compared to the wild type (Wuhan isolate) or the first isolated virus.18,19 RNA viruses tend to possess higher mutation rates than DNA viruses, due to the absence of the RNA polymerase proofreading capability. SARS-CoV-2 on the contrary possesses the specific gene referred to as ExoN or the non-structural protein 14 (nsp14) which allows the virus to lower the rate of mutation compared to other RNA viruses. The nsp14 allows correction to be done on the non-specific nucleotides incorporated during viral RNA synthesis.20,21
It is estimated that the nucleotide mutation rates of the whole genome and spike glycoprotein of SARS-CoV-2 are at 6.677 X 10-4 and 8.066 X 10-4 per nucleotide each year, respectively.22 The higher mutation rate of the spike compared to the other SARS-CoV-2 proteins results in the modification of the viral immunogenicity and pathogenicity. Mutations may also arise in the immunodominant receptor-binding domain (RBD) of SARS-CoV-2 spike protein, which subsequently renders the virus to develop the ability of host immune evasion.17 This has been described by Singh et al.23 where they observed resistance against the neutralisation governed by neutralising antibodies (NAbs), related to the mutations occurring in NTD. The reported amino acid substitutions of SARS-CoV-2 spikes in the simplified form consist of three main parts which are (1) the original amino acid, (2) the position of the amino acid and (3) the substituting amino acid. For example, the substitution of aspartic acid by glycine at position 614 of the spike protein, will be reported as D614G. The emergence of variants, especially the VOC has been proven to cause vaccine breakthrough infection apart from the reduction of vaccine efficacies. Vaccine breakthrough infections refer to the positive testing of either SARS-CoV-2 RNA or antigen from persons, indicating active COVID-19 infection who have completed the vaccination doses.
SARS-CoV-2 variants with D614G were first detected in February 2020, thus becoming one of the earliest reported variants and has now become common to nearly all sequenced SARS-CoV-2 genomes worldwide. The variants dominated the circulating SARS-CoV-2 variants, by June 2020, four months following its first isolation.24 Since then, many new variants have been reported globally, where all of these SARS-CoV-2 variants also possess the D614G mutation, which may indicate all these variants emerged from the ancestral D614G variant.25 The mutation involves the substitution of aspartic acid (D) to glycine (G) at amino acid 614 of the SARS-CoV-2 spike glycoprotein, resulting in increased transmissibility of the variant than the wild-type.
The variants are also classified into several classifications or groups by WHO and other health governing organisations, based on their characteristics and implications towards the public health. The classification involved variants of interest (VOI), variants of concern (VOC), variants of high consequences (VOHC) and variants under monitoring (VUM) or variants being monitored (VBM) (Table 1). Some of the major SARS-CoV-2 variants include B.1.1.7 (Alpha), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2) and Omicron (B.1.1.529). The spike mutations detected in these variants are responsible for reduced accuracy of diagnostic tools, increased transmissibility of the virus and higher risk of disease severity, compared to the wild-type.
Table (1):
List of major SARS-CoV-2 variants with their respective classifications by WHO.26,28-34.
| WHO Label | PANGO lineage | First Detection | Total Spike Mutations | Date of Designation / Reclassification | Classifications by CDC |
|---|---|---|---|---|---|
| Variants of Interest | |||||
| Lambda28 | C.37 | Peru (December 2020) | 13 | June 14, 2021 | N/A |
| Mu29 | B.1.621 | Colombia (January 2021) | 19 | August 30, 2021 | VBM |
| Variants of Concern | |||||
| Alpha | B.1.1.7 | United Kingdom (September 2020) | 10 | December 18, 2020 | VBM |
| Beta | B.1.351 | South Africa (September 2020) | 12 | December 18, 2020 | VBM |
| Gamma | P.1 | Brazil (December 2020) | 11 | January 11, 2021 | VBM |
| Delta | B.1.617.2 | India (December 2020) | 15 | May 11, 2021 | VOC |
| Omicron | B.1.1.529 | Multiple Countries (November 2021) | 37 | November 26, 2021 | VOC |
| Variants Under Monitoring (VUM) / Variants Being Monitored (VBM) / Formerly Monitored Variants | |||||
| None 30 | B.1.1.31 | Multiple Countries (January 2021) | 14 | June 2, 2021 | N/A |
| None 31 | C.1.2 | South Africa (May 2021) | 15 | September 1, 2021 | N/A |
| None 32 | B.1.640 | Multiple Countries (September 2021) | 23 | November 22, 2021 | N/A |
| Formerly monitored variants | |||||
| Epsilon 33 | B.1.427
B.1.429 |
USA (March 2020) | 6 | November 9, 2021 | VBM |
| Eta 33 | B.1.525 | Multiple Countries (December 2020) | 8 | December 22, 2021 | VBM |
| Iota 33 | B.1.526 | USA (November 2020) | 14 | December 22, 2021 | VBM |
| Kappa 33 | B.1.617.1 | India (October 2020) | 8 | December 29, 2021 | VBM |
| Zeta 33 | P.2 | Brazil (April 2020) | 4 | August 17, 2021 | VBM |
| Theta 34 | P.3 | Philippines (January 2021) | 10 | August 17, 2021 | N/A |
Variants of Interest (VOI)
Variants of interest (VOI) refers to the variants possessing specific mutation or amino acid changes in comparison to the reference (Wuhan) isolate. This encompasses the alteration in its antigenicity, virulence, and epidemiology which may lead to the possibility of negative consequences in the performance of diagnostic kits treatments and public health measures.26 The variants are also reported to be responsible for widespread transmission within a community leading to the formation of several COVID-19 clusters. There is also a possibility of these variants resulting in significant impacts on public health measures.
Variants of Concern (VOC)
Variants of concern (VOC) refer to the variants with substantial evidence of their notable impacts on the viral transmissibility and severity of the disease. There is also evidence on the reduction of neutralising capabilities of antibodies, lower efficacy and effectiveness of readily administered vaccines and results in the potential of compromising the detection of SARS-CoV-2 results. WHO has listed five VOCs, which are different from the list classified by CDC, since they are no longer responsible for the epidemiological conditions in the United States of America (USA).27
Variants of High Consequences (VOHC)
Variants of high consequences (VOHC) is the term used in classifying the variants, with consequential implications on the diagnosis, approved treatments, clinical disease and vaccines effectiveness. Vaccines may be negatively impacted with this variant, encompassing reduction in effectiveness and increase in breakthrough infections among vaccinated individuals. The variants may also lead to critically reduced protection by antisera induced by the vaccine in neutralizing the virus. The biggest threat of variants would be VOHC, which may include failure of many diagnostic tests and causing more severe disease and increased hospitalisations compared to the VOC. This variant so far, has not been described anywhere or governing organisations.
