ISSN: 0973-7510
E-ISSN: 2581-690X
https://dx.doi.org/10.22207/JPAM.13.1.56 | © The Author(s). 2019
Ciprofloxacin comprise a medically effective and generally utilized class of broad-spectrum antibiotics; as a result of the intensive use of this type of therapy, bacteria have become resistant, lead to minimize the usage of such bactericides in field of management as well as treatment of microbial illness. Pseudomonas aeruginosa one of important pathogens that case wide spectrum of infections, the evolution of ciprofloxacin-resistant Pseudomonas aeruginosa were notify. In this investigation 50 clinical isolates of Pseudomonas aeruginosa were collected, antibiotic susceptibility test for ciprofloxacin by using the Kirby–Bauer technique as well as MIC was performed to all these isolates. Specific primers sequences for detection of Topoisomerase II (gyrA) genes were used by conventional PCR then, PCR product were treated with Sac II enzyme at 37°C for 2 h, then restriction bands were recovered by gel electrophoresis. From Fifty Pseudomonas aeruginosa isolates enrolled in this study, 33(66%) show sensitive to Ciprofloxacin, 17(34%) reported resistant to Ciprofloxacin. RFLP-PCR showed that out of 17 isolates resistant to Ciprofloxacin mutation inDNA gyrase (GyrA) were detected in six (35.2 %) isolated while the reminder 11 (64.7%) has no mutation. The presence of mutations in Topoisomerases II genes mediate Ciprofloxacin defeat as therapy for Pseudomonas aeruginosa. The current venture, were made to demonstrate Topoisomerase II genes (GyrA mutations in Pseudomonas aeruginosa that could be responsible for molecular mechanism of Ciprofloxacin resistance.
Topoisomerase II genes, DNA gyrase, Ciprofloxacin, RFLP-PCR, SacII enzyme.
Pseudomonas aeruginosa causes hospital acquired infection due to its ubiquitous nature, capability to continue to exist in damp environments, what’s more, un-respond to numerous anti-infection agents. Quinolones are one of these drug group in which Pseudomonas aeruginosa resistant is being reported and has been identified as a motive of remedy failure1. On this group, Ciprofloxacin is the maximum antibiotic trends towards Gram-negative bacteria, for examples Salmonella species, Acinetobacter baumannii and Pseudomonas aeruginosa, Ciprofloxacin is a 2nd-genration agent, belong to the quinolones derivates. Which have ability to interfered with the bacterial DNA gyrase, it has bactericidal activity with a wide spectrum in opposition to Gram positive and Gram-negative microorganism2.
As a result, to critical use of Ciprofloxacin as one of the mighty agents in organization fluoroquinolones against P. aeruginosa has prompted the set off the improvement of resistant strains3. Quinolones has the ability to sort kind two topoisomerases, which can be crucial compounds in arrange the topological condition of DNA via its replication and transcription. DNA gyrase is made from two A and B fragments, these fragments encoded through the gyrA and gyrB genes4.
Topoisomerases are assuming crucial elements of DNA metabolism5. Quinolones conflict with the motion of DNA gyrase, a critical bacterial kind II DNA topoisomerase6.
Several researches have been exhibited that mutations imparting fluoroquinolone resistance in P. aeruginosa may attributed to amendment in DNA gyrase, hypothesizing that faded sensitivity to fluoroquinolone result from inhibition of DNA supercoiling in resistant isolates of Gram-negative pathogens and alteration in gyrA consider leading causes in this resistance7. This study aims to demonstrate Topoisomerase II genes (GyrA mutations in Pseudomonas aeruginosa that could be responsible for molecular mechanism of Ciprofloxacin resistance.
From April 2015 to March 2016, fifty clinical isolates of P. aeruginosa were collected from different infectious source in patients who admitted into Al-Khadhmiya Teaching Hospital by using cotton swabs, samples were culture immediately on Nutrient, Blood and Mac-Conkey agar at 37°C for 24 hours, the isolated colony were identified by biochemical test and Epi 20-E system.
