ISSN: 0973-7510
E-ISSN: 2581-690X
The rise of multidrug-resistant bacterial species in hospitals becomes a global challenge for surgeons who treat healthcare-associated infections. This study aimed to identify the pathogens involved in surgical site infections (SSI) as well as the prevalence of antibiotic resistant bacteria in the Nilgiris region. A hospital-based retrospective study was conducted for three years, at Microbiology Laboratory, the Govt. Medical College Hospital, where the clinical samples were collected, cultured, and identified. Antibiotic susceptibility was assessed using Kirby Bauer’s disc diffusion method. Out of 513 pus samples (from SSI), 242 (47%) have shown positive microbial growth. These isolates were evaluated for antimicrobial resistance using 20 antibiotics belonging to different groups. Staphylococcus aureus was found to be more prominent (69%), followed by Enterococcus species (14.5%) and Streptococcus species (10.3%). Other species like Proteus species, Klebsiella species, Escherichia coli, and Pseudomonas aeruginosa account for less than 2%. These results clearly indicate that Staphylococcus aureus was the leading cause of surgical site infections. Among the antibiotics studied, Staphylococcus aureus was found to be more resistant to Penicillin G (84%) followed by Ampicillin (23%). The high rate of antibiotic resistance highlighted the need for an antibiotic policy that encourages more rational use of antibiotics.
Surgical Site Infections, S. aureus, Antibiotic Resistance, Antibiotic Policy, Multidrug Resistance
Surgical site infections (SSIs) are the highly conventional type of nosocomial infection. These infections can range in intensity from annoying to life-threatening and can cause a lot of pain for patients. A high percentage of SSIs may be avoided and preventing them is a critical patient safety issue that necessitates collaboration with healthcare professionals.1 Surgical infection treatment remains an urgent concern as it is a primary reason for nosocomial mortality and morbidity.2 In surgical patient populations in countries with limited resources, SSIs are most prevalent, affecting up to 66 percent of patients who have undergone surgery which is nine times more when compared to industrialized countries.3 SSIs lengthen postoperative hospital stay, increased healthcare costs, and increase the rate of readmissions.4 Drug-resistant SSIs are also becoming a significant issue in emerging countries like India, owing to congested hospital environments, irrational antibiotic prescriptions, and inadequate infection prevention and control systems.5
Surgical site infections (SSIs)
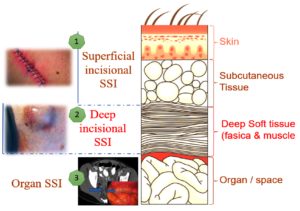
Establishing explicit definitions for SSI cases are crucial in assisting the effective surveillance of SSIs. The Centers for Disease Control and Prevention (CDC), USA, established the National Healthcare Safety Network (NHSN) to track quality measures, including SSIs, and has established commonly accepted criteria for SSIs.6 The classification of SSI is based on the extent of the infection involvement, which may be restricted to the subcutaneous and skin tissues (surface level incisional SSI), necessitate deeper soft tissue, like the superficial and deep and muscular layers (deep incisional SSI), or go beyond these anatomical limits (organ/space SSI). Figure 1 represents the types of SSI in different layers of body.7
Epidemiology
Surgical infection rates were quite high prior to the antisepsis period, making surgery extremely risky. Initial surgical methods had restricted success compared to innovative surgical operations due to the lack of appropriate pre-antibiotic therapy. The acceptance of the sterile technique had a substantial influence on the results. Semmelweis’ easy introduction of handwashing resulted in a reduction in puerperal sepsis mortality from 12 percent to 2%.8 The evolution of numerous facets of modern surgical treatment has resulted in considerable advancements. SSIs, however, remains to be a frequent postoperative problem, happening in 3% to 20% of surgical treatments. SSI rates can be much higher depending on the amount of risk variables present and can vary greatly depending on the operating strategy.9 SSI has a considerable effect on mortality as well as morbidity. However, given of the reliance on the quality of reporting and the heterogeneity of patient follow-up, proving the precise effect of SSI is challenging.
