ISSN: 0973-7510
E-ISSN: 2581-690X
Staphylococcus aureus mainly Methicillin Resistant Staphylococcus aureus(MRSA) is a life-threatening infection that occurring in food and caused a public health concern. This study designed to examine the prevalence of S. aureus and MRSA in different types of processed food. Food samples were screened for the recovered strains of S. aureus and MRSA, and they were examined for antimicrobial susceptibility and by molecular characterization of mecA and staphylococcal cassette chromosome mec(SCCmec). Detection of virulence factors like Panton-Valentine Leukocidin (PVL), Staphylococcus aureus protein A(spa) and Staphylococcal enterotoxins(SEs) by PCR using specific primers. Among the 150 collected processed food samples, 62.7% were contaminated by S. aureus bacteria, 56.4% of which were proved as MRSA. 17% of MRSA isolates were positive for mecA genes with the SCCmec type IVb and V (11.1% each) as the solely existing types of SCCmec. None of the MRSA isolates carried mecC or mecB genes. Most of MRSA isolates were multidrug resistance and 33.3% of MRSA-mecA positive isolates also carried vancomycin resistance genes (i.e., vanB). In addition, spa gene was found among 7.5% of MRSA isolates; none of which were positive for PVL gene. Further, there were variant presence of SEs among MRSA isolates and the highest presence was from type SEH (49.1%). Generally, our results confirmed that processed foods in Saudi Arabia (Riyadh) are potential vehicles for multidrug resistant S. aureus and MRSA transmission; which are serious public health risks, and underlined the need for good hygiene practices.
MRSA, processed foods, multidrug resistance, mecA, CA-MRSA, LA-MRSA
Staphylococcus aureus is a main source for food poisoning globally. In addition, it is a significant reason for human infection and it can be a reason for lethal diseases such as pneumonia, endocarditis, toxic shock syndrome, and sepsis1. Additionally, high levels of antimicrobial resistance to these bacteria pose a serious public health threat. Methicillin-Resistant Staphylococcus aureus (MRSA) forms around 13-74% of S. aureus’s infections2. The acquisition of mecA gen via the mobile staphylococcal cassette chromosome mec (SCCmec) in MRSA strains is the reason for the emergence of resistance to all b-lactam antibiotics3 . MRSA was classified to the three groups, due to the differences in the origin of the outbreaks, healthcare-associated MRSA (HA-MRSA) , community-associated MRSA (CA-MRSA), and livestock-associated MRSA (LA-MRSA)4. Epidemiological reports have declared variance among HA-MRSA, CA-MRSA, and LA-MRSA strains counting antimicrobial resistance profiles and SCCmec types. For instance, SCCmec types IV and V are detected among CA-MRSA strains, whereas SCCmec types I, II, and III are detected among HA-MRSA strains and the most common types between them is SCCmec V and SCCmec IV, respectively5. In addition, SCCmec XI type is discovered newly in humans and cattle, especially from mastitis in cow in UK and Denmark and it carries mecC gene, which initially knew as mecA LGA2516,7. Furthermore, PVL gene is a genetic marker for the MRSA presence was recognized for the first time in 2003 and it produces strains that may cause skin infection or serious diseases such as pneumonia. However, PVL gene can be predominantly seen in CA-MRSA more than in HA-MRSA and it is found in around 60% to 100% of CA-MRSA strains. Consequently, PVL develops the pathogenicity of CA-MRSA strains and the prevalence of PVL between strains is depending on bacteriophages8-10. Nonetheless, specific types of symptoms such as red and swollen skin frequently occur with MRSA infection, which is painful and filled with pus bump fever.
With regard to MRSA’s types, livestock is considered as a fertile source of staphylococci species generally and S. aureus especially, causing livestock-associated MRSA [LA-MRSA]. In addition, LA-MRSA can be occurred by direct contact with animals or utilization of animal products e.g., lamb, calve, and goat. Yet, the spread rate of S. aureus was lower in goat and calve (12.5% and 1.4%, respectively) than it in lamb (29.7%). LA-MRSA was discovered firstly in France and in the Netherlands then it was found in the pig population in countless countries of Europe. Further, as CA-MRSA and HA-MRSA, LA-MRSA clones are different between countries; e.g., clonal complex 398 (CC398) was detected in Europe and USA, whereas clonal complex 9 (CC9) was found in Asia3,6,11.
