ISSN: 0973-7510
E-ISSN: 2581-690X
This study was aimed at isolating endophytic bacteria from Zea mays L. that have potential as phytase enzyme producers. Isolation and screening were performed using the Phytase Selective Media (PSM) medium. Bacteria that grew and formed a clear zone around the colony on the media were indicated as phytase-producer bacteria. The isolates with the highest phytanic index were further characterized based on morphology and physiological properties, then molecularly identified based on a 16S-rRNA analysis. Based on the results of Zea mays L. endophytic bacterial screening, bacterial isolate HF.7, was shown to be qualitatively able to produce phytase and can be developed as a phytase producer. Based on morphological and biochemical characterization, HF.7 belonged to the Burkholderia genus. Further molecular identification with 16S-rRNA analysis revealed that HF.7 isolate had 99% similarity with Burkholderia lata.
Endophytic bacteria, Phytase, Phytic Acid, Zea mays
Phytic acid (C6H18O24P6) is the main form of phosphorus storage in seeds as well as other plant tissues of cereal plants and is an anti-nutrient compound in poultry-based feed ingredients. Phytic acid compounds are chelated because of their constituent minerals such as calcium (Ca), iron (Fe), zinc (Zn), magnesium (Mg), manganese (Mn) and copper (Cu), and proteins. Phytic acid– protein and phytic acid–mineral complexes are insoluble and cannot be digested by monogastric animals, due to a lack of phytic acid hydrolysis enzyme in the gastrointestinal tract, thereby decreasing the nutrient value of feed ingredients consumed.
The negative effects of the lack of availability of phosphorus and other minerals bound by phytic acid can be reduced it by degrading the phytic acid bonds by hydrolysis via the phytase enzyme. Phytase (Myo-inositol hexakisphosphate phosphohydrolase) can hydrolyze the phosphodiester bonds of phytic acid, producing inorganic phosphate and phosphate ester1.
Phytase can be obtained from various sources, such as plants, fungi, bacteria, and the rumen of ruminant livestock. Bacterial phytase has a higher activity than phytase from animals and plants. Bacterial cells are relatively easier and faster to grow, can be grown independent of season, and have a uniform quality. Based on these advantages, further research for a phytase resource is very important because each enzyme source has different characteristics. A previous study showed that that the plant-derived phytase had a lower specific activity than microbe-derived ones. It is expected that phytase derived from microbes can be applied in the production of poultry feed2.
Utilization of microbial phytase in the production of monogastric animal feed has been reported in different studies by Barabara et al.3. The utilization of phytase is effective in increasing phosphorus availability and thus reducing phosphorus pollution in the environment. Several phytase enzymes from bacterial strains have been isolated, cloned, sequenced, and expressed, for example, from Escherichia coli, Bacillus sp., B. amyloliquefaciens, Lactobacillus amylovorus, Selenomonas ruminantium, Klebsiella pneumonia, K. oxitoca, K. aerogenes, and K. terrigena2, 4-7
The endophytic bacteria are a group of unique bacteria that have a natural habitat within the plant tissue. Various of their secondary metabolites have been exploited, but their use is still limited in animal husbandry. The potential of endophytic bacteria of Zea mays L. as phytase producers are based on the presence of phytic acid in all of the plant’s organs, such that associated endophytic bacteria should produce phytase to obtain phosphorus for their metabolic needs. This study aims to isolate and identify endophytic bacteria from Z. mays in an attempt to obtain the potential diversity of potential phytase enzyme sources.