All isolates enrolled in this study were tested by disk diffusion methods to determine antimicrobial susceptibility to Ciprofloxacin (5mg), when the inhibition zone across the disks was formed it measured and comparison with the break points of clinical laboratory institute (CLSI)8.
The MIC became completed by way of a standard agar dilution approach and has been carried out for determination the lowest antibiotics concentration that prevents the growth of bacteria, the break points encouraged by using CLSI have been used for ciprofloxacin (susceptible £ 1 µg/mL; resistant ³four mg/mL). E. coli ATCC25922 isolates from central public health laboratory standard strain used as negative control. This study authorized by Institutional Review Board (IRB) in the College of Medicine /AL-Nahrain University, and conducted in the Microbiology Department on the College of Medicine Al-Nahrain University.
DNA Extractions
DNA extraction was done by a simple and rapid procedure boiling according to Reischl et al.9. Briefly one ml of an overnight bacterial growth became centrifuged, at 14000 rpm for two minutes, the supernatant was eliminated and pellet became suspended in 2 hundred ml lysis buffer containing 1% Triton X-100, 0.5% Tween 20, 10 mM Tris-HCl (pH 8.0), and 1 mM EDTA and incubated in a screw-cap reaction tube for 10 min in a boiling water bath, the suspension turned into centrifuged at 14000 rpm for two minutes, supernatant containing DNA turned into Eppendorf tube and used as a template for PCR assay targeting the Gyrase A genes.
PCR Amplification
Specific primers sequences targeting DNA gyrase according to Kureishi et al.10 were used for PCR amplification and examined the connection among gene mutations of topoisomerase II (gyrA) with resistance to Ciprofloxacin in P.aeruginosa, this genemake in Alpha DNA® (Canada) table (1).
Table (1):
Sequences of primer that use to detection II topoisomerase (GyrA gene).
Nucleotide sequences |
Reference |
Products |
||
|---|---|---|---|---|
Genes |
(5′——————- ► 3′ ) |
bP |
||
F |
GACGGCCTGAAGCCGGTGCAC |
416 |
||
Gyrase A |
R |
GCCCACGGCGATACCGCTGGA. |
(10) |
The primers have been diluted via adding nuclease free water consistent with the manufacturer commands. The master mix contents have been thawed at room temperature before use, all precautions have been taken to avoid any contamination. 2ml from forward and reverse primer for Gyrase A gene had been added in single pre-mixed PCR reaction tube, then DNA template equal to Five microliter had been added. Twelve and a 1/2 microliters of GoTaq® Green Master Mix was added for each reaction tube, the quantity changed into completed to 25ml with Deionized Nuclease –Free and tubes had been spun down to ensure mixing of the reaction components.
Non- DNA template in PCR mixture were used as negative control. The tubes have been positioned at PCR machine (Eppendorf Master Cycler) and the PCR software, with the proper cycling situations pre-installed, Amplification reaction for Gyrase A gene become as follows: 94°C for 3 min followed by 40 cycles of 94°C for 30 sec, 50°C for 30 sec, and 72°C for 60 sec, terminating in 72°C for 7 min, 10ml from PCR product was subjected to 1% (wt/vol) agarose gel electrophoresis with 0.5 mg /ml ethidium bromide, Five microliters of the 100bp DNA ladder marker turned into subjected to electrophoresis in a single lane. Amplicon visualization was done the use of a UV trans illuminator.
REFLP- PCR assay
In order to detect mutation in Gyrase A gene PCR -REFLP with Cfr42 I (SacII) enzyme (10 U/µL) were used according to manufacture instruction (Thermo Fisher Scientific). Briefly, The PCR product were treated with Sac II enzyme at 37°C for 2 hr. Restriction enzyme is inactivated by incubation at 65°C for 20 min. then restriction bands were recovered by gel electrophoresis.
Disc diffusion test showed that out of 50 Pseudomonas aeruginosa isolates enrolled in this study, 33(66%) were delicate to ciprofloxacin, 17(34%) were impervious to Ciprofloxacin.