However, MRSA infections spread vigorously throughout Saudi Arabia and reached 50% during 2011 in King Fahad Medical City in Riyadh city12. The number of studies about MRSA infection in Saudi Arabia is insufficient compared to the other countries around the world; although, MRSA in Saudi Arabia has been discovered in 1990s13. In spite of focusing on investigating and discussing HA-MRSA mainly, there is still a shortage in the number of studies on HA-MRSA in children and surprisingly it is found in males more than females13. Based on the prevalence of CA-MRSA disease, the quantity of patients significantly raised in Saudi Arabia from 9.9 per 10,000 people in 2001 to 67 per 10,000 people in 200813,14. Furthermore, during the time between 2000 to 2008, the spread rate of CA-MRSA along Eastern region of Riyadh city increased by six times12. Also, CA-MRSA samples collected from Qatif city, Saudi Arabia showed that CA-MRSA cases increased gradually from 23% in 2006 to 60% in 2015. In terms of LA-MRSA infections, there is still a lack of studies that investigate this type of MRSA15,16. The purpose of this study was to investigate the prevalence of two types of MRSA: CA-MRSA and LA-MRS in processed foods collected from the capital city of Saudi Arabia and their antibiotic resistance and molecular characteristics of each isolate based on the identification of MecA gene, SCCmec types, PVL, spa gene, and SEs by PCR technique.
Sample collection
A total of 150 samples of processed foods were collected during a period of August 2019 to March 2020 from different farms and local retail markets throughout Riyadh city, Kingdom of Saudi Arabia, which are namely: A, B, C, D, E, F, G, H, and I. Samples were smoked Turkey (N=50), sausages (N=50), and salami (N=50).
Bacterial Growth Conditions
After cutting processed foods samples into small pieces by sterilized scalpel and forceps, 0.5g of each were added to approximately 5ml of specific liquid nutrient medium (i.e., brain heart infusion [BHI]) that were later incubated for 24 hours at 37°C in order to cultivate the bacteria.
Isolation and identification of S. aureus
In order to examine S. aureus colonies phenotypically, a loop of each cultivated sample was streaked on the Mannitol salt agar (MSA) -as a selective media to differentiate and isolate such bacteria- which were incubated for overnight at 37°C. As a confirmation stage, DNAs test was also performed for all S. aureus isolates. To detect the growth of MRSA isolates, we assessed the performance of MRSA chromagar as compared to that of mannitol salt agar supplemented with oxacillin (MSAO) for the recovery of MRSA from food specimens.
Antimicrobial susceptibility testing
All MRSA isolates were screened for antimicrobial resistance using disk diffusion method for the following antibiotics classes; β-lactams (penicillin, ampicillin, and oxacillin), macrolides (erythromycin), aminoglycosides (gentamicin), tetracyclines (oxytetracycline), sulfonamides (trimethoprim-sulphamethoxazole), and glycopeptides (vancomycin), were examined.
DNA extraction
Two colonies on MRSA chromagar plates were picked and used to be inoculated in 5 ml of BHI and grown overnight at 37°C. DNA was isolated and purified using a QIAamp DNA mini kit according to the manufacturers’ instructions.
Detection of mecA, mecB , mecC and SCCmec typing of the mecA positive MRSA isolates
mecA, mecB, and mecC (also named mecALGA251) were identified by PCR using primers defined previously17,18 The mecA-positive MRSA isolates were also illustrated by SCCmec type using PCR using primers defined previously19.
Virulence factors of Staphylococcus aureus isolates
The incidence of virulence genes 14 enterotoxin genes (sea, seb, sec, sed, see, seg, seh, sei, sej, sen, seo, sem, sek, sel), Panton-Valentine Leukocidin (PVL), Staphylococcus aureus protein A (spa) and vancomycin resistance genes vanA and vanB were examined by PCR using primers defined previously20-26.
Prevalence of S. aureus and MRSA infection in food
From 150 food samples 94 samples (62.7%) were contaminated by S. aureus bacteria. The contaminated samples were from turkey (27.7%), salami (42.6%), and sausages (29.8%). Despite the fact that S. aureus bacteria were detected in raw sausages samples, two samples were cooked for two different period of times (5 and 10 minutes) and they were still positive for S. aureus and MRSA infection as well. From the total of the 94 S. aureus isolates from different types of processed food, 53 (56.4%) isolates were confirmed as MRSA. Of the 53 MRSA isolates, 3/53 (5.7%) were from Turkey, 19/53(35.8%) were from Salami, and 31/53 (58.5%) were from Sausages (Table 1). In addition, mecA, mecB and mecC gene were examined in all subjected methicillin resistant S. aureus (MRSA) isolates. The result indicated that 17% of MRSA isolates (9/53) were positive for mecA genes, while none of them were positive for mecC or mecB genes.