The significant sources of morbidity in surgical patients are microorganisms. The initial stage in the formation of an SSI is microbial contamination of the surgical site, which can occur from either endogenous or external sources. The patient’s epidermis, hollow viscera and mucous membranes, all contain endogenous bacteria. S. aureus, coagulase-negative staphylococci, Enterococcus, and Escherichia coli are the most frequent endogenous contributing pathogens. However, a lot will rely on the surgical procedure done. Whereas the Exogenous flora, such as surgical instruments, air, supplies, and operating personnel, may originate in the operating theater room. The most prevalent external microbes are streptococci and staphylococci. 10
It was discovered in the 1980s that surgical infections increased the hospital stays by 10 days.11 Even after 15 years, a new study described continued prolonged discharge from the hospital and the need for post-discharge care had grown.9 In-hospital mortality was 14.5% for patients with SSIs in a study of 288,906 individuals compared to 1.8% for patients without SSIs. According to estimates, SSIs cause more than 8000 fatalities per year in the United States.12 Surgical infections may be of much superior concern in emerging nations since surveillance levels of these surgical infections in research done by the International Nosocomial Infection Control Consortium have been greater for major surgical methods contrasted with CDC-NHSN rates.13
Antibiotic Resistance
In a study conducted at a tertiary care teaching hospital in India, it was found that the isolated SSI bacteria reacted differently to different drugs. Enterobacteriaceae species including E. coli and Klebsiella had a very high level of resistance to first-generation cephalosporins and penicillins. A significant level of resistance to tetracycline (46-51%), amoxicillin-clavulanic acid (38-43%), co-trimoxazole (37-45%), and gentamicin (45-48%) was also observed. Staphylococcus species showed significant resistance to these antibiotics. Low resistance was documented against macrolides (15–17%) and clindamycin (0–15%). However, 20% of enterococci were found to be vancomycin-resistant.14
In another study on antimicrobial resistance, carried out at a public hospital in Tanzania, 63% of the isolates were multidrug resistant. In contrast, a comparable investigation carried out in Uganda’s national hospital revealed that 78% of the bacterial isolates from surgical site infection were multidrug-resistant.15 Similar resistance-related differences in surgical outcomes are anticipated to emerge, or may already exist, in the United States, but have not been studied.16
Healthcare facilities are unable to receive accurate information about bacteria resistant to antimicrobial medicines due to insufficient SSI surveillance efforts. Antibiotics that are effective are costly and difficult to obtain, hence broad-spectrum antibiotics are frequently used which leads to alarming resistance rates.17 To create locally pertinent data and guide experiential treatment in areas wherever microbial identification and antibiotic susceptibility testing procedures and facilities are infrequent, a retrospective study was conducted to isolate and identify the causative pathogen involved in these surgical infections as well as the rate of antibiotic-resistant bacteria in the Nilgiris region.
Study site
District microbiology laboratory, Govt. Medical College Hospital, Ooty to understand the pathogenicity and resistance trends of SSI-causing clinical isolates. The teaching hospital is one of the pioneering tertiary care hospitals in the Nilgiris region, which serves more than 0.5 million people in and around the Nilgiris region. And the Microbiology Laboratory is the district-level Laboratory that tests the clinical samples obtained from different health centers from the Nilgiris region.
Study duration
Three years from January 2019 to December 2021.
Study design
This is a hospital-based, retrospective study conducted by retrieving the laboratory records of Three years from January 2019 to December 2021 at the District microbiology laboratory, Govt. Medical College Hospital, Ooty to understand the pathogenicity and resistance trends of SSI-causing clinical isolates.
Study criteria
In this retrospective study laboratory records were collected for the patients who are diagnosed with surgical site infection, aged between 18-75 years, with comorbid conditions (if any) were included, whereas pregnant women, pediatric age group, patients who are suffering from psychiatric and behavioral problems data were excluded in the study.
Sample collection
The pus samples were collected by sterile syringe aspiration and by sterile swabs from inpatients of different wards. Based on the degree of SSI samples have been collected from superficial or deep tissues of the infected site. Around 513 clinical samples were collected from the patients who developed an infection at the site of surgery. A wound was considered an SSI if it developed within 30 days after surgery and had at least one of the following symptoms: purulent discharge from the incision, redness, discomfort or pain, local edoema, foul odour, wound abscess, or fever.18
Media used
Clinical samples were inoculated on MacConkey agar, blood agar, and Mannitol salt agar, The samples were aseptically inoculated on blood agar (with 5% sheep blood) and MacConkey agar plates, incubated aerobically at 35°C–37°C for 24–48 hr.