Variants Being Monitored (VBM)/Variants Under Monitoring (VUM) or Formerly Monitored Variants
In addition to the previously mentioned SARS-CoV-2 variants classifications, there is another class of variants, referred to as the variants under monitoring (VUM) by WHO or variants being monitored (VBM) as addressed by CDC. VUM is also being assigned to previously classified VOC or VOI, yet is now responsible for insignificant impacts against the public health or prevention measures in the USA. The variants will continue to be monitored in order to determine if there is any threat they may impose, as there are insufficient data or ambiguous evidence to relate these variants with the possible impacts they may cause in the future. Formerly monitored variants refer to the variants that have been identified by WHO as the long circulating SARS-CoV-2 variants with no reported epidemiological impacts. It may also refer to the variants with no public health significance due to very low prevalence among the circulating variants. Previous VOI or VUM which meet these criteria may also result in the reclassification of them as the formerly monitored variants.
Impacts of SARS-CoV-2 Variants Emergence
The emergence of many SARS-CoV-2 variants, especially for the variants classified under VOC or VOI resulted in many impacts on many aspects such as the detection of the SARS-CoV-2, the viral transmissibility, disease severity and vaccine effectiveness.
Performance of SARS-CoV-2 Diagnostic Tests
RT-PCR has been the gold standard and is considered the most sensitive and specific in detecting the active SARS-CoV-2 infection status. The most commonly targeted genes of the viral nucleic acid will be the RNA-dependent RNA-polymerase (RdRp), open reading frame ORF1ab, envelope (E), spike glycoproteins (S), and sometimes, nucleocapsid (N) of SARS-CoV-2. The main focus of SARS-CoV-2 variants emergence is the changes they exert towards the genes targeted by molecular kits, which may compromise the sensitivity and specificity of the molecular detection. Genetic alteration of viral particles as the result of mutation may reduce the specificities and efficacies of the RT-PCR, leading to possible false-negative results.35 False-negative results in RT-PCR can occur due to the inefficient amplification and inaccurate probe binding due to the inability of the oligonucleotides to bind specifically and efficiently to the specific sites of the viral genome, due to the mutation.36
Mutations arising in nucleocapsid and spike glycoprotein of B.1.1.529 (Omicron) have caused malfunctioning or failure of detection of SARS-CoV-2 viral genomes in several commercial molecular test kits.37 Deletion of amino acids 69 and 70 (Δ69-70), which can be found in most Omicron and B.1.1.7 (Alpha) lineages is also accountable for the failure of some commercial diagnostic kits, targeting the spike proteins of SARS-CoV-2.38 This condition is often termed as the S-gene target failure (SGTF), where the failure of detection of SARS-CoV-2 occurs due to the mutations taking place in the spike proteins. There are also some mutations resulting in N-gene target failure (NGTF) in some molecular test kits. The mutations include the nucleotide substitution from cytosine to thyme at position 26, 340 and 29197 of the SARS-CoV-2 genome.39,40 In Omicron variant, there is a nine-nucleotides deletion in its N-gene, which affects the sensitivity and specificity of Revogene SARS-CoV-2 kit (Meridian Bioscience, USA). T23599G and C23604A in Omicron, also impacted the performance of the Line COVID-19 Assay kit (Applied DNA Sciences, USA).
The detection of highly conserved ORF1ab and N region in RT-PCR will be able to reduce the impact of variants emergence towards the failure in detection, when compared to the detection of mutation-prone S and ORF8 genes. Using multiple genes or target regions on the viral genome known as multiplex RT-PCR can also mitigate the impacts of the poor sensitivity and specificity in the detection of SARS-CoV-2 variants.37
Antigen-based detection approach has been reported to have a lower specificity for detection of Omicron variants than the other circulating VOCs. Omicron and Delta were only being detected in 49.2% and 65.6%, respectively of overall tests performed on seven commercial Ag-RDT kits, using positive clinical samples.41 The samples were previously determined to be of Omicron and Delta through RT-PCR analysis.
Transmissibility and Prevalence of Infections
In term of transmissibility, natural selection favours variants with the best fitness advantage, by enabling specific variants to displace and dominate the other circulating variants, within a population.42 The dominance of certain SARS-CoV-2 variants even resulted in pandemic waves or the phase in which a particular country experiences significantly high COVID-19 infection rates within a specific timeframe. The D614G variants, first detected in February 2020, were responsible for the first COVID-19 wave in most countries. The variant has caused a sudden escalation in detection, encountered for 78% of approximately 12, 000 sequences analysed in mid-May 2020 as compared to only 10% out of 1,000 sequences being deposited on March 1, 2020.25
India recorded 0.36% of additional confirmed COVID-19 cases from its total population on April 23, 2021, which was only the eighth week of the second wave of COVID-19 infections in India (that started around March 2021). Before the start of the second wave, India only reported a total prevalence of about 0.7% of its total population. The rapid surge of COVID-19 cases in India, was most probably due to the three circulating variants at that time, which are Alpha (B.1.1.7), Delta (B.1.617.2) and Kappa (B.1.617.1).43
Currently, many countries are facing the fourth wave of the pandemic, due to the dominance of Omicron. In late November 2021, Omicron variant was detected in less than 0.1% of total sequences submitted to GISAID in the preceding period of 60 days. The variant was only detected in several countries across four WHO regions, before being detected in 57 countries in all WHO regions in the second week of classification as VOC. Omicron then rapidly dominated the circulating variants, with a value of 96.7% of all sequences submitted to the database in the period of 30 days till February 8, 2022.44
Viral shedding of SARS-CoV-2 bearing D614G occurred at a fast rate, as reported in an animal study. It was as early as day two of exposure with D614G-infected hamsters with contrary results observed in exposure of the wild type-infected hamsters.45 This finding suggested aerosols and droplets of infected individuals with D614G can lead to more rapid infection, than the wild type. This mutation also allows efficient spikes glycoproteins deposition on the virions, leading to an increase in infectivity of human angiotensin-converting enzyme 2 (hACE2) receptors.46,47 This is because D614G mutation improves RBD exposures to the receptors thus leading to efficient attachment and virus-receptor binding. In addition, the efficient membrane fusion activity was also observed, through the abrogation of particular interactions between the non-mutated amino acid D614 with several other amino acids.48