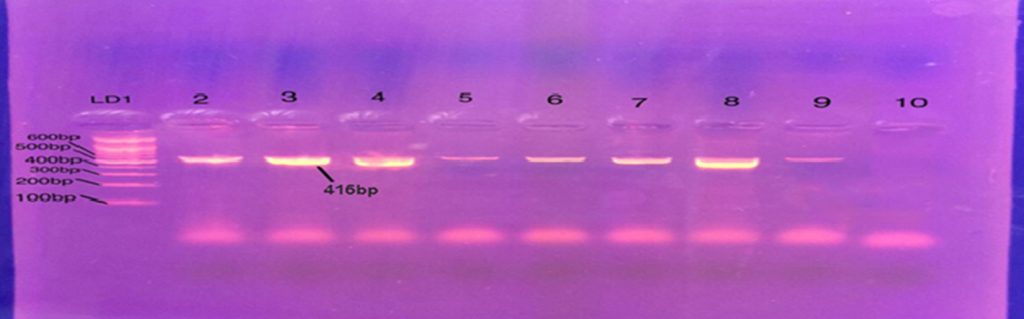
Determination of MIC to Ciprofloxacin reveals that out of 17 bacteria resistant to Ciprofloxacin, 8 isolates had MIC 64mg/mL. Three isolates with MIC 32mg/mL. five isolates with MIC 16 mg/mL Conventional PCR affirmed existence of Gyrase A genes in all resistant strain of P. aeruginosa for Ciprofloxacin fig. 1. RFLP-PCR methods showed that out of 17 isolates resistant to Ciprofloxacin mutation in type II topo-isomerase genes (GyrA) were found in 6 (35.2 %) isolated indicate by single band while the reminder 11 (64.7%) has no mutation indicate by double fragment fig. 2.
Pseudomonas aeruginosa are recognized as most important Gram-negative pathogen, remains a challenge to many doctors and researchers because of their ability to resist many of the antibiotics using different mechanisms, and one of these mechanisms’ mutation in DNA gyrase (type II Topoisomerase) which is essential enzyme contributory in chromosomal DNA replication and segregation.
In the present study, the percentage of Ciprofloxacin resistant (34%) is lower than the percentage recorded in Iraqi by Zeyad K. et al 2018 who reported 83.50% out of 103 Pseudomonas aeruginosa isolates11.
Other study conducted in India by Goel et al (2011) stated that increasing fluoro-quinolone Pseudomonas aeruginosa resistant 12. This difference may be explained by small size of Pseudomonas aeruginosa isolated in current study in compare with other studies, further more some Iraqi hospitals initiate stewardship antibiotic program surveillance which limited used of fluoroquinolone.
According to AL-Muhannak (2012), appropriate antibiotic dose and convenient medical procedures supervision can limit and sometime prevent bacterial resistant to quinolone, in our study 33(66%) were sensitive to Ciprofloxacin, which confirms AL-Muhannak (2012) observation13.
So, this result may be highlighting that bacteria responsible for hospital acquired infection such as Pseudomonas aeruginosa may develop resistant when steadily contact with a wide range of antibiotic, and as results to this events surveillance strategies in hospitalized patients may lead to minimized infections with resistant bacteria.
One of the major mechanisms used by bacteria to resist fluoroquinolone is mutation in topoisomerases genes, Noticeably in gyrA on codon 83 and 8714. This mutation lead to variation in amino acid which are found in regions that responsible for quinolone-resistance (QRDRs), since this area contains this type of topoisomerase15.
Have crucial role in the replications and segregations of DNA, furthermore these enzymes considered as a main target for antibiotic16.
Our finding that the out of 17 isolates resistants to Ciprofloxacin, 11 (64.7%) has no mutation, these results were consistent with Varug heseet al (2018)17.
This could be attributed to the presence of other mechanisms that make Pseudomonas aeruginosa resistant to fluoroquinolone antibiotic patterns such as mutation in other genes such as parE, parC gene efflux pump, changes in the target protein of antibiotic, furthermore minimize in the penetration ability of an antibiotic is also a resistance mechanism for many classes of antibiotics18.