Identification methods
The clinical isolates were characterized by means of regular bacteriological identification methods such as colony morphology, microscopic examination, and biochemical assays as per Bergey’s manual of systematic bacteriology, such as the MR-VP test, indole test, citrate utilization test, urease test, TSIA test, and dry spot agglutination test.19
Antibiotics susceptibility
All the clinical isolates were evaluated for antibiotic resistance/susceptibility using the revised Kirby-Bauer disc diffusion procedure using Muller Hinton agar medium and respective antibiotics discs according to the Clinical and Laboratory Standards Institute (CLSI) guidelines available during the study period.
Statistical analysis
Statistical assessment was made with the help of Microsoft Excel software to determine the association of SSI incidence in patients of various age groups and genders, and rates of antibiotic resistance.
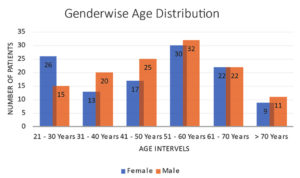
From the retrieved data it was observed that, out of 513 clinical samples obtained from different health centers of the Nilgiris region, 242 (47%) have shown positive microbial growth. Out of these 242 samples, 117 were female (48%) and 125 were male (52%). Similarly, the distribution of Surgical site infection in the different age groups analyzed. The age range 51–60 years had the highest infection rate (25.6%), followed by the age range 41–50 years (17.4%), the distribution of infection rates in different age groups are looking similar, except for the age group above 70 years, whose infection rate is around 8.3%, this may be due to low rates of Geriatric surgeries in the Nilgiris region because of risk factors and majority of SSI are nosocomial infection. These incidence rate of SSIs are quite similar to findings observed in the Mukagendaneza et al. study.20 The distribution of SSI age-wise is represented graphically in Figure 2.
Nature of Clinical Isolates
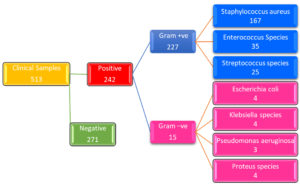
The Gram’s nature of clinical isolates shows that 94% are Gram-positive and 6% found to be Gram-negative. Among 242 clinical isolates, Staphylococcus aureus was found to be more prominent with 69% (n=167), followed by Enterococcus species and Streptococcus species with 14.5% (n=35) and 10.3% (n=25) respectively, along with a few other species like Proteus species, Klebsiella species, Escherichia coli, and Pseudomonas aeruginosa which account for less than 2%. Figure 3 shows the flowchart of clinical isolates and positive cultures, Gram’s nature, and respective clinical isolates with their number.
Figure 3. Flowchart showing number of samples, Clinical Isolates, Gram’s Nature, and respective number of isolates
Antimicrobial resistance
Antimicrobial resistance studies were performed on 20 antibiotics prescribed during the study period on these clinical isolates, Table 1 shows the antibiotic resistance patterns Gram positive clinical isolates obtained from clinical samples, whereas Table 2 shows Gram negative clinical isolates. Percentage of Resistance are calculated by Number of Resistant strains / Total number of Isolates tested X 100.
Table (1):
Antibiotic resistance Patterns of Gram positive Clinical Isolates of SSI.
