ISSN: 0973-7510
E-ISSN: 2581-690X
The abscess underneath the skin is a common disease, which seriously affects the quality and yield of goat breeding. The main pathogens that cause abscesses are well understood, but the microbial community yet remains relatively unexplored. To determine the population and diversity of the microorganisms in the abscess underneath the skin of goats, in this work, 5 pus samples randomly collected from different goat farms (Jiangsu Province, China) were subjected to metagenomics sequencing and bioinformatics analysis. The test data show that the microbial communities of each sample contain about 79~82 kinds of microorganisms. Interestingly, each sample contained similar microbial species, including 53~59 kinds of bacteria, 5~6 fungi, 3 viruses, and 16~18 parasites. The top 5 dominant bacteria are Staphylococcus aureus, Lactococcus garvieae, Helicobacter pylori, Streptococcus pneumoniae, and Klebsiella pneumoniae, with an average abundance value of 29.88 %, 8.2%, 6.16%, 3.5%, and 3.26%, respectively. The remaining microbial abundances ranged from 0.01% to 3%. Although each of these frequent microorganisms is a tiny part of the total community, they constitute a major portion of individual reads (~1/2). In the conclusion, Staphylococcus aureus is the most dominant but nonunique bacterium responsible for the abscess underneath the skin of the goat, and the microbial community in the subcutaneous abscess is highly diverse. Bacterial coinfection should play an important role.
Goat, Abscess, Microorganism, Diversity
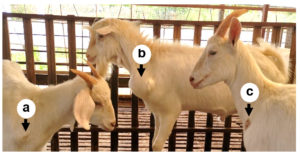
In recent years, sheep and goat raising in China has developed rapidly,1,2 but the breeding benefits are still affected by many digestive tract diseases, respiratory diseases, obstetric diseases, and various puzzling and incurable diseases.3 The abscess underneath the skin is a common disease in goats, especially in house feeding conditions. The incidence of the disease is 1% to 5% in many goat farms, and often more than 15% in some farms with poor sanitation. This disease is more found in goats, while sheep have a lower incidence. The main symptoms are subcutaneous swelling and maturation in the neck, chest, and abdomen (Figure 1), but generally did not show significant systemic symptoms. Early lump is like egg size and then gradually larger, round or oval, larger than the fist. In the late stages, the skin can rupture and ooze pus. The pus is thinner and light yellow-green in the early phase and later becomes sticky like bean residue. The ruptured region often can form a scab and clears without treatment, but in some cases can form a fistula because of the long flow of pus. This disease generally does not lead to the death of the goat but will affect its sales. The dominant bacteria that cause abscesses are well understood, but very few studies have described the microbial community of the abscess underneath the skin. To do a good job in the prevention and treatment, the pathogens of the disease should be revealed. Therefore, this work performed a metagenomic analysis of microbial communities on 5 samples from different goat farms in Jiangsu Province, China.
Sample Collection
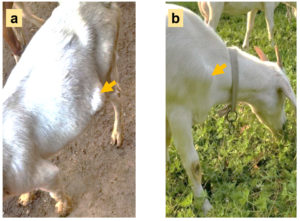
In August 2021, five pus samples were collected from 5 different goat farms (4 farms are house-feeding and another one is grazing locally) in Jiangsu Province, China. The number of goats in each farm is about 2000, and the incidence of subcutaneous abscess is 1% to 5%. The diseased goats with large subcutaneous abscess (Figure 2) were randomly picked out and the swollen areas were disinfected with iodophor diluent and alcohol (75%) after fixation. Following the abscess was cut open with a sterilized scalpel (Figure 3), about 2 ml of pus was sampled with a sterilized pipette and stored in a 5 ml EP tube. Following the EP tubes which contained pus were stored in the biosafety transport box (UN3373), all samples were sent to Suzhou Genomics Biotech Co., Ltd. (China) for metagenomics sequencing and bioinformatics analysis.