The materials used in this study were Z. mays plant organs, including roots, stems, leaves, and seeds that were 80–110 days old. Other materials used were Luria Bertani (LB) Media, sterile distilled water, Phytase Selective Medium, universal primer (63F and 1387R), DNA template, DNA extraction kit (PrestoTM Mini gDNA Bacteria Kit), proteinase K, lysozyme, Gram(+) buffer, Gram(-) buffer, GB buffer, W1 buffer, wash buffer, elution buffer, absolute ethanol (96%), Master Mix PCR Kit, Tris borate EDTA (TBE) 10x, DNA ladder 100 bp, loading dye, agarose, ddH2O, ethidium bromide, gloves and mask, spirtus, 70% alcohol, 96% alcohol, sodium hypochlorite, and the gram staining reagents crystal violet, iodine, acetone alcohol, safranin, and methylene blue.
Isolation and Screening of Endophytic Bacteria producing Phytase
As noted above, Z. mays plant organs, including roots, stems, leaves as well as Z. mays seeds aged 80–110 days were used. After cleaning with tap water, organs were sterilized by immersion in sodium hypochlorite solution for 2 minutes, then 70% ethanol and 96% ethanol for 2 minutes each, then rinsed with sterile distilled water twice. Samples were cultivated on the media; if no colonies grow, the colonies grown from the sample of plant organs can be confirmed as endophytic bacteria. Each sample was crushed with mortar and pestle to obtain 10 g of sample powder. Samples were cultivated in LB broth medium, and incubated in a rotary shaker for 1 × 24 hours. The cultivation yield was serially diluted until 10-8, the last three dilutions (10-6, 10-7,10-8 bacteria/mL) were inoculated on LB agar medium at 37 ÚC for 24 hours. The growing colonies were then purified and screened on PSM selective media at 37 ÚC for 24 hours. Bacteria that grew and also formed a clear zone around the colony were indicated as phytase-producing bacteria. Isolates with a high phytic index (ratio of the highest diameter of the clear zone and bacterial colony diameter) were then selected for further identification.
Characterization and Identification of Bacteria
Isolates with the highest phytic index were characterized by macroscopic and microscopic observations and biochemical tests. Macroscopic observations included observation of colony size, pigmentation, shape, elevation, surface, and margin. Microscopic observations included Gram staining. Bacterial biochemical tests included carbohydrate fermentation reactions, IMVIC test (Indole, Methyl Red, Voges-Proskauer, Simmon’s citrate), catalase test, H2S reducing test, and motility test.
The selected isolates were identified molecularly using a 16S rRNA gene marker. Bacterial DNA was extracted using a Geneaid kit. The DNA was amplified using PCR Universal 63f primer and 1387r reverse primer. The PCR cycle consisted of 3 stages including denaturation, annealing, and extension. The pre-denaturation stage was performed at 94 °C for 2 minutes, then denaturation at 94 °C for 1 minute, annealing at 58 °C for 45 seconds, extension at 72 °C for 90 seconds for 35 cycles, followed by end extension temperature of 72 °C for 5 minutes and a final hold at 4 °C.
PCR results were observed using electrophoresis tools using a 100 bp marker to analyze the DNA bands of the sample. The result of electrophoresis was sequenced to find out their DNA base order for phylogenetic tree construction based on sequence database from GenBank. Samples were sequenced in 1st BASE sequencing INT Malaysia. The sequences cluster was analyzed by the BLAST (Basic Local Alignment Search Tool) program from the NCBI (National Center for Biotechnology Information) online at the website (http:// www.ncbi.nlm.nih.gov). Based on the sequence result, the phylogenetic tree was constructed using Neighbor-Joining Method to track the genetic relationship with other phytase-producer bacterial sequences.
A total of 21 endophytic bacteria that were isolated from roots, stems, leaves and seeds of Z. mays plants were screened on PSM selective media, and ten high-phytate isolates were selected for identification and further screening. The phytic index of each isolate is presented in Table 1.
Table (1):
Phytic Index of Endophytic Bacteria from each organ of Zea mays L.