| Staphylococcus aureus (n = 167) | Enterococcus Species (n = 35) | Streptococcus species (n = 25) | ||||
|---|---|---|---|---|---|---|
| Number | Percentage | Number | Percentage | Number | Percentage | |
| Amikacin | 31 | 19 | 28 | 80 | 11 | 43 |
| Ampicillin | 39 | 23 | 19 | 56 | 17 | 67 |
| Ceftazidime | 31 | 19 | 14 | 40 | 0 | 0 |
| Cefixime | 24 | 14 | 0 | 0 | 0 | 0 |
| Cefotaxime | 24 | 14 | 6 | 17 | 4 | 16 |
| Cefoxitin | 56 | 33 | 0 | 0 | 25 | 100 |
| Ceftriaxone | 67 | 40 | 35 | 100 | 0 | 0 |
| Clindamycin | 46 | 27 | 21 | 60 | 17 | 67 |
| Ciprofloxacin | 54 | 33 | 35 | 100 | 0 | 0 |
| Cotrimoxazole | 32 | 19 | 6 | 17 | 4 | 17 |
| Erythromycin | 42 | 25 | 9 | 25 | 0 | 0 |
| Gentamycin | 23 | 14 | 14 | 39 | 5 | 18 |
| Imipenem | 30 | 18 | 9 | 25 | 7 | 29 |
| Linezolid | 37 | 22 | 12 | 33 | 13 | 53 |
| Meropenem | 13 | 08 | 5 | 13 | 4 | 15 |
| Penicillin-G | 140 | 84 | 26 | 75 | 0 | 0 |
| Piperacillin/ Tazobactam | 121 | 73 | 0 | 0 | 0 | 0 |
| Tetracycline | 41 | 24 | 18 | 50 | 13 | 54 |
| Teicoplanin | 46 | 27 | 23 | 67 | 0, 0 | 0 |
| Vancomycin | 89 | 53 | 23 | 67 | 0, 0 | 0 |
Table (2):
Antibiotic resistance Patterns of Gram negative Clinical Isolates of SSI.
| Escherichia coli (n = 4) |
Klebsiella species (n = 4) |
Pseudomonas aeruginosa (n=3) |
Proteus species (n = 4) |
|||||
|---|---|---|---|---|---|---|---|---|
| Number | Percentage | Number | Percentage | Number | Percentage | Number | Percentage | |
| Amikacin | 1 | 25 | 0 | 0 | 2 | 67 | 11 | 43 |
| Ampicillin | 0 | 0 | 0 | 0 | 3 | 100 | 17 | 67 |
| Ceftazidime | 0 | 0 | 4 | 100 | 0 | 0 | 0 | 0 |
| Cefixime | 4 | 100 | 0 | 0 | 0 | 0 | 0 | 0 |
| Cefotaxime | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 16 |
| Cefoxitin | 0 | 0 | 0 | 0 | 0 | 0 | 25 | 100 |
| Ceftriaxone | 0 | 0 | 4 | 100 | 0 | 0 | 0 | 0 |
| Clindamycin | 0 | 0 | 0 | 0 | 3 | 100 | 17 | 67 |
| Ciprofloxacin | 4 | 100 | 0 | 0 | 0 | 0 | 0 | 0 |
| Cotrimoxazole | 0 | 0 | 0 | 0 | 2 | 67 | 4 | 17 |
| Erythromycin | 0 | 0 | 3 | 75 | 0 | 0 | 0 | 0 |
| Gentamycin | 2 | 50 | 0 | 0 | 0 | 0 | 5 | 18 |
| Imipenem | 0 | 0 | 4 | 100 | 0 | 0 | 7 | 29 |
| Linezolid | 0 | 0 | 0 | 0 | 0 | 0 | 13 | 53 |
| Meropenem | 0 | 0 | 0 | 0 | 2 | 67 | 4 | 15 |
| Penicillin-G | 4 | 10 | 0 | 0 | 3 | 100 | 0 | 0 |
| Piperacillin/
Tazobactam |
0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Tetracycline | 3 | 75 | 2 | 50 | 0 | 0 | 13 | 54 |
| Teicoplanin | 1 | 25 | 0 | 0 | 0 | 0 | 0, 0 | 0 |
| Vancomycin | 0 | 0 | 4 | 100 | 3 | 100 | 0, 0 | 0 |
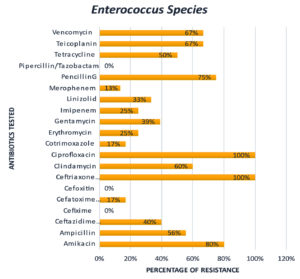
From these results, it’s clear that 84% of Staphylococcus aureus isolates are got resistance to Penicillin G, 73% of this got resistance towards Piperacillin/Tazobactam, and 53% towards Vancomycin. It is also observed that Enterococcus species got 100% resistance toward Ceftriaxone, Ciprofloxacin, and 80% resistance toward Amikacin.