Table (1):
The prevalence of S. aureus and MRSA among the examined samples in Riyadh, Saudi Arabia.
| Types of samples | No of samples positive for S. aureus | N/P (%) | No of samples positive for MRSA | N/P (%) |
|---|---|---|---|---|
| Turkey | 26 | 26/94 (27.7%) | 3 | 3/53 (5.7%) |
| Salami | 40 | 40/94 (42.6%) | 19 | 19/53 (35.8%) |
| Sausages | 28 | 28/94 (29.8%) | 31 | 31/53 (58.5%) |
| N is the total Number of samples and P is the total number of positive samples | ||||
Antibiotic resistance profiles of MRSA isolates
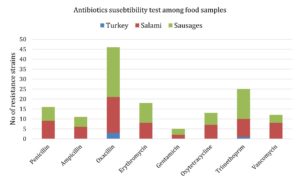
A total of 8 antimicrobial agents were examined for antimicrobial susceptibility in all MRSA isolates. Hence, the percentages of the bacterial resistance for each one of these antibiotics from all kinds of the examined samples are: 30.2% (16/53) to penicillin, 20.8% (11/53) to ampicillin, 86.8% (46/53) to oxacillin, 34% (18/53) to erythromycin, 9.4% (5/53) to gentamicin, 24.5% (13/53) to oxytetracycline, 47.2% (25/53) to trimethoprim-sulphamethoxazole, and 22.6% (12/53) to vancomycin. As regards to vancomycin resistance genes (i.e., vanA and vanB), vancomycin resistant isolates were tested and found that 33.3% (4/12) of them carried vanB gene but none of them were carried vanA gene and their origins were from salami (2), and sausages (2). Noticeably, oxacillin was the most commonly detected antibiotic among sausage samples, while trimethoprim-sulphamethoxazole resistance was detected at lower frequencies among turkey samples in MRSA isolates in comparison with other types of food isolates (i.e., salami origin) (Fig. 1 and Table 2). However, most of MRSA isolates were resistant to more than three antibiotics. For instances, 13.2% (7/53) of such isolates were resistant to 5-7 antibiotics (five from salami, and two from sausages), 43.4% (23/53) of them were resistant to 2-4 antibiotics (16 from sausages, six from salami, and one from turkey origin) and 5.7% (3/53) of them were resistant to all eight antibiotics (one from salami, and two from sausages). On the other hand, 64.2% (34/53) of the isolates were sensitive to 5-7 antibiotics, 11.3% (6/53) of them were sensitive to 2-4 antibiotics, and 7.5% (4/53) of them were sensitive to the all eight antibiotics.
Table (2):
Antibiotics susceptibility test among food samples collected from Riyadh, Saudi Arabia.
| Antibiotic Classes | Antibiotics | Turkey | Salami | Sausages | N/P (%) |
|---|---|---|---|---|---|
| β-lactams | penicillin | 0 | 9 | 7 | 30.2 |
| ampicillin | 0 | 6 | 5 | 20.8 | |
| oxacillin | 3 | 18 | 25 | 86.8 | |
| Macrolides | erythromycin | 0 | 8 | 10 | 34 |
| Aminoglycosides | gentamicin | 0 | 2 | 3 | 9.4 |
| Tetracyclines | oxytetracycline | 0 | 7 | 6 | 24.5 |
| Sulfonamides | trimethoprim | 1 | 9 | 15 | 47.2 |
| Glycopeptides | vancomycin | 0 | 8 | 4 | 22.6 |
Molecular typing of MRSA isolates
The positive mecA isolates were exposed to identify five different types of SCCmec: type I, II, III, IV, and V. consequently, SCCmec types between seven out of nine isolates that carried mecA gene (from salami and sausages origins) could not be detected, whereas only two isolates had a single type which are SCCmec type V and IVb and they were from sausages origin, Table 3.