Figure 2. The diseased goats for sampling (photoed by Cheng D). a: An abscess located in the ventral side; b: An abscess located in front of the forelimb.
DNA Extraction
To investigate the microbial species contained in the abscess underneath the skin, genomic DNA from each sample was extracted by using HiPure Tissue DNA Kit (Shanghai Magen Biotech Co., Ltd, China). The extracted DNA concentration was tested by using Thermo NanoDrop 2000 Spectrophotometer (America), and the ratio of OD260/280, as well as that of OD260/230, was used to determine the purity of DNA.
Library Preparation and Metagenomic Sequencing
Following 200 μg genomic DNA was randomly fragmented by Covaris S220 Focused ultrasonicator (America) to an average size of 300-350bp, the sheared fragments were treated with End Prep Enzyme Mix (Beijing Biolab Technology Co., Ltd, China) for end repairing, 5’ phosphorylation and 3’ adenylation, to add adaptors to both ends. Size selection of Adaptor-ligated DNA was then performed by DNA Cleanup beads. Each sample was then amplified by PCR for 8 cycles using P5 and P7 primers, with both primers carrying sequences that can anneal to Flowcell to perform bridge PCR and P7 primer carrying a six-base index allowing for multiplexing. The PCR products were cleaned up and validated using an Agilent 2100 Bioanalyzer. The qualified libraries were sequenced pair-end PE150 on the Illumina HiseqXten/Novaseq/MGI2000 System.
Data Analysis
Raw shotgun sequencing reads were trimmed using Cutadapt (v1.9.1). Low-quality reads, N-rich reads, and adapter-polluted reads were removed. Then host contamination reads were removed. Samples were each assembled de novo to obtain separate assemblies. Whole-genome de novo assemblies were performed using Megahit (v1.1.3) with different k-mer. The best assembly result of the Scaffold, which has the largest N50, was selected for the subsequent analysis.
Prodigal (v3.02) software was to predict coding genes and then integrated the gene sequences of all samples. Use the sequence clustering software MMseq2 for further de-redundancy processing. By default, identity 95% and coverage 95% were used for clustering. Using the alignment software, SoapAligner (v2.21), we aligned the clean reads to construct nonredundant gene sets and obtained the number of reads aligned by unigene in each sample. Then, based on the number of reads and gene length in each unigene alignment, the abundance information of unigene in each sample was calculated.
Diamond (v0.8.15.77) was used to search the protein sequences of the unigenes in the NR database, CAZy database, eggNOG database, CARD database and KEGG database with E<10-5. The matched result with the best score was selected for annotation. To explore the microbial composition of the samples, we used Diamond to align the unigene sequences with the NR database and obtained the species annotation results of each sequence through the taxonomic annotation information corresponding to each sequence in the NR database. The abundance of a species in one sample is equal to the sum of the gene abundance annotated for the species.
The microbial species of the abscess were analyzed by using the Venn diagram (venn) method (Venn (cloudtutu.com)). The relative abundance maps of families, genera, and species of the microbial community were analyzed by Microsoft Excel. The Information on the microbial community was compared by using the PCOA (bray) method (PCOA (cloudtutu.com)) and the correlation coefficient between the microbial communities of the 5 goats was analyzed by Microsoft Excel.
Validity of Genomic DNA
The quality of the sequencing is not only affected by the sequencer itself and the sequencing reagent, etc., but also by the amount of the sample. Therefore, each pus sample in this research was provided more than 2 g, which was referred to the requirements for feces samples. The extracted DNA of each sample has an obvious main band, no degradation, and no impurities such as RNA and protein (OD260/280≥1.5, OD260/230≥1.0).