N501Y mutation involves the substitution of amino acid asparagine (N) by tyrosine (Y) at position 501 of the SARS-CoV-2 spike. It was first detected during the emergence of B.1.1.7 (Alpha) variant in September 2020, results in approximately 70 to 80% better transmissibility, due to the better binding affinity and faster attachment of the virus to the hACE2 receptor, in addition to the slower dissociation rate of RBD from the receptors when compared to the wild type. 49,50 The mutation grants fitness advantage for the variant to continuously replicate in the upper airway of hamster model and primary human airway epithelial cells, thus permitting efficient transmission.51
Mutations involving the amino acid phenylalanine (P) at position 681 found in Omicron and Delta variants, is deduced to be the factor of greater fitness of the variant, in replication efficiency. This mutation occurs at the furin cleavage site of spike glycoprotein, which results in a more efficient entry of the virus into the host cells.52 In addition, Omicron P681H mutation, H being histidine is accompanied by two more mutations taking place near the furin cleavage site which are H655Y and N679K, where K being (Lysine) which are also expected to increase the furin cleavage efficiency. This combination results in the high transmissibility of the Omicron variant, in comparison with the other variants.53,54
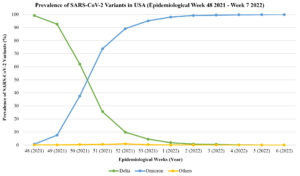
In Europe, an estimated prevalence of 69.4% for Omicron was reported based on the sequencing done from December 20, 2021, to January 9, 2022, by 23 European countries.55 The emergence of Omicron challenges the Delta dominance, and this can be observed from the reported number of cases caused by these variants in the USA (Figure).56 A study by Shah et al. concluded that at least 11 out of 15 RBD mutations of the Omicron variant, lead to the enhancement of binding affinity of the virus towards the ACE2 receptors.57 These mutations include some of the common substitutions determined in other VOC, like changing amino acids to T478 (Delta), K417 (Beta, Gamma and Delta) and E484 (Beta and Gamma). The mutations on the RBD Omicron variant include Q493R, N501Y, S371L, S373P, S375F, Q498R, and T478K.58,59
The infection rate of Omicron was also proven to be higher than any other VOCs, based on a study involving the pseudoviruses which are constructed from lentivirus and introduced with spike proteins of Beta, Gamma, Delta and Omicron. Omicron showed a two to four-fold of infectivity compared to the Delta and wild-type, suggesting an increase in infectivity through enhanced affinity for ACE2.60 The Omicron variant is responsible for the rapid increase of COVID-19 infections in Tshwane, South Africa, surpassing 27,000 cases within a time span of only 33 days, but then declining steadily since then.61
Delta (B.1.617.2) variant is one the most carefully monitored and focused variants, due to its high transmissibility. It is assumed to be able to spread up to 13 persons, from a single infected individual. Majority of these cases are reported in United Kingdom with a figure exceeding 1.73 million of confirmed cases, as of March 7, 2022.62 Considering all mostly reported variants globally, B.1.617.2 was estimated to own the highest mean effective reproduction number of 97%. The findings indicated that B.1.617.2 has a high transmissibility which may be the reason behind its previous dominance.63 In Malaysia, the Ministry of Health reported that, B.1.617.2 rapidly dominated the circulating variants in the country, within a short period of time. The variant was responsible for only six positive cases at the end of May 2021, before steadily increasing to 1,612 of a cumulative total of infections on September 25, 2021.64,65 As of February 11, 2022, Malaysia has reported at least 6, 504 cases (85.7%) caused by B.1.617.2 from a confirmed total of 7, 586 cases caused by various VOC and VOI.66 Rapid replication of B.1.617.2 variant with an estimated viral load of approximately 1, 000 times higher than the wild-type strain, with the brief incubation period may be the factors of expeditious transmission of this lineage.67
In a prevalence study performed by Noureddine et al.,68 a drastic increase of positive cases caused by SARS-CoV-2 variants was reported, from only 40% on January 11, 2021, to 96.5% on February 15, 2021. The finding is most probably due to the B.1.1.7 (Alpha) which was dominating the circulating variants, then in Lebanon. Earlier, B.1.1.7 variant also dominated the circulating variant sequenced in Italy, from a prevalence of only 3.5% in December 2020 to a concerning value of 86.7% in March 2021.69
In addition, there is a mutation which allows the virus to be spread between animals and humans, allowing zoonotic transmission of SARS-CoV-2. For instance, the Y453F mutation at RBD, can be detected in several variants, primarily emerging in Denmark between minks and humans.70 Infection of farmers with SARS-CoV-2 variants presented with specific mutations, distinctively related to minks have also been reported in Netherlands, Poland and USA.71 Despite capturing universal attention due to its transmissibility towards humans, the mutation was proven to only escalate the affinity of RBD against ACE2 receptor, with no significant impacts towards neutralising antibodies from both infected and vaccinated people.72
Severity of COVID-19
WHO and The National Institute of Health (NIH) USA has provided a guideline in classifying the severity of disease in COVID-19, encompassing of five clinical stages – (1) asymptomatic, (2) mild illness, (3) moderate illness, (4) severe illness and (5) critical illness (Table 2).73
Table (2):
Summary of stages of SARS-CoV-2 infections based on the severities.73
Stage of Infection |
Explanation |
|---|---|
Asymptomatic or Presymptomatic |
Individuals who are tested positive for SARS-CoV-2 but showing no symptoms of COVID-19. |
Mild Illness |
Individuals with several symptoms of COVID-19 (fever, cough, sore throat, headache, muscle pain, malaise, loss of taste and smell) but having no pneumonia. |
Moderate Illness |
Individuals showing lower respiratory disease during clinical assessment and oxygen saturation (SpO2) ≥ 94%. |
Severe Illness |
Individuals with SpO2 value lower than 94%, respiratory rate of more than 30 breaths per minute or lung infiltrate of more than 50%. |
Critical Illness |
Individuals with respiratory failure, septic shock and involvement of multiple organs. |
The severity of the disease can also be related to the risks or rate of hospitalisation which indicates the needs for further treatment or monitoring by the healthcare workers compared to the asymptomatic or normal infection. Admission into intensive care units (ICU) can also be an indication of severity, in which the patients may require more intensive care and monitoring due to the progression of their disease with more serious clinical symptoms. A study performed by Abdullah et al.61 in a hospital in the City of Tshwane, South Africa, recorded low disease severity of COVID-19 in the period of Omicron dominance. The average length of stay in the hospital was shorter and admission to ICU in addition to total reported fatalities during the time were lower, compared to the period of dominance by other variants, namely Delta.