Table (3):
The discovery of mecA gene (147bp), SCCmec types II, III, IVa,IVb, IVd, and V among food smples in Riyadh, Saudi Arabia.
| Samples types | Strains ID | MecA gene | SCCmec II | SCCmec III | SCCmec IV a | SCCmec IV b | SCCmec IV d | SCCmec V |
|---|---|---|---|---|---|---|---|---|
| Salami | 56 | + | – | – | – | – | – | – |
| 57 | + | – | – | – | – | – | – | |
| 61 | + | – | – | – | – | – | – | |
| 63 | + | – | – | – | – | – | – | |
| 73 | + | – | – | – | – | – | – | |
| Sausages | 86 | + | – | – | – | + | – | – |
| 87 | + | – | – | – | – | – | – | |
| 88 | + | – | – | – | – | – | – | |
| 97 | + | – | – | – | – | – | + |
SCCmec= staphylococcal cassette chromosome mec
Distribution of virulence genes
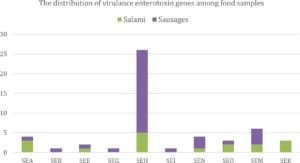
In the tested MRSA samples (53 isolates), Staphylococcus aureus protein A (spa) gene was found between 4 out of 53 (7.5%) and none of them were positive to the Panton Valentine leukocidin (PVL) gene. Yet, a total of 53 methicillin resistant Staphylococcus aureus isolates were examined for the existence of virulence genes. Of the 14 investigated enterotoxin genes SE (i.e., Staphylococcal enterotoxin A [SEA], B [SEB], C [SEC], D [SED], E [SEE], G18, H [SEH], I [SEI], J [SEJ], N [SEN], O [SEO], M [SEM], K [SEK], and L[SEL] 27), there were variant presence of these types among such isolates. With regard to the presence of 14 investigated enterotoxin genes between examined samples, the highest proportion was from SEH (49.1%) followed by SEM (11.3%), SEA and SEN (7.5% each), SEO and SEK (5.7% each), SEE (3.8%), and then SEB, SEG, and SEI (1.9% each). Whilst, none of SEC, SED, SEJ, and SEL were existed in any of MRSA isolates (Fig. 2). Overall, MRSA isolates that were derived from turkey samples did not carry any of the 14 investigated enterotoxin genes.
Fig. 2. The distribution of enterotoxin genes among food samples collected from Riyadh, Saudi Arabia.
Staphylococcal enterotoxin (SEA, SEB, SEC, SED, and SEE) and their origins were from salami (4/6 [66.7%]) and sausages (2/6 [33.3%]) samples. The majority of classic staphylococcal enterotoxins (i.e., SEA, SEB, SEC, SED, and SEE) were between salami samples. Moreover, 6 of 53 MRSA isolates (11.3%) harbored the genes of the enterotoxin gene cluster (egc) (SEG, SEI, SEN, SEO, and SEM) and their origins were from salami and sausages but the highest presence of egc was from sausages samples (i.e., 66.7%). Moreover, some described combinations of the virulence genes of S. aureus were spotted. The SEC-SEL gene combination, typical of the SaPIbov pathogenicity island, was not detected in any of the isolates, as shown in Fig. 2 and Table 4.
Table (4):
The distribution of virulence enterotoxin genes among food samples collected from Riyadh, Saudi Arabia.
Samples types |
SEA |
SEB |
SEE |
SEG |
SEH |
SEI |
SEN |
SEO |
SEM |
SEK |
|---|---|---|---|---|---|---|---|---|---|---|
Turkey |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
Salami |
3 |
0 |
1 |
0 |
5 |
0 |
1 |
2 |
2 |
3 |
Sausages |
1 |
1 |
1 |
1 |
21 |
1 |
3 |
1 |
4 |
0 |
N/P (%) |
7.5 |
1.9 |
3.8 |
1.9 |
49.1 |
1.9 |
7.5 |
5.7 |
11.3 |
5.7 |
SEA=staphylococcal enterotoxin A, SEB= staphylococcal enterotoxin B, SEE= staphylococcal enterotoxin E, SEG= staphylococcal enterotoxin G, SEH= staphylococcal enterotoxin H, SEI= staphylococcal enterotoxin I, SEN= staphylococcal enterotoxin N, SEO= staphylococcal enterotoxin O, SEM= staphylococcal enterotoxin M, SEK= staphylococcal enterotoxin K
Because of CA-MRSA is reported to be less serious outcomes than HA-MRSA, HA-MRSA was investigated more than CA-MRSA and LA-MRSA locally16,28. Apart from samples origins, S. aureus has been known to be an important reason for causing animal illnesses like LA-MRSA (which can transmit to humans via livestock over handling, close contact, or animal-derived food consumption) and food poisoning; especially when such bacteria are multidrug resistant and have the ability to produce toxins like enterotoxins11,29-32.