Diversity of Microorganisms in the Abscess Underneath the Skin of the Goat
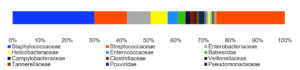
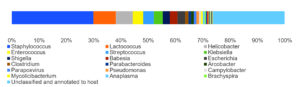
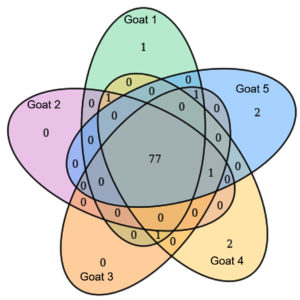
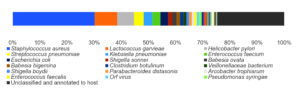
By analyzing the species classificational information corresponding to each sequence, the taxonomic level before the first branch is used as the species classification of the sequence. No removal of the host genome was performed at this time, so some genes were annotated to host or closely related species, such as Bovidae, Cervidae, Gekkonidae, Bos mutus, Bos taurus, Capra hircus, Bos indicus, Bubalus bubalis, and so on. After removing the host animal and closely related species, the microorganisms are mainly distributed in 12 families (Figure 4) and 18 genera (Figure 5). A total of 79~82 kinds of organisms were identified in the 5 cases (Figure 6), which suggested that the microbial community in the abscess underneath the skin of goats is highly diverse. Interestingly, each sample contained similar microbial species, including 53~59 species of bacteria, 5~6 fungi, 3 viruses, and 16~18 parasites. The top 5 dominant bacteria are Staphylococcus aureus, Lactococcus garvieae, Helicobacter pylori, Streptococcus pneumoniae, and Klebsiella pneumonia (Figure 7).
Figure 6. The microbial species in the 5 cases were analyzed by using the Venn diagram (venn) method.
Abundance of Microorganisms in the Abscess Underneath the Skin of Goat
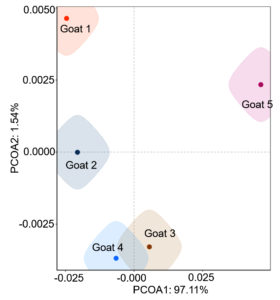
Identification of microbial species is key to disease diagnosis. Among the 5 samples tested, the microbial communities were almost identical (Figure 8, Table 1), for the correlation coefficients between the microbial communities of the 5 goats were over 99% (Table 2). There are 14 species of bacteria with an abundance of more than 1%, and 24 species with an abundance of 0.05% to 1%, and the remaining ones with an abundance of less than 0.05% respectively.
Table (1):
The abundance of microorganisms in each sample (%).
Taxon |
Goat 1 |
Goat 2 |
Goat 3 |
Goat 4 |
Goat 5 |
|---|---|---|---|---|---|
|
29.18 |
29.20 |
29.88 |
29.70 |
31.42 |
2. Lactococcus garvieae |
9.03 |
8.81 |
8.01 |
8.34 |
6.79 |
3. Helicobacter pylori |
6.78 |
6.58 |
6.00 |
6.37 |
5.08 |
4. Streptococcus pneumoniae |
3.45 |
3.44 |
3.50 |
3.46 |
3.66 |
5. Klebsiella pneumoniae |
3.16 |
3.20 |
3.27 |
3.27 |
3.42 |
6. Enterococcus faecium |
3.14 |
3.13 |
2.75 |
2.97 |
2.40 |
7. Escherichia coli |
2.62 |
2.72 |
2.96 |
2.73 |
2.95 |
8. Shigella sonnei |
1.71 |
1.70 |
1.55 |
1.59 |
1.25 |
9. Babesia ovata |
1.51 |
1.49 |
1.55 |
1.52 |
1.59 |
10. Babesia bigemina |