In another study, Ulloa et al.74 reported that the severity of disease due to Omicron infections is lower when compared to the Delta variant. Rates of hospitalisation and death were higher in cases of Delta with 2.8% and 0.5%, respectively compared to Omicron infections with values of only 0.3% and 0.03%, respectively. The lower severity of COVID-19 due to Omicron variants, might also be related to the increasing global vaccination rate against COVID-19, during the time of Omicron dominance.75 According to an ex vivo experiment conducted using lung cell cultures, Omicron variant was observed to be replicating at a lower rate than the wild type and the Delta variant. This suggested that the Omicron variant might lead to a less severe disease.76
B.1617.2 (Delta) variant tends to result in more hospitalisation and progression of severe COVID-19 cases at 2.3% and 5.7%, respectively, despite a lesser total of infections compared to the other variants.77 In Scotland, B.1.617.2 infections led to a higher possibility of patients being admitted and treated at hospital, in comparison with B.1.1.7 variant, with a significant difference of around 58% between those two lineages.78 The risk of admission also increases with the number of underlying comorbidities. The surge of hospitalisation from 7% to 14% and increased reported mortality from 8% to 16% due to COVID-19, were also reported in 13 regions in the USA due to B.1.617.2 domination.79 In Qatar, 123 out of 451 patients infected with Delta variant, were hospitalised and 22 of them were admitted to the intensive care unit (ICU). In the same study, the Beta variant recorded a lower risk of hospitalisation (90 out of 451) and admission into ICU (14 out of 451).80 An increase in threats of progression or development of symptomatic disease were also observed, even after complete dose of BNT162b2.81
P681R mutation, may be crucial in causing the severe diseases by Delta variant. Based on an in vitro study involving B.1.617.2-infected cells, it was observed that P681R-bearing D614G variant which possesses the spike of Delta variant, resulted in larger syncytia formation as compared to common D614G variant. Syncytia in COVID-19 refers to the multinucleated large cell mass formed, due to the fusion of pneumocytes.82 In the same study, Syrian hamsters infected with pseudovirus harbouring the P681R mutation were observed to suffer from weight loss at a higher degree than the D614G-infected hamsters, at a rate of 4.7% to 6.9% on day 7 post infection. These findings indicate that P681R might be the main mutation resulting in increased disease severity caused by Delta, compared to the other variants.83
A higher hospitalisation risk of 42% caused by B.1.17 (Alpha) variant was concluded in Denmark, where a huge proportion involving patients aged more than 60 years old, with no difference in risks between genders.84 Conversely, hospitalised females due to B.1.1.7 infections were prone to higher risks of both mortality and requirement of intensive treatment unit (ITU), based on the findings of a separate study conducted in the United Kingdom.85 Huge proportion of disease progression, into the obligation of supplemental oxygen, demand of invasive ventilation and deaths, were also revealed in infections by B.1.17 86. B.1.351 (Beta) variant, was assumed to be the determinant for the increased death to infection ratio recorded in the urban community of South Africa, between July 2020 and March 2021.87
COVID-19 Vaccine Effectiveness and Breakthrough Infections
Emergence of variants can also impact the effectiveness of globally administered COVID-19 vaccines. Complete vaccination status did not guarantee a total elimination of being a transmitter, as proven by Kleynhans et al.87 There was no obvious difference in the median cycle threshold (Ct) value of RT-PCR between samples of vaccinated and unvaccinated individuals infected with B.1.617.2 (Delta) variant.88 Delta variant was justified to be the main cause for the infection, even after complete COVID-19 vaccination. The variant was responsible for almost 91% of the infections out of 63 confirmed breakthrough infections 89. In spite of the possibility of infection after completion of vaccination, severity of disease due to breakthrough infection is much milder compared to infections among unvaccinated individuals. The vaccinated COVID-19 patients were less likely to require invasive mechanical ventilation and recorded low rate of death. 24.7% of the cases involving unvaccinated individuals, resulted in death which accounted for 93.8% of the total death reported.90
The effectiveness of mRNA-based vaccines was critically lowered to only 30.0% and 35.6% after one dose, for ChAdOx1 nCoV-19 and BNT162b2, respectively, against particular SARS-CoV-2 variants (Table 3).
Table (3):
Summary of adjusted vaccines effectiveness against the B.1.1.7 (Alpha) and B.1.617.2 (Delta) variant infections.91
| Vaccines | Adjusted vaccine effectiveness (%) | |
|---|---|---|
| B.1.1.7 (Alpha) | B.1.617.2 (Delta) | |
| BNT162b2 Dose 1 Dose 2 |
47.5 93.7 |
35.6 88.0 |
| ChAdOx1 Dose 1 Dose 2 |
48.7 74.5 |
30.0 67.0 |
Sheikh et al.92 reported that a single dose of ChAdOx1 was proven to offer better protection against Delta variants, compared to a single dose of BNT162b2, which recorded a higher death rate for single dose recipients. However, complete dose of BNT162b2 recorded a lower mortality rate than ChAdOx1. Apart from Delta, the South Africa-originating B.1.351 (Beta) variant was also responsible for the significant reduction of ChAdOx1 vaccines efficacy, from 75.0% against the non-variants to only 10.4% against the variants.93 In the contrary, another study deduced that the presence of several variants in the community, like B.1.351 (Beta), B.1.429, B.1.427 (Epsilon), and B.1.526 (Iota), resulted in no reduction in efficacy of ChAdOx1 nCoV-19 vaccines. The effectiveness and efficacies of the vaccines was also proven to be higher in recipients ageing more than 65 years old.94 Ad26.COV2.S which implements the application of non-replicating viral vectors for the vaccination was also proven to be effective against some of the SARS-CoV-2 variants (Table 4).95
Table (4):
Efficacy of Ad26.COV2.S against B.1.351 and P.2 variants of SARS-CoV-2 (WHO).95
| Variant | Countries | Vaccine effectiveness (%) | |
|---|---|---|---|
| Moderate to critical | Severe or Critical | ||
| B.1.351 (Beta) | South Africa | 64.0 | 81.7 |
| P.2 (Zeta) | Brazil | 68.1 | 87.6 |
Gazit et al.81 suggested that complete vaccinations with BNT162b2 were exposed to a higher risk of breakthrough infections by B.1.617.2 compared to the possibility of reinfections. In a study involving 8, 678 vaccinated personnel in Asan Medical Centre, South Korea, only four individuals were detected for SARS-CoV-2 reinfection, where all of them received ChAdOx1 nCoV-19 vaccine 96. A study reported that only 18% of the total of 969 patients admitted to Yale New Haven Health System hospital, had received at least one dose of COVID-19 vaccines. 54 out of 103 patients already received two doses of either BNT162b2 or mRNA-1273, or one dose of Ad26.COV2.S (Table 5).97
Table (5):
Summary of vaccine breakthrough infections among fully vaccinated patients at Yale New Haven System hospital between March 21 and July 1, 2021.97
Infection or Disease Status |
Counts |
Total |
|---|---|---|
Asymptomatic |
25 |
|
Mild |
4 |
|
Moderate |
11 |
|
Severe / Critical (Deaths) |
14 (3) |
54 |
Pre-existing comorbidities (Severe / Critical) |
||
Overweight |
9 |
|
Cardiovascular disease |
12 |
|
Lung disease |
7 |
|
Malignancy |
4 |
|
Type-2 diabetes |
7 |
|
Immunosuppressive medications |
4 |
On the other hand, 2.61% out of 1, 497 vaccinated healthcare workers at Sheba Medical Centre, Israel, were confirmed with vaccine breakthrough infection, in which a huge proportion of the cases were female (64%) at an average age of 42 years old. Thirty-three isolates from the healthcare workers are classified as variants, with 85% of them coming from B.1.1.7 variant.98 In Dane County, USA, a sum of 1, 472 from 371, 000 fully vaccinated residents were tested positive for SARS-CoV-2, where a diminutive fraction of 25 persons were hospitalised, accounting for three fatalities.99 As of epidemiological week 52, of 2021, approximately 851, 711 and 1180 individuals from the USA were reported with COVID-19 vaccines breakthrough infections and deaths post complete vaccinations, respectively.100 In Malaysia, 1063 of vaccine breakthrough cases resulting in category 3, 4 and 5 of COVID-19 infection were reported in epidemiological week 8 of 2022 (February 21 to February 27, 2022). Of these recorded cases, 549 (52.99%) and 227 (21.91%) cases were of the two-doses of vaccines recipients and booster dose recipients, respectively.101-107
Of all COVID-19 vaccine breakthrough infections, the one reported in India which involved a 61-years old female healthcare worker should gain the most attention. In this case, the patient was confirmed for reinfection with Alpha variant, after approximately eight months and 24 days, post-infection and complete vaccination, respectively. The patient was then diagnosed with the second vaccine breakthrough infection, this time with the Delta variant.108
The latest addition to the VOC family by the B.1.1.529 (Omicron) variant did also exert negative consequences towards the acquired immunity through vaccinations. Vaccine breakthrough infections by the variant, have been reported in several people who even completed their booster dose.109 In a study spanned across 49 states of the United States of America, 18.6% (2441 cases) and 55.3% (7245 cases) from a total of 13, 098 confirmed infections by Omicron variant, were detected among three and two vaccine doses recipients, respectively.110 E484K mutation, is being highlighted as the potential major mutation leading to the breakthrough infections. This was suggested from a study, where the mutation was detected in most of the sequenced virus causing the breakthrough infections in Maryland, USA.111 Currently, many researchers have already embarked on the development of a pan-sarbecovirus vaccine which contains antigens other than the spike proteins with the hope that it will work against all types of SARS-CoV-2 variants in the future.112
This review describes the information related to SARS-CoV-2 variants, focusing on their characteristics, mutations, and also the consequences. Despite already being circulating for more than two years, regular mutations lead to the emergence of numerous SARS-CoV-2 variants, as part of viral evolution. The impacts include poor performance of SARS-CoV-2 diagnostic kits, high transmissibility and prevalence of infections, increased severity of disease and reduced COVID-19 vaccines effectiveness leading to reports of breakthrough infections. However, it is crucial to conclude that the impacts might have been mitigated with the vaccinations. Further research is still required to find solutions on how to predict and control the emergence of variants so that the infections can stop spreading. This is important for the subsequent measures which will be implemented in order to minimise the impacts of the SARS-CoV-2 variants’ emergence.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This study was supported by Universiti Teknologi MARA, Research Grant Scheme 600-RMC/LESTARI SDG-T 5/3 (184/2019).