From these results, it is evident that none of these clinical isolates got resistant to Piperacillin / Tazobactam, except 73% of Staphylococcus aureus species. Majority of the clinical isolates namely Staphylococcus aureus – 19%, Enterococcus Species – 80%, Streptococcus species – 43%, Escherichia coli – 25%, Proteus species – 50%, and Pseudomonas aeruginosa – 67% got resistance to Amikacin.
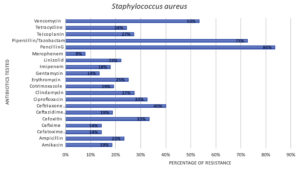
Among the 242 clinical isolates, majority of the isolates found to be Staphylococcus aureus which counts for 167. Figure 4, graphically represents the resistance patterns of Staphylococcus aureus, through which it is clearly evident that Staphylococcus aureus acquired resistance majority of the antibiotics.
Figure 5 and 6 graphically represents the resistance patterns of the other two major clinical isolates of Surgical site infection namely Enterococcus Species and Streptococcus species, respectively. These results clearly indicate that these two bacteria also attained multidrug resistance. From Figure 5 it is clear that Enterococcus Species is still susceptible to Piperacillin/Tazobactam, Cefoxitin and Cefixime. But it got 100% resistance to Ciprofloxacin and Ceftriaxone.
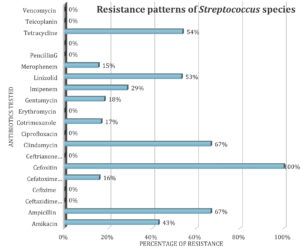
From Figure 6, it is clearly understood that Streptococcus species got resistance towards many of the antibiotics and attained 100% resistance to Cefoxitin. It is still susceptible to the antibiotics like Vancomycin, Teicoplanin, Penciling G, Erythromycin, Ciprofloxacin, Cefixime etc.
Antimicrobial-resistant surgical site infections are developing a substantial health care issue in the intensive care unit and other areas of health care. It also increases length of stay, healthcare expenditures, mortality, and morbidity. The epidemiology and antibiotic resistance characteristics of Nosocomial infections revealed differences amongst hospitals around the world.21 Majority of the infections are caused by bacteria that are multi-drug resistant (MDR). This study determined the fraction of SSIs caused by various clinical isolates from surgical site infection specimens.
In this study 47% of patients who underwent surgery during the study period got infected, i.e., 242 patients out of 513 tested. Comparatively this number is lower than a comparable study performed by Yehouenou et al.22 in six public hospitals in Benin (West African nation). Many of the clinical isolates identified in our study were found to be MDR. Several variables might have led to the high prevalence of MDR. According to Magiorakos et al. 23 MDR species means any organism which has attained resistance to at least one antibiotic from three or more antibiotic groups.
The first is most likely related to an absence of antibiotic resistance observation and stewardship programs. There is enough evidence to show that such programs help in both identifying the pattern of resistance and avoiding the development of antibiotic resistance by increasing antibiotic usage. The second explanation might be due to a lack of a thorough antibiotic policy on the usage of antimicrobial agents. Instead, buying antibiotics, especially broad-spectrum antibiotics, from independent drug stores and pharmacies without a prescription is common. Deficiency of diagnostic laboratory facilities before the prescription of antibiotics by medical professionals who lack an antibiogram or evidence of the etiologic agent may be the third reason.
Staphylococcus aureus (69%) and Enterococcus species (15%) were the most prevalent isolates in SSIs. These findings are consistent with past research that has linked S. aureus to Surgical site infections.22,24 S. aureus is a commensal bacterium of the skin that may readily infect a surgical wound.25 According to Alverdy et al.5 Staphylococcus aureus is the bacterium most usually linked with a prosthetic-related hip SSI, and it is assumed to emanate from the skin near to the surgery site.
