ISSN: 0973-7510
E-ISSN: 2581-690X
https://dx.doi.org/10.22207/JPAM.13.1.37 | © The Author(s). 2019
Glaciers are the cryospheric niches which support concealed microbial life. They inhabit broad- spectrum culturable and non-culturable bacterial diversity. There is virtually very little information on the psychrophilic/psychrotolerant bacterial diversity found in the glaciers in India. Indian Himalayas are regarded as the world heritage of flora and fauna. As it houses many largest glaciers in its lap, a new venture into glaciers has been started. Microbiological investigation of the glaciers in North-East India will help us to have an insight into the hidden treasure of microflora. We are providing the first report on the Psuedomonas sp. diversity from Kanchengayao glacier, North Sikkim, India. It is one of the most dominant genera isolated from glacier ice samples. This genus is one of the most medically and ecologically important groups of Gamma-proteobacteria present in environment. In the present study, the diversity of Pseudomonas species isolated from ice core sample was carried out based on the phenotypic and genotypic analysis. It was found that the glacier was abundant in Pseudomonas azotoformans; Pseudomonas poae; Stenotrophomonas maltophilia; Pseudomonas fluorescens; Pseudomonas reactants; Pseudomonas hibiscicola and Pseudomonas synxantha. Interestingly, the antibiotic susceptibility test showed that all the isolates were resistant to Ampicillin (10mcg) but all were sensitive to Streptomycin (10mcg), 19 isolates were resistant to Vancomycin (30mcg) and six were resistant against Tetracycline (30mcg) whereas majority of the isolates showed intermediate response. The antibiotic resistance found in this unexplored area is an important study and first of its kind reported from this glacier.
Pseudomonas, Kanchengayao glacier, Sikkim, 16S rRNA, antibiotic resistance.
In 1887 Forster had discovered bacteria which can replicate at 0°C. After a few years, Schmidt-Nielsen for the first time coined the term ‘psychrophilic’ to define the bacteria living under extremely cold conditions1,2. As only minimum growth temperature was believed in this early definition some confuse arose due to the lack of comprehensible definition between cold-loving and cold-tolerant adaptation types3. After the isolation of true psychrophiles in 1960s, cold adapted bacteria were placed into the steno-thermic and eurythermic category and finally Morita coined them into psychrophilic and psychrotrophic / psychrotolerant micro-organisms2. True psychrophilic bacteria shows growth at or below 0°C, optimally grows around 15°C to 20°C, whereas psychrotolerant can grows and tolerate temperature above 20°C2 .
Temperature governs the abiotic and biotic factors which in turn directly dictate the protein machinery and adaptive features of the cryospheric microflora4-6. Cold-dwelling microbes have evolved many such traits that adapt their growth at low temperatures7-8. Both the psychrophilic and psychrotolerant micro-organisms influence the ecology of cryosphere portions of the earth9. They play major roles in the biodegradation and recycling of organic matter as well as inorganic nutrients9.
The potential for biotechnological applications of psychrophilic and psychro-tolerant bacteria are receiving escalating attention nowadays. Psychrophiles are potential source of novel enzymes for industrial applications and some pigments produced by these microbes are also used as food additive10. One of the important habitats of psychrophiles are glaciers11. However, the majority of the glaciers studies demonstrate only broad-spectrum bacterial diversity on the surface of the galciers, and virtually very little information exists on which particular psychrophilic or psychrotolerant bacterial species diversity actually reside in those glaciers in the depth zone.
Pseudomonas is one of the most dominant genera reported from many glaciers12-14. Along with glaciers, they are present in other cold environments and very often in intimate association with plants and animals. It is one of the most versatile and ecologically important groups of gamma-proteobacteria present in many environments and it plays a very significant role in nitrogen and carbon cycles15. Therefore their species diversity studies in glacier ecosystems are crucial in relation to climate change because if there is a sudden deficit of these species diversity in a glacier, then we can co-relate the effects of climate change in glaciers9.