No. |
Isolates Code |
Phytic Index (PI) |
|---|---|---|
1 |
Root 10-6 HF.1 |
1.00 |
2 |
Root 10-6 HF.5 |
1.09 |
3 |
Root 10-7 HF.7 |
1.37 |
4 |
Stem 10-6 HF.2 |
0.90 |
5 |
Stem 10-7 HF.2 |
1.09 |
6 |
Leaf 10-6 HF.2 |
1.21 |
7 |
Leaf 10-6 HF.3 |
1.36 |
8 |
Seed 10-6 HF.1 |
0.95 |
9 |
Seed 10-6 HF.3 |
0.93 |
10 |
Seed 10-8 HF.1 |
0.98 |
Based on Table 1, the highest phytic index was obtained from a bacterial isolate from Z. mays root, which is similar to previous studies. This is influenced by the physiology of plant roots that play a role in absorbing nutrients and naturally have a special mechanism in terms of nutrient distribution (phosphorus) that binds to phytic acid for plant growth. Phytic acid is hydrolyzed to be absorbed as phosphorus before entering the stems and leaves. The ability of endophytic microorganisms to produce secondary metabolites (enzymes) in accordance with the physiology of host plants8.
All of the selected isolates were Gram-negative cocci. This finding is similar to some studies suggesting that the bacteria producing phytase are generally Gram-negative. Biochemical test sequences, as well as the morphological characteristics and properties of Gram isolates, lead to the characteristics of the genus Burkholderia. Biochemical test results of selected isolates are presented in Table 2. The isolate of the highest phytase-producing bacteria, isolate HF.7, produced circular colonies on LB media, yellowish white color, raised colony elevation, entire edge, and crimped surface.
Table (2):
Biochemical test results of endophytic phytase-producer bacterial isolates from Zea mays L.
| No | Biochemistry Test | Test Result | |
|---|---|---|---|
| 1 | TSIA (Triple Sugar Iron Agar) Test | – | |
| 2 | H2S Test | – | |
| 3 | Motility Test | – | |
| 4 | Catalase Test | + | |
| 5 | Indole Test | – | |
| 6 | Methyl Red Test | – | |
| 7 | Voges Proskauer Test | – | |
| 8 | Citrate Test | + | |
| 9 | Carbohydrate Fermentation | Lactose | – |
| Mannitol | + | ||
| Glucose | + | ||
| Genus | Burkholderia | ||
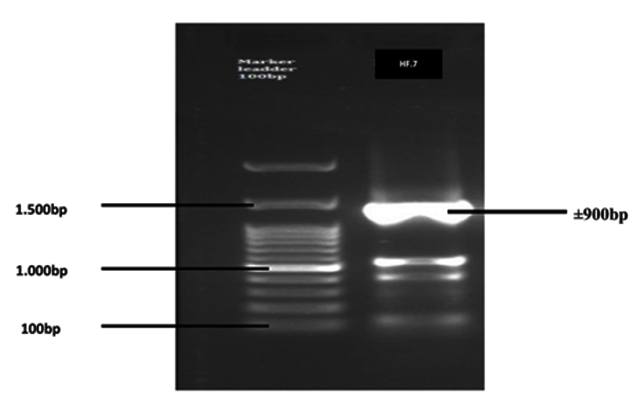
The results of electrophoresis of the 16S-rRNA gene amplification product from the HF.7 bacterial isolate are presented in Figure 1.