In the current study, Staphylococcus aureus is found to be Multidrug-resistant, with 84% of these species got resistance towards Penicillin-G, 73% for Piperacillin/Tazobactam, 53% for Vancomycin, 40% for Ceftriaxone and 33% for Cefoxitin and Ciprofloxacin. MDR bacteria are the bacteria that were found resistant to at least one antibiotic from three or more distinct antibiotic groups. While S. aureus high resistance to penicillin has been recorded in Uganda and Nepal.15,24 Whereas low levels of resistance were observed with Meropenem and Gentamycin in 8% and 14 % of S. aureus species respectively. The developing pattern of microbial resistance, along with concerns about the sensitivity and efficacy of currently available anti-MRSA medications, restricts the therapeutic choices accessible to patients with SSIs. Alalevonadifloxacin and Levonadifloxacin are innovative benzoquinolizine anti-MRSA medicines which have just been licensed in India to treat gramme positive ‘super-bugs’.26
India, like many other countries, lacks a well-controlled antibiotic-prescription system, making antibiotic abuse particularly simple.27 Our study findings subsequently constitute an urgent call to monitor and optimize antibiotic usage in that setting. Our first suggestion is to use a multidisciplinary approach to SSI management that includes doctors, infection-control professionals, microbiologists, and pharmacists. Second, enhancing laboratory and diagnostic services at the local and national levels would guarantee efficient antibiotic resistance surveillance. Finally, to reduce the transmission of MDR, we propose rigorous adherence to appropriate infection-prevention control methods, notably hand hygiene and inanimate surface cleaning.
Microbial isolates from Surgical Site Infections in the Nilgiris region majorly consisted of Gram positive Staphylococcus aureus pathogens and these were mostly resistant to frequently used antimicrobial drugs. Several other species including Gram negative isolates shown similar trend of resistance towards many antibiotics. This causes concerns regarding the effectiveness of the antimicrobial medications now used for surgical prophylaxis and therapy, and it may help determine the best course of treatment for SSIs in the hospital settings of the Nilgiris region.
This study helps identify how common drug-resistant bacteria are in surgical site infections in the Nilgiris region. None of these clinical isolates had a wild-type phenotype that was sensitive. These results highlight the urgent need for high-activity medications, the prudent use of antibiotics, and rigorous adherence to great hand hygiene standards in order to stop the development of multidrug-resistant bacteria in the Nilgiris region. Building surveillance programmes that reduce the prevalence of surgical site infections is essential, despite the fact that antimicrobial resistance research in India is still in its early phases.
ACKNOWLEDGMENTS
The authors would like to thank JSS College of Pharmacy in Ooty and JSS Academy of Higher Education & Research in Mysuru, who provided the researchers with all the tools they needed to complete this study. Authors are also grateful to the doctors, consultants, and microbiologists from Govt. Medical College & Hospital, Ooty, who provided the essential inputs for retrospective data collection.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This study was supported by the Department of Science and Technology (DST), New Delhi, with the WOS-B fellowship (DST/WOS-B/HN-5/2021).
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee, Institutional Review Board of the University of Gondar, India with approval number JSSCP/IEC/06/2019-20 dated 10th January 2019.
- Seidelman J, Anderson DJ. Surgical Site Infections. Infect Dis Clin North Am. 2021;35(4):901-929.
Crossref - Young PY, Khadaroo RG. Surgical Site Infections. Surg. Clin North Am. 2014;94(6):1245-1264.
Crossref - Singh R, Singla P, Chaudhary U. Surgical site infections: classification, risk factors, pathogenesis, and preventive management. Int J Pharm Res Health Sci. 2014;2(3):203-214.
- Chavan AR, Kelkar V. Study of healthcare-associated infections in surgical unit in a newly established tertiary care hospital of Nanded, Maharashtra, India. Int J Sur Open. 2017;9:30-35.
Crossref - Alverdy JC, Hyman N, Gilbert J. Re-examining causes of surgical site infections following elective surgery in the era of asepsis. Lancet Infect Dis. 2020;20(3):e38-e43.
Crossref - Horan T, Gaynes R, Martone W, Jarvis W, Graceemori T. CDC definitions of nosocomial surgical site infections, 1992: A modification of CDC definitions of surgical wound infections. Am J Infect Control. 1992;20(5):271-274.
Crossref - Mangram AJ, Horan TC, Pearson ML, Silver LC, Jarvis WR. Guideline for Prevention of Surgical Site Infection, 1999. Am J Infect Control. 1999;27(2):97-134.
Crossref - Nespoli A, Geroulanos S, Nardone A, Coppola S, Nespoli L. The History of Surgical Infections. Surg Infect. 2011;12(1):3-13.
Crossref - Klevens RM, Edwards JR, Richards CL, et al. Estimating Health Care-Associated Infections and Deaths in U.S. Hospitals, 2002. Pub Health Rep. 2007;122(2):160-166.