In this paper, the diversity of psychro-tolerant Pseudomonas species isolated from Kanchengayao glacier ice, North Sikkim, India was studied based on the phenotypic and genotypic analysis. This is the first ever report of Pseudomonas species diversity from Kanchen-gayao glacier of Himalayan valley located at North Sikkim, India.
Sampling Site
GPS MAP 78S, which is an automated Global Positioning device GPS was used for mapping and to determining the coordinates and land elevation of glacier study sites16 .
Sample collection and preparation
The ice core samples from the Accu-mulation zone of Kanchengayao glacier was aseptically collected in a sterile thermal cryo sampling box. These sterile sampling containers were sealed tightly and were immediately brought to the laboratory16-19. For microbiological analysis, the ice core samples were cut aseptically with a sterilized sawtooth knife and around 5 mm annulus of glacial ice core was discarded20. The remaining inner core was first rinsed with cold ethanol (95%) and later on washed with cold sterile water. The ice samples were allowed to melt in the sterile beaker placed at 4°C cold incubator21. As described by Zhang all the handling procedures carried out aseptically below 20°C using positive pressure laminar flow hood22.
Physicochemical analysis of glacial ice core samples
The glacial ice core samples were melted at 4°C cold incubator. Physical parameters such as temperature, conductivity, pH, dissolved oxygen (DO) and turbidity were measured with the help of Multi Water Quality Checker U-50 Series Instrument (Horiba, Japan). Chemical constituents like magnesium, calcium, nitrite, nitrate, sulfate, fluoride, alkalinity, ortho-phosphate, total dissolved solids, silica, iron, and sodium was analyzed by Multi- Parameter Water Testing Kit, WTO15 (Himedia, India). Piper diagram was plotted to draw the analysis23.
Isolation of bacteria
Bacterial isolation were carried on Pseudomonas Isolation Agar (Peptic digest of animal tissue 20g/L; MgCl2 1.4g/L; K2SO4 10g/L; Triclosan 0.025g/L; Glycerol 20mL; distilled water 1L). Briefly, 200µL of thawed inner ice core samples were spread plated and incubated at 15°C for 3 weeks. Also, bacterial enrichment was carried out by incubating 100µL water sample in 50 mL Pseudomonas isolation broth in 250mL conical flask and then incubated at 15°C for 14 days in incubator cum shaker at 120rpm. After enrichment, bacterial isolation was carried on Pseudomonas isolation agar as mentioned above. Morphologically different colonies were selected and sub-cultured by streak plate method16-19. A total of 22 pure bacterial isolates were isolated from Kanchengayao glacier samples and stored at -80°C in 40% glycerol.
Characterization of bacterial isolates
The morphological, physiological and biochemical characteristics of the selected isolates were evaluated16. The morphological characters such as colony morphology, Gram-staining, spore staining were performed. The growth of isolates were checked in LB broth at different temperature i.e., 5°C, 10°C, 15 °C, 20°C, 30°C and 40°C in an incubator. Similarly, NaCl dependent growth profile was determined by incubating bacterial isolates in LB media containing different concentrations of NaCl (0%, 1% 2%, 4%, 6%, 8% and 10%) at 15°C incubator. For 0% NaCl concentration in LB media following media composition was used (Casein enzymic hydrolysate: 10g/L; Yeast Extract: 5g/L; d.w. 1L) and pH dependent growth profile were also checked at different range pH (4, 6, 8 and 10) at 15°C in an incubator16-19.