1.26 |
1.25 |
1.31 |
1.26 |
1.35 |
11. Clostridium botulinum |
1.19 |
1.19 |
1.23 |
1.21 |
1.29 |
12. Veillonellaceae bacterium DNF00626 |
1.13 |
1.15 |
1.04 |
1.05 |
0.85 |
13. Shigella boydii |
1.11 |
1.13 |
1.02 |
1.05 |
0.82 |
14. Parabacteroides distasonis |
1.09 |
1.05 |
0.95 |
1.03 |
0.81 |
15. Arcobacter trophiarum |
0.94 |
0.93 |
0.83 |
0.88 |
0.71 |
16. Enterococcus faecalis |
0.92 |
1.00 |
0.98 |
0.73 |
0.84 |
17. Orf virus |
0.92 |
0.92 |
0.97 |
0.95 |
0.99 |
18. Pseudomonas syringae |
0.85 |
0.85 |
0.88 |
0.86 |
0.93 |
19. Campylobacter coli |
0.70 |
0.70 |
0.74 |
0.73 |
0.77 |
20. Mycolicibacterium malmesburyense |
0.68 |
0.68 |
0.70 |
0.67 |
0.73 |
21. Anaplasma phagocytophilum |
0.63 |
0.65 |
0.65 |
0.67 |
0.67 |
22. Acinetobacter baumannii |
0.40 |
0.40 |
0.43 |
0.42 |
0.44 |
23. Oenococcus oeni |
0.39 |
0.39 |
0.36 |
0.36 |
0.30 |
24. Streptococcus agalactiae |
0.36 |
0.38 |
0.42 |
0.38 |
0.47 |
25. Brachyspira hampsonii |
0.32 |
0.32 |
0.33 |
0.32 |
0.34 |
26. Eggerthia catenaformis |
0.28 |
0.28 |
0.30 |
0.29 |
0.31 |
27. Dictyocaulus viviparus |
0.21 |
0.25 |
0.26 |
0.25 |
0.23 |
28. Clostridioides difficile |
0.17 |
0.18 |
0.18 |
0.19 |
0.19 |
29. Corynebacterium diphtheriae |
0.17 |
0.17 |
0.18 |
0.18 |
0.18 |
30. Neisseria meningitidis |
0.16 |
0.17 |
0.17 |
0.17 |
0.16 |
31. Brachyspira murdochii |
0.15 |
0.15 |
0.14 |
0.15 |
0.11 |
32. Haemonchus contortus |
0.13 |
0.14 |
0.13 |
0.14 |
0.15 |
33. Arcobacter defluvii |
0.10 |
0.10 |
0.11 |
0.11 |
0.11 |
34. Helicobacter equorum |
0.09 |
0.10 |
0.09 |
0.09 |
0.08 |
35. Penicillium italicum |
0.04 |
0.05 |
0.05 |
0.05 |
0.04 |
36. Candida intermedia |
0.04 |
0.04 |
0.03 |
0.04 |
0.03 |
37. Chlamydia trachomatis |
0.03 |
0.04 |
0.04 |
0.03 |
0.04 |
38. Enzootic nasal tumour virus of goats |
0.03 |
0.03 |
0.04 |
0.04 |
0.04 |
39. Mycobacterium tuberculosis |
0.03 |
0.03 |
0.03 |
0.04 |
0.04 |
40. Pseudomonas plecoglossicida |
0.03 |
0.03 |
0.03 |
0.03 |
0.03 |
41. Microbacterium esteraromaticum |
0.03 |
0.03 |
0.02 |
0.03 |
0.03 |
42. Monosiga brevicollis |
0.02 |
0.03 |
0.03 |
0.03 |
0.04 |
43. Staphylococcus pseudintermedius |
0.02 |
0.02 |
0.02 |
0.02 |
0.02 |
44. Bacillus sp VT-16-64 |
0.02 |
0.02 |
0.02 |
0.02 |
0.02 |
45. Onchocerca flexuosa |
0.02 |
0.02 |
0.02 |
0.02 |
0.02 |
46. Neisseria polysaccharea |
0.02 |
0.02 |
0.02 |
0.02 |
0.02 |
47. Lodderomyces elongisporus |
0.02 |
0.02 |
0.02 |
0.02 |
0.02 |
48. Trypanosoma congolense |
0.01 |
0.02 |
0.02 |
0.02 |
0.03 |
49. Human betaherpesvirus 6A |
0.01 |
0.02 |
0.02 |
0.02 |
0.03 |
50. Burkholderia pseudomallei |
0.01 |
0.02 |
0.02 |
0.02 |
0.02 |
51. Schizophyllum commune |
0.01 |
0.01 |
0.02 |