DATA AVAILABILITY
Not applicable.
ETHICS STATEMENT
Not applicable.
- Zhang J, Garrett S, Sun J. Gastrointestinal symptoms, pathophysiology, and treatment in COVID-19. Genes Dis. 2021;8(4):385-400.
Crossref - WHO Coronavirus (COVID-19) Dashboard. World Health Organization. 2022. Accessed 04 March 2022. https://covid19.who.int/
- V’kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19(3):155-170.
Crossref - Yuki K, Fujiogi M, Koutsogiannaki S. COVID-19 pathophysiology: A review. Clin Immunol. 2020;215:108427.
Crossref - Xu X, Yu C, Qu J, et al. Imaging and clinical features of patients with 2019 novel coronavirus SARS-CoV-2. Eur J Nucl Med Mol Imaging. 2020;47(5):1275-1280.
Crossref - Parasher A. COVID-19: Current understanding of its Pathophysiology, Clinical presentation and Treatment. Postgrad Med J. 2021;97(1147):312-320.
Crossref - Yadav R, Chaudhary JK, Jain N, et al. Role of Structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19. Cells. 2021;10(4):821.
Crossref - Fang F, Chen Y, Zhao D, et al. Recommendations for the Diagnosis, Prevention, and Control of Coronavirus Disease-19 in Children – The Chinese Perspectives. Front Pediatr. 2020;8:553394.
Crossref - Habibzadeh P, Mofatteh M, Silawi M, Ghavami S, Faghihi MA. Molecular diagnostic assays for COVID-19: an overview. Crit Rev Clin Lab Sci. 2021;58(6):385-398.
Crossref - Real-World Performance of COVID-19 Rapid Antigen Tests. American Society for Microbiology. 2021. Accessed 12 January 2022. https://asm.org/Articles/2021/December/Real-World-Performance-of-COVID-19-Rapid-Antigen-T
- Ong DSY, Fragkou PC, Schweitzer VA, Chemaly RF, Moschopoulos CD, Skevaki C. How to interpret and use COVID-19 serology and immunology tests. Clin Microbiol Infect. 2021;27(7):981-986.
Crossref - Amanat F, Krammer F. SARS-CoV-2 Vaccines: Status Report. Immunity. 2020;52(4):583-589.
Crossref - COVID-19 Vaccines Work. Centers for Disease Control and Prevention. 2021. Accessed 05 January 2022. https://www.cdc.gov/coronavirus/2019-ncov/vaccines/effectiveness/work.html
- Pfizer and BioNTech Conclude Phase 3 Study of COVID-19 Vaccine Candidate, Meeting All Primary Efficacy Endpoints. Pfizer. 2020. Accessed October 1, 2021. https://www.pfizer.com/news/press-release/press-release-detail/pfizer-and-biontech-conclude-phase-3-study-covid-19-vaccine
- Frost SDW, Magalis BR, Kosakovsky Pond SL. Neutral Theory and Rapidly Evolving Viral Pathogens. Mol Biol Evol. 2018;35(6):1348-1354.
Crossref - Fauver JR, Petrone ME, Hodcroft EB, et al. Coast-to-Coast Spread of SARS-CoV-2 during the Early Epidemic in the United States. Cell. 2020;181(5):990-996.e5.
Crossref - Harvey WT, Carabelli AM, Jackson B, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19(7):409-424.
Crossref - Lauring AS, Hodcroft EB. Genetic Variants of SARS-CoV-2 – What Do They Mean? JAMA. 2021;325(6):529-531.
Crossref - Bechtold P, Wagner P, Hosch S, et al. Rapid Identification of SARS-CoV-2 Variants of Concern Using a Portable peakPCR Platform. Anal Chem. 2021;93(49):16350-16359.
Crossref - Pachetti M, Marini B, Benedetti F, et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J Transl Med. 2020;18(1):179.
Crossref - Ogando NS, Zevenhoven-Dobbe JC, van der Meer Y, Bredenbeek PJ, Posthuma CC, Snijder EJ. The Enzymatic Activity of the nsp14 Exoribonuclease Is Critical for Replication of MERS-CoV and SARS-CoV-2. J Virol. 2020;94(23):e01246-20.
Crossref - Wang S, Xu X, Wei C, et al. Molecular evolutionary characteristics of SARS-CoV-2 emerging in the United States. J Med Virol. 2022;94(1):310-317.
Crossref - Singh Y, Fuloria NK, Fuloria S, et al. N-terminal domain of SARS CoV-2 spike protein mutation associated reduction in effectivity of neutralizing antibody with vaccinated individuals. J Med Virol. 2021;93(10):5726-5728.
Crossref - Yurkovetskiy L, Wang X, Pascal KE, et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell. 2020;183(3):739-751.e8.
Crossref - Korber B, Fischer WM, Gnanakaran S, et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182(4):812-827.e19.
Crossref - Tracking SARS-CoV-2 Variants. World Health Organization. 2022. Accessed 24 February 2022. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
- SARS-CoV-2 Variants Classifications and Definitions. Centers for Disease Control and Prevention. 2021. Accessed 22 February 2022. https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html
- Kimura I, Kosugi Y, Wu J, et al. SARS-CoV-2 Lambda variant exhibits higher infectivity and immune resistance. bioRxiv. 2021.