For protease enzymatic activities SM Agar (Skim Milk powder 28g/L; Tryptone 5g/L; Yeast Extract 2.5g/L; Dextrose 1g/L; Agar 25g/L; distilled water 1L) was used for the demonstration of coagulation and proteolysis24. The proteolytic bacteria hydrolyzed casein to form soluble nitrogenous compounds indicated as clear zone surrounding the colonies. Amylase enzyme activities were detected by flooding the surface of 48 hours old pure culture on Starch Agar media (Meat extract 3g /L; Peptic digest of animal tissue 5g /L; Starch 2g/L; Agar 25g/L; distilled water 1L) with Iodine solution (Iodine crystals 0.34g; KI 0.66g; distilled water 100mL)25. Amylase positive organisms showed clear halo zone around the colony. Size of the clear halo zone is directly proportional to the starch hydrolyzing activity of the strain under study. Gelatinase hydrolysis was detected on Gelatin Agar (Gelatin 30g/L; Casein enzymic hydrolysate 10g/L; NaCl 10g/L; Agar 25g/L; distilled water 1L). Gelatin positive organisms showed clear halo zone of inhibition around the colony. Size of the clear zone is directly proportional to the gelatin hydrolyzing activity of the strain under study.
Antibiotic resistance profiling of the bacterial isolates
All the bacterial isolates were tested for their response to various antibiotics by disc diffusion method26. The isolates were tested using antibiotic discs for their response to four antibiotics: Ampicillin (10mcg), Vancomycin (30mcg), Tetracycline (30mcg) and Streptomycin (10mcg). All the antibiotic tests were performed in Muller Hinton Agar (MHA) at 15°C incubator. The results were recorded as diameter of the inhibition zone formed which was interpreted from the Zone Size Interpretative Chart.
PCR amplification, sequencing, and analysis of 16S rDNA
The 16S rRNA gene was amplified using universal bacterial primers 27F (5’- AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’-CGGTTACCTTGTTACGACTT-3’)13. For amplification of PCR product, following cycle conditions were used: 94°C for 5 min; 35 cycles of 1min at 95°C, 1min at 55°C, and 2 min at 72°C; and a final extension step of 10 min at 72°C. QIAquick PCR purification kit (Qiagen, Germany) was utilized for purifying the PCR products and subsequently PCR products were sequenced by ABI Applied Biosystems (3500 Genetic Analyzer) using each universal primer i.e., 27F and 1492R. The 16S rRNA gene sequences were aligned with reference gene sequences of related taxa using Clustal W software27. A phylogenetic tree was constructed by MEGA 7 software28 using the neighbor-joining method29.