Hence, turkey, salami, and sausages have been checked for the first time for S. aureus and MRSA in this study; which were collected from different huge supermarkets around Riyadh city. This current study has found approximately 62.7% recorded presence of S. aureus between the tested samples and it was significantly higher than it (18%) in Islam et al. (2019) study in Dhaka, Bangladesh29. Further, more than half of such contaminated samples (56.4%) from processed food considered to be CA-MRSA infection and it occurred through the food preparing process or even hygienic handling circumstances, according to Kluytmans (2010)33. Moreover, turkey samples recorded as the lowest percentage of CA-MRSA (5.7%) followed by salami (35.8%) then sausages (58.5%) from all different collecting places. This may be due to the lower content of fat comparing with the rest of our processed food samples, considering to be rich nutrient environment for bacterial growth, as what confirmed by Raji et al. (2016) when poultry retail meat from Riyadh city were examined16.
According to study done by El-Ghareeb et al. (2019), they examined 3 out of 20 isolates of camel meat collected from random supermarkets in Al-Hasa city and they were described as MRSA (15%), which proved our findings about CA-MRSA infection among examined raw camel meat sausages samples (6% [3/50])34. Furthermore, MRSA infection was still apparent among the four cooked samples of sausages from camel and lamb meat for two different periods (5 and 10 minutes) what supported Kluytmans (2010) study about appropriate cooking for longer time can reduce the risk of their consumption33 .
MecA gene that is responsible for the resistance to methicillin and located on SCCmec was discovered for the first time in Riyadh city among processed food. In fact, MecA gene was found between 9 out of 53 of MRSA isolates (17%) and the ratios of which were diverse between samples (i.e., 55.6% from salami and 44.4% from sausages samples). In addition, our study found that about 86.8% of the positive mecA isolates were resistant to Oxacillin which was confirmed by Osman et al. (2017) about the negative mecA strains are resistant to Oxacillin rather than methicillin or could be a novel of mecA homologue that was linked with cattle35. Moreover, a study was done by Elhaaan et al, stated that it is extremely suggested to consider alternate mechanisms for β-lactams resistance that may compete with mecA gene to detect the emergence of MRSA in the community36.
Antibiotic resistance profiles of MRSA isolates
The susceptibility test of a fifty-three MRSA isolates in this study was estimated for eight antibiotics from different classes and the resistance was detected against Oxacillin (86.8%), which was higher comparing with Gentamicin (9.4 %). It is worth noting that 13.2% of MRSA isolates were sensitive to Oxacillin. This result confirmed the study of Saeed et al. (2014) who stated that Methicillin resistant Staphylococcus aureus (MRSA) is phenotypically presenting minimum inhibitory concentration (MIC) of oxacillin greater than 2 mg/L. Nevertheless, lately, cefoxitin/oxacillin-susceptible mecA-positive S. aureus (OS-MRSA) has been described globally37.
Such giant resistance; in addition, was stated by Osman et al. (2017) to the over using of antibiotics in order to promote the animal growth or treatment35. The widespread of multidrug resistant phenotypes among MRSA isolates around Saudi Arabia generally was accounted by Adam and Abomughaid (2018) for the significant number of pilgrims whom may carry these such drug resistant bacteria38. Surprisingly, this study uncovered 22.6% (12/53) were resistant to Vancomycin and 33.3% (4/12) were carried vanB gene (from salami [2], and sausages [2]); which is still using as a drug of choice to cure MRSA infections, according to Gade and Qazi (2013)39.
The mec A carrier (i.e., SCCmec) type I, II, III, IV, and V were also screened in this study among the positive mec A samples in order to achieve the goal of the epidemiology study for the MRSA isolates. It was found two types among two samples from salami and sausage (types IVb and V). In addition, this study completely support the studies done by Deurenberg, and Stobberingh (2008) and Moussa et al. (2012) which stated SCCmec types V and IV are coupled with community transmission and CA-MRSA and with Moussa et al. (2012) results about coupling SCCmec type III with HA-MRSA due to the absence of such type between LA-MRSA strains currently40,41.