0.02 |
0.03 |
52. Lysobacter antibioticus |
0.01 |
0.01 |
0.02 |
0.02 |
0.01 |
53. Bacillus anthracis |
0.01 |
0.01 |
0.02 |
0.01 |
0.02 |
54. Clostridium symbiosum |
0.01 |
0.01 |
0.02 |
0.01 |
0.01 |
55. Campylobacter jejuni |
0.01 |
0.01 |
0.01 |
0.01 |
0.02 |
56. Salmonella enterica |
0.01 |
0.01 |
0.01 |
0.01 |
0.01 |
57. Vibrio parahaemolyticus |
0.01 |
0.01 |
0.01 |
0.01 |
0.01 |
58. Bacteroides sp An51A |
0.01 |
0.01 |
0.01 |
0.01 |
0.01 |
59. Haemophilus influenzae |
0.01 |
0.01 |
0.01 |
0.01 |
0.01 |
60. Naegleria gruberi |
0.01 |
0.01 |
0.01 |
0.01 |
0.01 |
61. Chlamydia abortus |
0.01 |
0.01 |
0.01 |
0.01 |
0.01 |
62. Mucor circinelloides |
0.01 |
0.01 |
0.01 |
0.01 |
0.01 |
63. Burkholderia multivorans |
0.01 |
0.01 |
0.01 |
0.01 |
0.01 |
64. Symbiodinium microadriaticum |
0.01 |
0.01 |
0.01 |
0.01 |
0.01 |
65. Enterobacter roggenkampii |
0.01 |
0.01 |
0.01 |
0.01 |
0.01 |
66. Brugia malayi |
0.01 |
0.01 |
0.01 |
0.01 |
0.01 |
67. Trichinella britovi |
0.01 |
0.01 |
0.01 |
0.01 |
0.01 |
68. Trichinella patagoniensis |
0.01 |
0.01 |
0.01 |
0.01 |
0.01 |
69. Trypanosoma brucei |
0.01 |
0.01 |
0.01 |
0.01 |
0.01 |
70. Salpingoeca rosetta |
0.01 |
0.01 |
0.01 |
0.01 |
0.01 |
71. Coniophora puteana |
0.01 |
0.01 |
0.01 |
0.01 |
0.01 |
72. Herbaspirillum sp VT-16-41 |
0.01 |
0.01 |
0.01 |
0.01 |
0.01 |
73. Trichinella sp T9 |
0.01 |
0.01 |
0.01 |
0.01 |
0.01 |
74. Besnoitia besnoiti |
0.01 |
0.01 |
0.01 |
0.01 |
0.01 |
75. Trichinella pseudospiralis |
0.01 |
0.01 |
0.01 |
0.01 |
0.01 |
76. Enterobacter mori |
0.01 |
0.01 |
0.01 |
0.01 |
0.01 |
77. Ciceribacter sp F8825 |
0.01 |
0.01 |
0.01 |
0.01 |
0.01 |
78. Clohesyomyces aquaticus |
0.01 |
0.01 |
– |
0.01 |
– |
79. Mycolicibacterium fortuitum |
0.01 |
– |
0.01 |
0.01 |
|
80. Enterobacter hormaechei |
0.01 |
– |
0.01 |
– |
0.01 |
81. Butyricicoccus pullicaecorum |
0.01 |
– |
– |
– |
– |
82. Trypanosoma cruzi |
– |
0.01 |
0.01 |
0.01 |
0.01 |
83. Bacillus cereus |
– |
– |
– |
0.01 |
– |
84. Neisseria subflava |
– |
– |
– |
0.01 |
– |
85. Trichuris suis |
– |
– |
– |
– |
0.01 |
86. Pyricularia oryzae |
– |
– |
– |
– |
0.01 |
Thereinto, Staphylococcus aureus is the most dominant bacterium (29.88% on average), followed by Lactococcus garvieae (8.20%), Helicobacter pylori (6.16%), Streptococcus pneumoniae (3.50%) and Klebsiella pneumoniae (3.26%). Although each of the remaining microorganisms is a tiny part of the total community, they constitute a major portion of individual reads (~1/3). Some viruses, fungi, and parasites have also been detected, but their abundance is low and their involvement in pathogenicity is debatable. More detailed data are shown in Table 1.