Physicochemical analysis
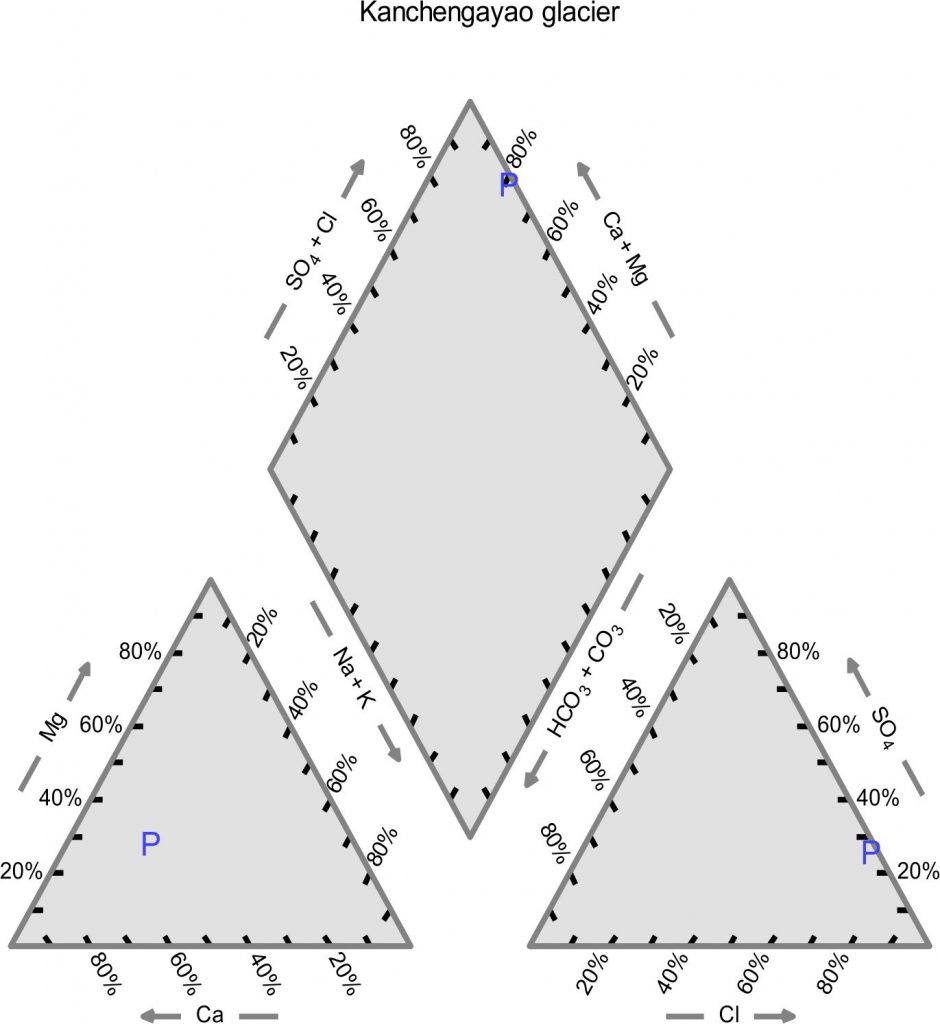
The physicochemical analysis of Kanchengayao glacier ice core samples, suggested that all the elements were present in very low concentration (Table 1). The physio-chemical parameters were plotted as piper diagram for classification on the basis of chemical composition23. Piper diagram divides water into four basic types conferring to their location near the four corners of the diamond. Water samples which are rich in (Ca2+ + Mg2+) and (Cl– + SO42-) are considered as permanent hardness and placed at the top of diamond, whereas water samples which are rich in (Ca2+ + Mg2+) and HCO3 – are considered as temporary hardness and placed near the left corner of diamond. Water samples mainly composed of alkali carbonates (Na+ + K+) and (HCO3 – + CO32-) are plotted at the lower corner whereas near the right-hand side of the diamond may be reflected as saline (Na+ + K+) and (Cl– + SO42). The piper diagram suggested that the Kanchengayao glacier ice was Ca-Cl type (Fig. 1).
Table (1):
Physicochemical Analysis of glacier sample
Name of the Chemical test |
Millipore Water
(Control) |
Glacier sample |
|---|---|---|
pH |
6.9 |
7.2 |
Total Dissolved Solid (TDS) (mg/L) |
BDL |
16 |
Fluoride Test (ppm) |
BDL |
0.5 |
Sulfate Test (ppm) |
BDL |
10 |
Nitrate Test (ppm) |
BDL |
10 |
Chloride Test (ppm) |
BDL |
20 |
Total Hardness Test (ppm) |
BDL |
100 |
Orthophosphate Test (ppm) |
BDL |
0.0 |
Arsenic Test (ppm) |
BDL |
0.05 |
Alkalinity Test (ppm) |
BDL |
200 |
Nitrite Test (ppm) |
BDL |
15 |
Iron Test (ppm) |
BDL |
0.3 |
Silica Test (ppm) |
BDL |
0.0 |
Free Chlorine Test (ppm) |
BDL |
0.0 |
Turbidity (NTU) |
BDL |
≤ 10 |
Dissolve Oxygen (DO) (ppm) |
BDL |
0.36 |
Fluoride (ppm) |
BDL |
0.5 |
Conductivity (μS/cm) |
BDL |
0.12 |
Magnesium (ppm) |
BDL |
05 |
Sodium (ppm) |
BDL |
07 |
Calcium (ppm) |
BDL |
14 |
The data was measured by AQUA CHECK kit (HiMedia, Mumbai, India) where BDL refers to “Below Detection Level”; ppm= parts per million.
Isolation and biochemical characterization of psychrotolerant Pseudomonas sp.