Panton-Valentine leukocidin (PVL), which is the major virulence factor, was not spotted in this study in all investigated processed food samples. Hence, our result was conflicted with Deurenberg and Stobberingh (2008) about using PVL as an indicator for the presence of CA-MRSA, but on the other hand, it was matching with Herrera et al., (2016) who stated that most of LA-MRSA isolates do not carry PVL42. Following this reason, other virulence factors such as spa gene were examined and formed 7.5% of the total number of LA-MRSA and CA-MRSA isolates. Besides, fourteen variant Staphylococcal enterotoxins were investigated as factors causing Staphylococcal food poisoning and health hazards for consumers27,43.
Positive MRSA isolates in our study found to be harbored one or more genes from classic staphylococcal enterotoxins (SEA, SEB, SEC, SED, and SEE) and the enterotoxin gene cluster (egc [SEG, SEI, SEN, SEO, and SEM]). In addition, our result confirms that SEH (49.1%) was the most spread enterotoxin followed by SEM (11.3%), 7.5% from each one of SEA and SEN, 5.7% from each one of SEO and SEK, SEE (3.8%), and 1.9% from each one of SEB, SEG, and SEI.
From the above, this is the first comprehensive investigation of the prevalence of S. aureus and MRSA in prossesed foods from Riyadh city, Saudi Arabia. This study declared a relatively high ratios of S.aureus and MRSA contamination in such foods (62.7% and 56.4%, respectively). Importantly, two cooked sausages samples were still contaminated with such bacteria and infection and none of MRSA isolates were positive for PVL gene. Whilst, nine of MRSA isolates were positive for MecA gene and spa gene was among four of which. High amount of antimicrobial resistance also found between the isolates ,therefore, our data approved the possible role of processed foods in the transmission of multidrug resistant S.aureus strains and successful MRSA in Riyadh , Saudi Arabia.
ACKNOWLEDGMENTS
My thanks must go to Princess Nourah bint Abdulrahman University for its financial support as well as the Health Sciences Research Center/core facilities for allowing me to complete the analysis of my research samples in their cutting-edge laboratory.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
MJA and AS contributed to the design and implementation of the research, to the analysis of the results and to the writing of the manuscript. M.A conceived of the presented idea.
FUNDING
This research was funded by Deanship of Scientific Research at Princess Nourah bint Abdulrahman University (Grant number PR-1441-5).
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Otto M. Staphylococcus colonization of the skin and antimicrobial peptides. Expert Rev Dermatol. 2010;5(2):183-195.
Crossref - Narayanan N, Adams CD, Kubiak DW, et al. Evaluation of treatment options for methicillin-resistant Staphylococcus aureus infections in the obese patient. Infect Drug Resist. 2019;12:877-891.
Crossref - Yang X, Zhang J, Yu S, et al. Prevalence of Staphylococcus aureus and Methicillin-Resistant Staphylococcus aureus in Retail Ready-to-Eat Foods in China. Front Microbiol. 2016;7:816.
Crossref - Dweba CC, Zishiri OT, El Zowalaty ME. Methicillin-resistant Staphylococcus aureus: livestock-associated, antimicrobial, and heavy metal resistance. Infect Drug Resist. 2018;11:2497-2509.
Crossref - Shah MS, Qureshi S, Kashoo Z, et al. Methicillin resistance genes and in vitro biofilm formation among Staphylococcus aureus isolates from bovine mastitis in India. Comp Immunol Microbiol Infect Dis. 2019;64:117-124.
Crossref - Butaye P, Argudin MA, Smith TC. Livestock-Associated MRSA and Its Current Evolution. Curr Clin Microbiol Rep. 2016;3(1):19-31.
Crossref - Vindel A, Trincado P, Cuevas O, Ballesteros C, Bouza E, Cercenado E. Molecular epidemiology of community-associated methicillin-resistant Staphylococcus aureus in Spain: 2004-12. J Antimicrob Chemother. 2014;69(11):2913-2919.
Crossref - Loewen K, Schreiber Y, Kirlew M, Bocking N, Kelly L. Community-associated methicillin-resistant Staphylococcus aureus infection: Literature review and clinical update. Can Fam Physician. 2017;63(7):512-520.
- Motamedi H, Rahmat Abadi SS, Moosavian SM, Torabi M. The Association of Panton-Valentine leukocidin and mecA Genes in Methicillin-Resistant Staphylococcus aureus Isolates From Patients Referred to Educational Hospitals in Ahvaz, Iran. Jundishapur J Microbiol. 2015;8(8):e22021.