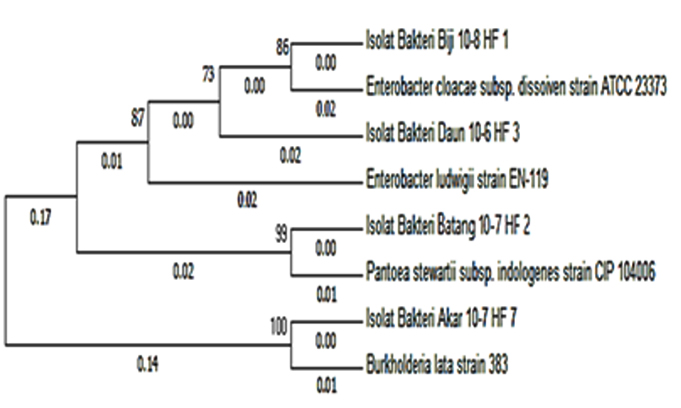
Based on the similarity between sample sequence and GenBank data, isolate of HF.7 bacteria has 99% similarity with Burkholderia lata. The results of the phylogenetic analysis showed that isolate HF.7 had the closest genetic relationship with Burkholderia lata strain 383 with 0.01 genetic distance (close) and 100 bootstrap value (Figure. 2). The bootstrap value is the parameter of the model data quality. If the bootstrap is low, then sequences should be excluded from the analysis to obtain a reliable phylogeny tree9.
Fig. 2. Phylogenetic tree of phytase-producing bacteria with Neighbor-Joining Method based on 16S-rRNA analysis
The Burkholderia genus was reported as an important endophytic bacterium in rice, Zea mays and sugarcane10. The Burkholderia genus was also reported as a phytase-producing bacteria11. Thus, B. lata has the potential to be further developed as phytase source by optimizing their production in vitro.
Based on the results of Zea mays endophytic bacteria screening, bacterial isolate HF.7, qualitatively had the ability as a phytase producer and had potential to be developed as phytase source. Morphological and biochemical characterization indicated HF.7 could be classified in the Burkholderia genus. Molecular identification analysis further indicated that HF.7 isolate had 99% similarity with B. lata.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
- Sari, E.S., Sajidan, Sugiyarto. Identifikasi bakteri penghasil phytase dan karakterisasi phytase dari Kawah Sikidang Dieng. El-Vivo., 2013; 1: 13-23.
- Sajidan, A., Farouk, A., Greiner, R., Jungblut, P., Muller, E.C., Borriss, R. Molecular and physiological characterization of a 3-phytase from soil bacterium Klebsiella sp ASR1. Appl. Microbiol. Biotechnol., 2004; 65: 110-118.
- Thomas, N.S (ed): What are Endophytes in Microbial Root Endophytes. Berlin: Springer-Verlag. 2013; pp 387.
- Shimizu, M. Purification and characterization of phytase from Bacillus subtilis (natto) N-77. Biosci. Biotechnol. Biochem., 1992; 56: 1266-1269.
- Greiner, R., Haller, E., Konietzny, U., Jany, K.D. Purification and characterization of a phytase from Klebsiella terrigena. Arch. Biochem. Biophys., 1997; 341: 201-206.
- Greiner, R., Konietzny, U., Jany, K.D. Purification and characterization of two phytases from Escherichia coli. Arch. Biochem. Biophys., 1993; 303: 107-113.
- Yanke, L.J., Selinger, L.B., Cheng, K.J. Phytase activity of Selenomonas ruminantium: a preliminary characterization. Lett. Appl. Microbiol., 1999; 29: 20-25.
- Radji, M. Peranan bioteknologi dan mikroba endofit dalam pengembangan obat herbal. Majalah Ilmu Kefarmasian., 2005; 3: 113-126.
- Dharmayanti, I.N.L.P. Filogenetika molekuler: metode taksonomi organisme berdasarkan sejarah evolusi. Makalah Wartazoa. 2011; 21: 1-10.
- Manzila, I., Priyatno, T.P., Fathin, M.F., Ambarsari, L., Suryadi, Y., Samudera, I.M., Susilowati, D.N. Karakterisasi Â-1,3-1,4-glukanasebakteri endofitik Burkholderia cepacia isolat 76 asal tanaman padi. Berita Biologi. 2015; 14: 143-153.
- Graminho, E.R., Naoki, T., Nakamura, A. Hoshino, T. Purification, biochemical characterization, and genetic cloning of the phytase produced by Burkholderia sp. strain a13. J. Gen. Appl. Microbiol. 2015; 61: 15-23.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.