Table (2):
Correlation coefficient analysis between the microbial communities of the 5 pus samples.
Animal |
Goat 1 |
Goat 2 |
Goat 3 |
Goat 4 |
Goat 5 |
|---|---|---|---|---|---|
Goat 1 |
1 |
||||
Goat 2 |
0.999944 |
1 |
|||
Goat 3 |
0.998748 |
0.999168 |
1 |
||
Goat 4 |
0.999493 |
0.999710 |
0.999765 |
1 |
|
Goat 5 |
0.993805 |
0.994752 |
0.998042 |
0.996754 |
1 |
This research aimed to reveal the microbial community in the abscess underneath the skin of goats based on analyzing the abundance of various microorganisms. The tests identified Staphylococcus aureus as the main pathogen of this disease but did not exclude the involvement of other pathogens, such as Lactococcus garvieae, Helicobacter pylori, Streptococcus pneumoniae, Klebsiella pneumoniae, Enterococcus faecium, Escherichia coli, etc.
Among the remaining microorganisms and parasites, many are more likely to be animal skin flora or environmental microbes and take advantage of the opportunity of suppuration to invade tissues and constitute a major portion of the microbial community. Whether they play a role in the disease, and what role they play in the disease, remains a mystery to reveal.
Clinically, this disease is often misdiagnosed as Pseudotuberculosis (superficial form) by veterinarians, but Corynebacterium Pseudotuberculosis was not found in 5 samples. The main symptom of pseudotuberculosis is the enlarged and firm lymph nodes with purulent lymphadenitis,4-6 while the subcutaneous abscess in the study was usually free of lymph node lesions.
Lactococcus garvieae was first found in bovine mastitis,7 and can also be isolated from diseased fish.8,9 More importantly, it can also cause various infections in humans, such as sepsis, cardiometritis and osteomyelitis, septic hip arthritis and liver abscess,10 but no infections have been reported in goats. Helicobacter pylori is the only microbial species known to survive in the human stomach and is a class of carcinogens published by the World Health Organization’s International Agency for Research on Cancer.11,12 Shigella sonnei and Shigella boydii are the most common pathogens of bacterial dysentery in humans and primates.13,14 Some zoonotic bacteria (such as Lactococcus garvieae, Helicobacter pylori, and Shigella) are harbored in goat infection, but what role they play is unknown. Remarkably, this leads us to wonder if goats are the reservoirs of these microbes and pose a risk to humans.
This small pilot study suggested that S. aureus is the most dominant but nonunique bacterium responsible for the abscess underneath the skin of a goat, and the microbial community in the subcutaneous abscess is highly diverse. Bacterial coinfection should also play an important role. It remains difficult to precisely predict which species of bacteria might be found on a particular goat, but predicting which species are most frequent (or rare) seems more straightforward, at least for those species living in a subcutaneous abscess. Demonstrating the impact of bacteria on this disease may be of value in demonstrating the need to implement preventive healthcare strategies, and subsequently improve animal health and welfare.