Crossref - Kong EF, Johnson JK, Jabra-Rizk MA. Community-Associated Methicillin-Resistant Staphylococcus aureus: An Enemy amidst Us. PLoS Pathog. 2016;12(10):e1005837.
Crossref - Mama OM, Gomez-Sanz E, Ruiz-Ripa L, Gomez P, Torres C. Diversity of staphylococcal species in food producing animals in Spain, with detection of PVL-positive MRSA ST8 (USA300). Vet Microbiol. 2019;233:5-10.
Crossref - Monecke S, Skakni L, Hasan R, et al. Characterisation of MRSA strains isolated from patients in a hospital in Riyadh, Kingdom of Saudi Arabia. BMC Microbiol. 2012;12:146.
Crossref - Alrabiah K, Al Alola S, Al Banyan E, Al Shaalan M, Al Johani S. Characteristics and risk factors of hospital acquired – Methicillin-resistant Staphylococcus aureus (HA-MRSA) infection of pediatric patients in a tertiary care hospital in Riyadh, Saudi Arabia. Int J Pediatr Adolesc Med. 2016;3(2):71-77.
Crossref - Moussa IM, Hessan AM. Rapid detection of community acquired- methicillin resistance Staphylococcus aureus recovered from King Saudi Arabia. Afr J Microbiol. 2010;4(24):2804-2810.
- Al-Hamad AM, Alfaraj AA, Altowaileb J, et al. Incidence and antibiotic susceptibility of MRSA infections in a Saudi Arabian Hospital: a 10-year surveillance study. J Infect Dev Ctries. 2018;12(6):454-461.
Crossref - Raji MA, Garaween G, Ehricht R, Monecke S, Shibl AM, Senok A. Genetic Characterization of Staphylococcus aureus Isolated from Retail Meat in Riyadh, Saudi Arabia. Front Microbiol. 2016;7:911.
Crossref - Najar-Peerayeh S, Jazayeri Moghadas A, Behmanesh M. Antibiotic Susceptibility and mecA Frequency in Staphylococcus epidermidis, Isolated From Intensive Care Unit Patients. Jundishapur J Microbiol. 2014;7(8):e11188.
Crossref - Becker K, van Alen S, Idelevich EA, et al. Plasmid-Encoded Transferable mecB-Mediated Methicillin Resistance in Staphylococcus aureus. Emerg Infect Dis. 2018;24(2):242-248.
Crossref - McClure-Warnier JA, Conly JM, Zhang K. Multiplex PCR assay for typing of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Vis Exp. 2013;79:50779.
Crossref - Soares Casaes Nunes R, Mere Del Aguila E, Paschoalin VM. Safety Evaluation of the Coagulase-Negative Staphylococci Microbiota of Salami: Superantigenic Toxin Production and Antimicrobial Resistance. Biomed Res Int. 2015;2015:483548.
Crossref - Puah SM, Chua KH, Tan JA. Virulence Factors and Antibiotic Susceptibility of Staphylococcus aureus Isolates in Ready-to-Eat Foods: Detection of S. aureus Contamination and a High Prevalence of Virulence Genes. Int J Environ Res Public Health. 2016;13(2):199.
Crossref - Lee YD, Moon BY, Park JH, Chang HI, Kim WJ. Expression of enterotoxin genes in Staphylococcus aureus isolates based on mRNA analysis. J Microbiol Biotechnol. 2007;17(3):461-467.
- Chiang YC, Chang LT, Lin CW, Yang CY, Tsen HY. PCR primers for the detection of staphylococcal enterotoxins K, L, and M and survey of staphylococcal enterotoxin types in Staphylococcus aureus isolates from food poisoning cases in Taiwan. J Food Prot. 2006;69(5):1072-1079.
Crossref - Khairalla AS, Wasfi R, Ashour HM. Carriage frequency, phenotypic, and genotypic characteristics of methicillin-resistant Staphylococcus aureus isolated from dental health-care personnel, patients, and environment. Sci Rep. 2017;7(1):7390.
Crossref - Kariyama R, Mitsuhata R, Chow JW, Clewell DB, Kumon H. Simple and reliable multiplex PCR assay for surveillance isolates of vancomycin-resistant enterococci. J Clin Microbiol. 2000;38(8):3092-3095.
Crossref - McClure JA, Conly JM, Lau V, et al. Novel multiplex PCR assay for detection of the staphylococcal virulence marker Panton-Valentine leukocidin genes and simultaneous discrimination of methicillin-susceptible from -resistant staphylococci. J Clin Microbiol. 2006;44(3):1141-1144.