Crossref - Uriu K, Kimura I, Shirakawa K, et al. Neutralization of the SARS-CoV-2 Mu Variant by Convalescent and Vaccine Serum. N Engl J Med. 2021;385(25):2397-2399.
Crossref - Tegally H, Ramuth M, Amoako D, et al. A Novel and Expanding SARS-CoV-2 Variant, B.1.1.318, Dominates Infections in Mauritius. ppmedrxiv. 2021.
Crossref - Scheepers C, Everatt J, Amoako DG, et al. Emergence and phenotypic characterization of C.1.2, a globally detected lineage that rapidly accumulated mutations of concern. medRxiv. 2021.
Crossref - Colson P, Delerce J, Burel E, et al. Emergence in Southern France of a new SARS-CoV-2 variant of probably Cameroonian origin harbouring both substitutions N501Y and E484K in the spike protein. medRxiv. 2021.
Crossref - Monajjemi M, Sayiner HS, Kandemirli F, Mollaamin F. An overview on lambda, epsilon, kappa, iota and zeta variants of covid-19 and its probability to merge with delta & delta plus, why it is a concern. Biointerface Res Appl Chem. 2022;12(5):6895-6914.
Crossref - Tablizo FA, Kim KM, Lapid CM, et al. Genome sequencing and analysis of an emergent SARS-CoV-2 variant characterized by multiple spike protein mutations detected from the Central Visayas Region of the Philippines. medRxiv. 2021.
Crossref - Genetic Variants of SARS-CoV-2 May Lead to False Negative Results with Molecular Tests for Detection of SARS-CoV-2 – Letter to Clinical Laboratory Staff and Health Care Providers. U.S. Food and Drug Administration. 2021. Accessed 03 January 2022. https://www.fda.gov/medical-devices/letters-health-care-providers/genetic-variants-sars-cov-2-may-lead-false-negative-results-molecular-tests-detection-sars-cov-2
- Janik E, Niemcewicz M, Podogrocki M, Majsterek I, Bijak M. The Emerging Concern and Interest SARS-CoV-2 Variants. Pathog. 2021;10(6):633.
Crossref - SARS-CoV-2 Viral Mutations: Impact on COVID-19 Tests. U.S. Food and Drug Administration. 2021. Accessed 03 January 2022. https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-viral-mutations-impact-covid-19-tests#:~:text=The%20presence%20of%20mutations%20in,design%20
differences%20of%20each%20test. - Science Brief: Omicron (B.1.1.529) Variant. Centers for Disease Control and Prevention. 2021. Accessed 05 January 2022. https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scientific-brief-omicron-variant.html
- Lopez D, Roberts J, Bourgeois M, et al. Infection clusters can elevate risk of diagnostic target failure for detection of SARS-CoV-2. PLoS One. 2022;17(2):e0264008.
Crossref - Ziegler K, Steininger P, Ziegler R, Steinmann J, Korn K, Ensser A. SARS-CoV-2 samples may escape detection because of a single point mutation in the N gene. Euro Surveill. 2020;25(39):2001650.
Crossref - Bekliz M, Perez-Rodriguez F, Puhach O, et al. Sensitivity of SARS-CoV-2 antigen-detecting rapid tests for Omicron variant. medRxiv. 2022.
Crossref - Davies NG, Abbott S, Barnard RC, et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372(6538):eabg3055.
Crossref - Singh J, Rahman SA, Ehtesham NZ, Hira S, Hasnain SE. SARS-CoV-2 variants of concern are emerging in India. Nat Med. 2021;27(7):1131-1133.
Crossref - COVID-19 Weekly Epidemiological Update. Edition 78. World Health Organization. 2022. Accessed 27 February 2022. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19—8-february-2022
- Hou YJ, Chiba S, Halfmann P, et al. SARS-CoV-2 D614G Variant Exhibits Enhanced Replication ex vivo and Earlier Transmission in vivo. bioRxiv. 2020.
Crossref - Zhang L, Jackson CB, Mou H, et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun. 2020;11(1):6013.
Crossref - Zhou B, Thao TTN, Hoffmann D, et al. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature. 2021;592(7852):122-127.
Crossref - Hu J, He CL, Gao QZ, et al. D614G mutation of SARS-CoV-2 spike protein enhances viral infectivity. bioRxiv. 2020.
Crossref - Leung K, Shum MH, Leung GM, Lam TT, Wu JT. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. [published correction appears in Euro Surveill. 2021;26(3):]. Euro Surveill. 2021;26(1):2002106.
Crossref - Tian F, Tong B, Sun L, et al. N501Y mutation of spike protein in SARS-CoV-2 strengthens its binding to receptor ACE2. Frank A, Faraldo-Gomez JD, eds. Elife. 2021;10:e69091.
Crossref - Liu Y, Liu J, Plante KS, et al. The N501Y spike substitution enhances SARS-CoV-2 transmission. bioRxiv. 2021.
Crossref - Liu Y, Liu J, Johnson BA, et al. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. bioRxiv. 2021.
Crossref - Hossain MG, Tang Y, Akter S, Zheng C. Roles of the polybasic furin cleavage site of spike protein in SARS-CoV-2 replication, pathogenesis, and host immune responses and vaccination. J Med Virol. 2022;94(5):1815-1820.
Crossref - Peacock TP, Brown JC, Zhou J, et al. The SARS-CoV-2 variant, Omicron, shows rapid replication in human primary nasal epithelial cultures and efficiently uses the endosomal route of entry. bioRxiv. 2022.
Crossref - Weekly Epidemiological Update: Omicron variant of concern (VOC) -week 2 (data as of 20 January 2022) EU/EEA. European Centre for Disease Prevention and Control. 2022. Accessed 10 February 2022. https://www.ecdc.europa.eu/en/news-events/weekly-epidemiological-update-omicron-variant-concern-voc-week-2-data-20-january-2022
- COVID Data Tracker; Variant Proportions. Centers for Disease Control and Prevention. 2022. Accessed 02 March 2022. https://covid.cdc.gov/covid-data-tracker/#variant-proportions
- Shah M, Woo HG. Omicron: A Heavily Mutated SARS-CoV-2 Variant Exhibits Stronger Binding to ACE2 and Potently Escapes Approved COVID-19 Therapeutic Antibodies. Front Immunol. 2022;12:830527.
Crossref - Kumar S, Thambiraja TS, Karuppanan K, Subramaniam G. Omicron and Delta variant of SARS-CoV-2: A comparative computational study of spike protein. J Med Virol. 2022;94(4):1641-1649.
Crossref - Pascarella S, Ciccozzi M, Bianchi M, Benvenuto D, Cauda R, Cassone A. The electrostatic potential of the Omicron variant spike is higher than in Delta and Delta-plus variants: A hint to higher transmissibility? J Med Virol. 2022;94(4):1277-1280.
Crossref - Garcia-Beltran WF, St. Denis KJ, Hoelzemer A, et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022;185(3):457-466.e4.