ACKNOWLEDGMENTS
The authors would like to thank the five farms that allowed sample collection for use in this study.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
DC conceived and designed the study. DC, CC, XC and SZ performed the experiment. DC and JT wrote the paper. All authors read and approved the final manuscript.
FUNDING
This study was funded by the Earmarked Fund for Jiangsu Agricultural Industry Technology System (JATS[2021]492), and partially funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) (2018).
DATA AVAILABILITY
The datasets used and/or analysed in this study are available from the corresponding author on reasonable request.
ETHICS STATEMENT
This study did not require official or institutional ethical approval. The animals were handled according to high ethical standards and national legislation. The sample collection in these farms occurred under the direction of the farms’ usual vet as part of their routine veterinary herd health programme; therefore, no changes in animal treatment or handling occurred as a result of the sample collection required for this study.
- Li J, Jin H. Overview of China’s meat sheep industry in 2020 and development trends and suggestions for the future. Chi J Ani Sci. 2021; 57: 223–228. http://qikan.cqvip.com/Qikan/Article/Detail?id=7104141425 (in Chinese)
- Quan F, Pan J, He Q et al. Domestic Research and Developmental Prospects of Sheep and Goats for Meat Production. Chi Herb Sci. 2021; 41:57–60. http://qikan.cqvip.com/Qikan/Article/Detail?id=7103781708 (in Chinese)
- Cheng D, Zhang H. Color atlas of meat sheep feeding management and disease prevention. 1st ed. Liu W, editor. Chi Agri Press, Beijing, 2020. ISBN 978-7-109-25973-7 (in Chinese)
- Baird GJ, Fontaine MC. Corynebacterium pseudotuberculosis and its role in ovine caseous lymphadenitis. J Comp Pathol. 2007;137(4):179-210.
Crossref - Almeida S, Sousa C, Abreu V et al. Exploration of Nitrate Reductase Metabolic Pathway in Corynebacterium pseudotuberculosis. Int J Genomics. 2017;2017:9481756.
Crossref - Ruiz H, Ferrer LM, Ramos JJ et al. The Relevance of Caseous Lymphadenitis as a Cause of Culling in Adult Sheep. Animals. 2020;10(11):1962.
Crossref - Rodrigues MX, Lima SF, Higgins CH, Canniatti-Brazaca SG, Bicalho RC. The Lactococcus genus as a potential emerging mastitis pathogen group: A report on an outbreak investigation. J Dairy Sci. 2016;99(12):9864-9874.
Crossref - Vendrell D, Balcazar JL, Ruiz-Zarzuela I et al. Lactococcus garvieae in fish: a review. Comp Immunol Microbiol Infect Dis Comp Immunol Microbiol Infect Dis. 2006;29(4):177-198.
Crossref - Meyburgh CM, Bragg RR, Boucher CE. Lactococcus garvieae: an emerging bacterial pathogen of fish. Dis Aquat Organ Dis Aquat Organ. 2017;123(1):67-79.
Crossref - Gibello A, Galan-Sanchez F, Blanco MM et al. The zoonotic potential of Lactococcus garvieae: An overview on microbiology, epidemiology, virulence factors and relationship with its presence in foods. Res Vet Sci Res Vet Sci. 2016;109:59-70.
Crossref - Veldhuyzen van Zanten SJ, Sherman PM. Helicobacter pylori infection as a cause of gastritis, duodenal ulcer, gastric cancer and nonulcer dyspepsia: a systematic overview. CMAJ. 1994;150:177-185.
- Arif M, Syed S. Association of Helicobacter pylori with carcinoma of stomach. J Pak Med Assoc. 2007;57(7):337-341. PMID: 17867254
- Torraca V, Holt K, Mostowy S. Shigella sonnei. Trends Microbiol Trends Microbiol. 2020;28(8):696-697.
Crossref - Peng JP, Yang J, Jin Q. Research progress in Shigella in the postgenomic era. Sci China Life Sci Sci China Life Sci. 2010;53(11):1284-1290.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.