In conclusion, this study has uncovered that the changes in Topoisomerases II (gyrA) play crucial role in the mechanism of quinolone resistance among the locally separate P. aeruginosa in Al-Khadhmiya Teaching Hospital Baghdad Iraqi.
None
The authors declare no conflict of interest.
- Krebs CJ. Ecology: The Experimental Analysis of Distribution and Abundance. Harper and Row Publisher, New York 1978.
- Zaid A, de Wet PF, Djerbi M, Oihabi A. in Date palm cultivation, Diseases and pests of date palm, edZaid A. (Food and Agriculture Organization Plant Production and Protection Paper no. 156. Food and Agriculture Organization of the United Nations, Rome, Italy) 2002; 227-281.
- Al-Jboory IJ. Survey and identification of the biotic factors in the date palm environmentand its application for designing IPM-Program of date palm pests in Iraq. Univ. Aden J. Nat. and Appl. Sci., 2007; 11(3): 3-10.
- Hameed MA. Inflorescence rot disease of date palm caused by Fusarium proliferatum in Southern Iraq. African Journal of Biotechnology, 2012 ; 11(35): 8616-8621.
- Al-Ani HY, El-Behadili A, Majeed HA, Majeed M. Reaction of date palm cultivars to inflorescence rot and persistency and spreading of the disease. Phytopathol. Mediterranean, 1971; 10: 57-62
- Abdullah SK, Asensio L, Monfort E, Gomez-Vidal S, Palma-Guerrero J, Salinas J, Lopez-Llorca LV, Jansson HB, Guarro J.Occurrence in Elx, SE Spain of inflorescence rot disease of date palms caused by Mauginiella scaettae. J. Phytopathol., 2005; 153: 417-422.
- Hassen KA . Testing the effect od salt, cinder and some fungicides against the influence rot disease on date-palm in center of Iraq (Diyala province). Diyala Journal for pure sciences, 2010; 6(2): 509-515.
- Hameed MA. Susceptibility of different cultivars of data palm (phoenix dactylifera L.) to Mauginiella scaettae the causal agent of inflorescnce rot. Basrah Journal for data palm research, 2005; 4(1-2): 37-53.
- Dherbi M. Diseases of the date palm (Phoenix dactylifera L.) FAO 1983; 114.
- Al Ghilan AJK. Isolation and identification of Mauginiella scaettae Cav. Which causes data palms inflorescencr rot disease in some areas of Thi-Qar province and its sensitivity to some plant extracts. master thesis university of Thi-Qar, college of education of pure science 2012.
- Alexoplulos CD, Mims CW and Blackwel M. Introductory Mycology, John Wiley and Sons, Incorporation 1996; 869.
- Barnett HL and Hunter BB Illustrated genera of imperfect fungi. 4th ed. The American Phyto-pathology Society, 1998; 218.
- Ellis MB. Dematiaceous, Hyphomycetes. Commonw. Mycol. Inst. Kew, England, 1971; 608.
- Al-Sharidi AM and Al-Shahwan IM. Fungi Associated with Rot Diseases of Inflorescence and Fruit of Date Palm in Riyadh Region. Saudi Arabia. Arab J. Pl. Prot., 2003; 21: 84-89.
- Stockwell MP, Clulow J and Mahony MJ . Sodium Chloride Inhibits the Growth and Infective Capacity of the Amphibian Chytrid Fungus and Increases Host Survival Rates. Plos One, 2012; 7(5):1-7.
- Amir H, Amir A and Riba A. Role de la microfloredans laresitance a la fusariose- vasculaire induitepar la salinitedans un sol depalmeriae. Soil Biol. Biochem., 1996; 28: 113-122.
- Fixen PE. Crop response to chloride. Adv. Agron., 1993; 50: 107-150.
- Yigit A and Korukluoglu M . The effect of potassium sorbate, NaCl and pH on the growth of food spoilage fungi. Annals of Microbiology, 2007; 57(2): 209-215.
- ReidTC , Hausbeck MK and Kizilkaya K. Effect of Sodium Chloride on commercial Asparagus and of Alternative Forms of chloride salt on Fusarium Crown and Root Rot. Plant Disease, 2001; 85(12):1271-1275.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.