Crossref - Fessler AT, Kadlec K, Hassel M, et al. Characterization of methicillin-resistant Staphylococcus aureus isolates from food and food products of poultry origin in Germany. Appl Environ Microbiol. 2011;77(20):7151-7157.
Crossref - Iyer AP, Baghallab I, Albaik M, Kumosani T. Nosocomial infections in Saudi Arabia caused by methicillin resistance Staphylococcus aureus (MRSA). Clin Microbial. 2014;3(3).
- Islam MA, Parveen S, Rahman M, et al. Occurrence and Characterization of Methicillin Resistant Staphylococcus aureus in Processed Raw Foods and Ready-to-Eat Foods in an Urban Setting of a Developing Country. Front Microbiol. 2019;10:503.
Crossref - Bhedi KR, Nayak JB, Brahmbhatt MN, Roy A, Mathakiya RA, Rajpura RM. Detection and molecular characterization of methicillin-resistant Staphylococcus aureus obtained from poultry and poultry house environment of Anand district, Gujarat, India. Int J Curr Microbiol Appl Sci. 2018;7:867-872.
Crossref - Cuny C, Layer F, Hansen S, Werner G, Witte W. Nasal Colonization of Humans with Occupational Exposure to Raw Meat and to Raw Meat Products with Methicillin-Susceptible and Methicillin-Resistant Staphylococcus aureus. Toxins (Basel). 2019;11(4)190.
Crossref - Wang W, Lin X, Jiang T, et al. Prevalence and Characterization of Staphylococcus aureus Cultured From Raw Milk Taken From Dairy Cows With Mastitis in Beijing, China. Front Microbiol. 2018;9:1123.
Crossref - Kluytmans JAJW. Methicillin-resistant Staphylococcus aureus in food products: cause for concern or case for complacency? Clin Microbiol Infect. 2010;16(1):11-15.
Crossref - El-Ghareeb W, Almathen F, Fayez M, Alsultan R. Methicillin resistance Staphylococcus aureus (MRSA) in camel meat: Prevalence and antibiotic susceptibility. Slovenian Veterinary Research. 2019;56(22):249-256.
Crossref - Osman K, Alvarez-Ordonez A, Ruiz L, et al. Antimicrobial resistance and virulence characterization of Staphylococcus aureus and coagulase-negative staphylococci from imported beef meat. Ann Clin Microbiol Antimicrob. 2017;16(1):35.
Crossref - Elhassan MM, Ozbak HA, Hemeg HA, Elmekki MA, Ahmed LM. Absence of the mecA Gene in Methicillin Resistant Staphylococcus aureus Isolated from Different Clinical Specimens in Shendi City, Sudan. Biomed Res Int. 2015;2015:895860.
Crossref - Saeed K, Ahmad N, Dryden M, et al. Oxacillin-susceptible methicillin-resistant Staphylococcus aureus (OS-MRSA), a hidden resistant mechanism among clinically significant isolates in the Wessex region/UK. Infection. 2014;42(5):843-847.
Crossref - Adam KM, Abomughaid MM. Prevalence of Methicillin-resistant Staphylococcus aureus in Saudi Arabia Revisited: A Meta-analysis. Open Public Health J. 2018;11(1)584-591.
Crossref - Gade ND, Qazi MS. Fluoroquinolone therapy in Staphylococcus aureus infections: where do we stand? J Lab Physicians. 2013;5(2):109-112.
Crossref - Deurenberg RH, Stobberingh EE. The evolution of Staphylococcus aureus. Infect Genet Evol. 2008;8(6):747-763.
Crossref - Moussa IMI, Kabli SA, Hemeg HA, Al-Garni SM, Shibl AM. A novel multiplex PCR for molecular characterization of methicillin resistant Staphylococcus aureus recovered from Jeddah, Kingdom of Saudi Arabia. Indian J Med Microbiol. 2012;30(3)-296-301.
Crossref - Herrera FC, Garcia-Lopez ML, Santos JA. Short communication: Characterization of methicillin-resistant Staphylococcus aureus isolated from raw milk fresh cheese in Colombia. J Dairy Sci. 2016;99(10):7872-7876.
Crossref - Jin W, Yamada K. Staphylococcal enterotoxins in processed dairy products. Academic Press. 2016.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.