Crossref - Abdullah F, Myers J, Basu D, et al. Decreased severity of disease during the first global Omicron variant COVID-19 outbreak in a large hospital in Tshwane, South Africa. Int J Infect Dis. 2022;116:38-42.
Crossref - Variants: Distribution of Case Data, 7 march 2022. GOV.UK. 2022. Accessed 20 March 2022. https://www.gov.uk/government/publications/covid-19-variants-genomically-confirmed-case-numbers/variants-distribution-of-case-data-2-march-2022
- Campbell F, Archer B, Laurenson-Schafer H, et al. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro Surveill. 2021;26(24):2100509.
Crossref - Kenyataan Akhbar KPK 31 Mei 2021 – Situasi Semasa Jangkitan Penyakit Coronavirus 2019 (COVID-19) di Malaysia. From the Desk of the Director-General of Health Malaysia. 2021. Accessed 03 December 2021. https://kpkesihatan.com/2021/05/31/kenyataan-akhbar-kpk-31-mei-2021-situasi-semasa-jangkitan-penyakit-coronavirus-2019-covid-19-di-malaysia/
- Kenyataan Akhbar KPK 25 September 2021 – Situasi Semasa Jangkitan Penyakit Coronavirus 2019 (COVID-19) di Malaysia. From the Desk of the Director-General of Health Malaysia. 2021. Accessed 03 December 2021. https://kpkesihatan.com/2021/09/25/kenyataan-akhbar-kpk-25-september-2021-situasi-semasa-jangkitan-penyakit-coronavirus-2019-covid-19-di-malaysia/
- Kenyataan Akhbar KPK 11 Februari 2022 – Situasi Semasa Jangkitan Penyakit Coronavirus 2019 (COVID-19) di Malaysia. From the Desk of the Director-General of Health Malaysia. 2022. Accessed 14 February 2021. https://kpkesihatan.com/2022/02/11/kenyataan-akhbar-kpk-11-februari-2022-situasi-semasa-jangkitan-penyakit-coronavirus-2019-covid-19/
- Li B, Deng A, Li K, et al. Viral infection and transmission in a large, well-traced outbreak caused by the SARS-CoV-2 Delta variant. medRxiv. 2021.
Crossref - Noureddine FY, Chakkour M, El RozA, et al. The emergence of SARS-CoV-2 variant(s) and its impact on the prevalence of COVID-19 cases in Nabatieh region, Lebanon. medRxiv. 2021.
Crossref - Lai A, Bergna A, Menzo S, et al. Circulating SARS-CoV-2 variants in Italy, October 2020-March 2021. Virol J. 2021;18(1):168.
Crossref - Devaux CA, Pinault L, Delerce J, Raoult D, Levasseur A, Frutos R. Spread of Mink SARS-CoV-2 Variants in Humans: A Model of Sarbecovirus Interspecies Evolution. Front Microbiol. 2021;12:675528.
Crossref - Animals and COVID-19. Centers for Disease Control and Prevention. 2021. Accessed 09 October 2021. https://www.cdc.gov/coronavirus/2019-ncov/daily-life-coping/animals.html
- Bayarri-Olmos R, Rosbjerg A, Johnsen LB, et al. The SARS-CoV-2 Y453F mink variant displays a pronounced increase in ACE-2 affinity but does not challenge antibody neutralization. J Biol Chem. 2021;296.
Crossref - Clinical Spectrum of SARS-CoV-2 Infection. National Institute of Health. 2021. Accessed 11 February 2022. https://www.covid19treatmentguidelines.nih.gov/overview/clinical-spectrum/#:~:text=Patients%20with%20COVID%2D19%20are,or%
20lung%20infiltrates%20%3E50%25. - Ulloa AC, Buchan SA, Daneman N, Brown KA. Estimates of SARS-CoV-2 Omicron Variant Severity in Ontario, Canada. JAMA. 2022.
Crossref - The Omicron Variant: Sorting Fact from Myth. World Health Organization Regional Office for Europe. 2022. Accessed 12 February 2022. https://www.who.int/europe/news/item/19-01-2022-the-omicron-variant-sorting-fact-from-myth
- Chan M, Hui K, Ho J, et al. SARS-CoV-2 Omicron Variant Replication in Human Respiratory Tract Ex Vivo. Research Square. 2021:1-20.
Crossref - Twohig KA, Nyberg T, Zaidi A, et al. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: a cohort study. Lancet Infect Dis. 2022;22(1):35-42.
Crossref - Sheikh A, McMenamin J, Taylor B, Robertson C. SARS-CoV-2 Delta VOC in Scotland: demographics, risk of hospital admission, and vaccine effectiveness. Lancet. 2021;397(10293):2461-2462.
Crossref - Scobie HM, Johnson AG, Suthar AB, et al. Monitoring Incidence of COVID-19 Cases, Hospitalizations, and Deaths, by Vaccination Status – 13 U.S. Jurisdictions, April 4-July 17, 2021. MMWR Morb Mortal Wkly Rep. 2021;70:1284-1290.
Crossref - Butt AA, Dargham SR, Chemaitelly H, et al. Severity of Illness in Persons Infected With the SARS-CoV-2 Delta Variant vs Beta Variant in Qatar. JAMA Intern Med. 2022;182(2):197-205.
Crossref - Gazit S, Shlezinger R, Perez G, et al. Comparing SARS-CoV-2 natural immunity to vaccine-induced immunity: reinfections versus breakthrough infections. medRxiv. 2021.
Crossref - Lin L, Li Q, Wang Y, Shi Y. Syncytia formation during SARS-CoV-2 lung infection: a disastrous unity to eliminate lymphocytes. Cell Death Differ. 2021;28(6):2019-2021.
Crossref - Saito A, Irie T, Suzuki R, et al. SARS-CoV-2 spike P681R mutation, a hallmark of the Delta variant, enhances viral fusogenicity and pathogenicity. bioRxiv. 2021.
Crossref - Bager P, Wohlfahrt J, Fonager J, et al. Risk of hospitalisation associated with infection with SARS-CoV-2 lineage B.1.1.7 in Denmark: an observational cohort study. Lancet Infect Dis. 2021;21(11):P1507-1517.
Crossref - Stirrup O, Boshier F, Venturini C, et al. SARS-CoV-2 lineage B.1.1.7 is associated with greater disease severity among hospitalised women but not men: multicentre cohort study. BMJ Open Respir Res. 2021;8(1):e001029.
Crossref - Pascall DJ, Mollett G, Blacow R, et al. The SARS-CoV-2 Alpha variant causes increased clinical severity of disease. medRxiv. 2021.
Crossref - Kleynhans J, Tempia S, Wolter N, et al. Longitudinal SARS-CoV-2 seroprevalence in a rural and urban community household cohort in South Africa, during the first and second waves July 2020-March 2021. medRxiv. 2021.
Crossref - Christensen PA, Olsen RJ, Long SW, et al. Delta variants of SARS-CoV-2 cause significantly increased vaccine breakthrough COVID-19 cases in Houston, Texas. medRxiv. 2021.