Crossref - Owens CD, Stoessel K. Surgical site infections: epidemiology, microbiology and prevention. J Hosp Infect. 2008;70(Suppl 2):3-10.
Crossref - Cruse PJE, Foord R. The Epidemiology of Wound Infection: A 10-Year Prospective Study of 62,939 Wounds. Surg Clin North Am. 1980;60(1):27-40.
Crossref - DiPiro JT, Martindale RG, Bakst A, Vacani PF, Watson P, Miller MT. Infection in surgical patients: effects on mortality, hospitalization, and postdischarge care. Am J Health Syst Pharm. 1998;55(8):777-781.
Crossref - Rosenthal VD, Richtmann R, Singh S, et al. Surgical Site Infections, International Nosocomial Infection Control Consortium (INICC) Report, Data Summary of 30 Countries, 2005-2010. Infect Control Hosp Epidemiol. 2013;34(6):597-604.
Crossref - Deka S, Kalita D, Mahanta P, Baruah D. High Prevalence of Antibiotic-Resistant Gram-Negative Bacteria Causing Surgical Site Infection in a Tertiary Care Hospital of Northeast India. Cureus. 2020;12(12):e12208.
Crossref - Hope D, Ampaire L, Oyet C, Muwanguzi E, Twizerimana H, Apecu RO. Antimicrobial resistance in pathogenic aerobic bacteria causing surgical site infections in Mbarara regional referral hospital, Southwestern Uganda. Sci Rep. 2019;9(1).
Crossref - Long DR, Alverdy JC, Vavilala MS. Emerging paradigms in the prevention of surgical site infection: the patient microbiome and antimicrobial resistance. Anesthesiology. 2022;137(2)252-262.
Crossref - Rihana BP, Wadhwani A, Balasubramaniam V, Ponnusankar S. Antimicrobial Resistance and Implications: Impact on Pregnant Women with Urinary Tract Infections. J Pure Appl Microbiol. 2022;16(2):769-781.
Crossref - Lakoh S, Yi L, Sevalie S, et al. Incidence and risk factors of surgical site infections and related antibiotic resistance in Freetown, Sierra Leone: a prospective cohort study. Antimicrob Resist Infect Control. 2022;11(1):39.
Crossref - Cheesbrough M. District Laboratory Practice in Tropical Countries. Part 2. Cambridge University Press; 2006.
Crossref - Mukagendaneza MJ, Munyaneza E, Muhawenayo E, et al. Incidence, root causes, and outcomes of surgical site infections in a tertiary care hospital in Rwanda: a prospective observational cohort study. Patient Saf Surg. 2019;13(10).
Crossref - Asres GS, Hailu MH, Woldearegay GM. Prevalence of Multidrug Resistant Bacteria in Postoperative Wound Infections at Tikur Anbessa Specialized Hospital, Addis Ababa, Ethiopia. Arch Med. 2017;09(4):1-9.
Crossref - Yehouenou CL, Kpangon AA, Affolabi D, et al. Antimicrobial resistance in hospitalized surgical patients: a silently emerging public health concern in Benin. Ann Clin Microbiol Antimicrob. 2020;19(1):54.
Crossref - Magiorakos A-P, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268-281.
Crossref - Chaudhary R, Thapa SK, Rana JC, Shah PK. Surgical Site Infections and Antimicrobial Resistance Pattern. J Nepal Health Res Counc. 2017;15(2):120-123.
Crossref - Reta A, Wubie M, Mekuria G. Nasal colonization and antimicrobial susceptibility pattern of Staphylococcus aureus among pre-school children in Ethiopia. BMC Res Notes. 2017;10(1):746.
Crossref - Gupta BB, Soman KC, Bhoir L, Gadahire M, Patel B, Ahdal J. The Burden of Methicillin Resistant Staphylococcus aureus in Surgical Site Infections: A Review. J Clin Diagnostic Res. 2021;1(15):PE01-PE06.
Crossref - Patnool RB, Wadhwani A, Balasubramanian V, Ponnusankar S. Need for the Implementation of Antibiotic Policy in India: An Overview. Int J Curr Sci Res Rev. 2021;13(05):168-178.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.