Crossref - Ayass MA, Zhang J, Zhu K, et al. The Impact of New SARS-CoV-2 Variants on Vaccine Breakthrough: A Pilot Study on Spreading Infection in the Communities. medRxiv. 2021.
Crossref - Tenforde MW, Patel MM, Ginde AA, et al. Effectiveness of Severe Acute Respiratory Syndrome Coronavirus 2 Messenger RNA Vaccines for Preventing Coronavirus Disease 2019 Hospitalizations in the United States. Clin Infect Dis. 2022.
Crossref - Lopez Bernal J, Andrews N, Gower C, et al. Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. N Engl J Med. 2021;385(7):585-594.
Crossref - Sheikh A, Robertson C, Taylor B. BNT162b2 and ChAdOx1 nCoV-19 Vaccine Effectiveness against Death from the Delta Variant. N Engl J Med. 2021;385(23):2195-2197.
Crossref - Madhi SA, Baillie V, Cutland CL, et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 Vaccine against the B.1.351 Variant. N Engl J Med. 2021;384(20):1885-1898.
Crossref - Falsey AR, Sobieszczyk ME, Hirsch I, et al. Phase 3 Safety and Efficacy of AZD1222 (ChAdOx1 nCoV-19) Covid-19 Vaccine. N Engl J Med. 2021;385:2348-2360.
Crossref - Interim recommendations for the use of the Janssen Ad26.COV2.S (COVID-19) vaccine. World Health Organization. 2021. Accessed 04 October 2021. https://www.who.int/publications/i/item/WHO-2019-nCoV-vaccines-SAGE-recommendation-Ad26.COV2.S-2021.1
- Jung J, Sung H, Kim SH. Covid-19 Breakthrough Infections in Vaccinated Health Care Workers. N Engl J Med. 2021;385(17):1629-1630.
Crossref - Juthani P V, Gupta A, Borges KA, et al. Hospitalisation among vaccine breakthrough COVID-19 infections. Lancet Infect Dis. 2021;21(11):1485-1486.
Crossref - Bergwerk M, Gonen T, Lustig Y, et al. Covid-19 Breakthrough Infections in Vaccinated Health Care Workers. N Engl J Med. 2021;385:1474-1484.
Crossref - Breakthrough Infections and Impact of the Delta Variant. Public Health Madison & Dane County. 2021. Accessed 03 October 2021. https://www.publichealthmdc.com/blog/breakthrough-infections-and-impact-of-the-delta-variant
- Rates of COVID-19 Cases or Deaths by Age Group and Vaccination Status and Booster Dose. Centers for Disease Control and Prevention. 2022. Accessed 03 March 2022. https://data.cdc.gov/Public-Health-Surveillance/Rates-of-COVID-19-Cases-or-Deaths-by-Age-Group-and/d6p8-wqjm
- Kenyataan Akhbar KPK 28 Februari 2022 – Situasi Semasa Jangkitan Penyakit Coronavirus 2019 (COVID-19). From the Desk of the Director-General of Health Malaysia. 2022. Accessed 03 March 2022. https://kpkesihatan.com/2022/02/28/kenyataan-akhbar-kpk-28-februari-2022-situasi-semasa-jangkitan-penyakit-coronavirus-2019-covid-19/
- Kenyataan Akhbar KPK 27 Februari 2022 – Situasi Semasa Jangkitan Penyakit Coronavirus 2019 (COVID-19). From the Desk of the Director-General of Health Malaysia. 2022. Accessed 03 March 2022. https://kpkesihatan.com/2022/02/27/kenyataan-akhbar-kpk-27-februari-2022-situasi-semasa-jangkitan-penyakit-coronavirus-2019-covid-19/
- Kenyataan Akhbar KPK 26 Februari 2022 – Situasi Semasa Jangkitan Penyakit Coronavirus 2019 (COVID-19). From the Desk of the Director-General of Health Malaysia. 2022. Accessed 03 March 2022. https://kpkesihatan.com/2022/02/26/kenyataan-akhbar-kpk-26-februari-2022-situasi-semasa-jangkitan-penyakit-coronavirus-2019-covid-19/
- Kenyataan Akhbar KPK 25 Februari 2022 – Situasi Semasa Jangkitan Penyakit Coronavirus 2019 (COVID-19). From the Desk of the Director-General of Health Malaysia. 2022. Accessed 03 March 2022. https://kpkesihatan.com/2022/02/25/kenyataan-akhbar-kpk-25-februari-2022-situasi-semasa-jangkitan-penyakit-coronavirus-2019-covid-19/
- Kenyataan Akhbar KPK 24 Februari 2022 – Situasi Semasa Jangkitan Penyakit Coronavirus 2019 (COVID-19). From the Desk of the Director-General of Health Malaysia. 2022. Accessed 03 March 2022. https://kpkesihatan.com/2022/02/24/kenyataan-akhbar-kpk-24-februari-2022-situasi-semasa-jangkitan-penyakit-coronavirus-2019-covid-19/
- Kenyataan Akhbar KPK 23 Februari 2022 – Situasi Semasa Jangkitan Penyakit Coronavirus 2019 (COVID-19). From the Desk of the Director-General of Health Malaysia. 2022. Accessed 03 March 2022. https://kpkesihatan.com/2022/02/23/kenyataan-akhbar-kpk-23-februari-2022-situasi-semasa-jangkitan-penyakit-coronavirus-2019-covid-19/
- Kenyataan Akhbar KPK 22 Februari 2022 – Situasi Semasa Jangkitan Penyakit Coronavirus 2019 (COVID-19). From the Desk of the Director-General of Health Malaysia. 2022. Accessed 03 March 2022. https://kpkesihatan.com/2022/02/22/kenyataan-akhbar-kpk-22-februari-2022-situasi-semasa-jangkitan-penyakit-coronavirus-2019-covid-19/
- Shastri J, Parikh S, Aggarwal V, et al. Severe SARS-CoV-2 Breakthrough Reinfection With Delta Variant After Recovery From Breakthrough Infection by Alpha Variant in a Fully Vaccinated Health Worker. Front Med. 2021;8:737007.
Crossref - Kuhlmann C, Mayer CK, Claassen M, et al. Breakthrough infections with SARS-CoV-2 omicron despite mRNA vaccine booster dose. Lancet. 2022;399(10325):625-626.
Crossref - Accorsi EK, Britton A, Fleming-Dutra KE, et al. Association Between 3 Doses of mRNA COVID-19 Vaccine and Symptomatic Infection Caused by the SARS-CoV-2 Omicron and Delta Variants. JAMA. 2022;327(7):639-651.
Crossref - Feder KA, Patel A, Vepachedu VR, et al. Association of E484K and L452R spike protein mutations with SARS-CoV-2 infection in vaccinated persons—Maryland, January – May 2021. medRxiv. 2021;71(11):2053-2056.
Crossref - WHO consultation on COVID vaccines research: Why do we need a pan-sarbecovirus vaccine? World Health Organization. 2022. Accessed 30 May 2022. https://www.who.int/news-room/events/detail/2022/01/28/default-calendar/who-consultation-on-covid-vaccines-research-why-do-we-need-a-pan-sarbecovirus-vaccine
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.