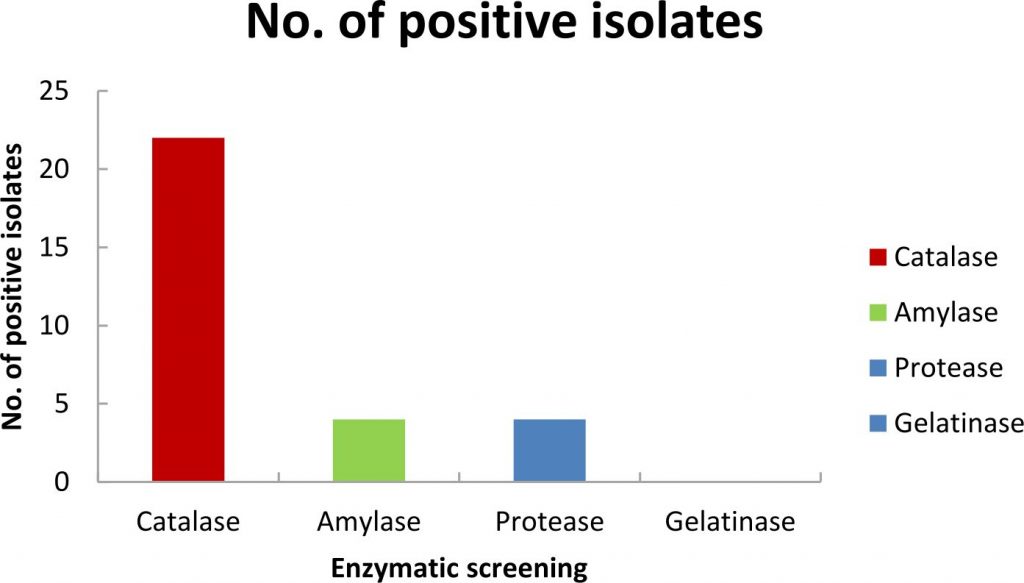
Incubation in Pseudomonas Isolation Agar at 15°C for 3 weeks at 15°C helped us to isolate psychrotolerant Pseudomonas sp. from glacier ice core samples. Twenty two pure bacterial isolates, isolated from enrichment method and streak plate method, were preserved in 40% glycerol at -80°C. Based on morphological characteristics, these bacterial isolates were selected for further analysis, i.e., Gram-staining, spore staining, colony morphology, growth temperature, NaCl and pH tolerance (Table 2). The bacterial isolates were all rod-shaped, non-spore formers and mostly produced white cream color colonies as shown in (Table 2). The Gram-staining results showed that all the isolates were Gram-negative. Among 22 bacterial isolates, during the initial screening for enzymatic activity of the isolates, it was observed that four isolates (KGG15, KGG22, KGG60, KGG62) were positive for amylase production, four isolates (KGG17, KGG25, KGG28, KGG59) were positive for protease production and all the isolates were catalase positive as shown in Fig. 2. Carbohydrate fermentation test showed that majority of the isolates were able to ferment simple sugars like dextrose, whereas few isolates were able to ferment sugars like dulcitol, lactose, mannose, arabinose, raffinose, and ribose, however, most of the bacterial isolates were unable to ferment fructose, sucrose, and xylose as shown in Table 3.
Table (2):
Morphological characterization of the bacterial isolates
| Isolates | Growth on agar plates | Morphology on the basis of staining | |||||
|---|---|---|---|---|---|---|---|
| Colony color | Margin | Elevation | Form | Simple Staining | Gram Staining | Spore Staining | |
| KGG2 | Yellow | Erose | Flat | Irregular | Rod | -ve | × |
| KGG6 | White | Undulate | Flat | Circular | Rod | -ve | × |
| KGG13 | White | Entire | Flat | Circular | Rod | -ve | × |
| KGG14 | White | Entire | Flat | Circular | Rod | -ve | × |
| KGG15 | White | Undulate | Flat | Circular | Rod | -ve | × |
| KGG16 | White | Erose | Flat | Circular | Rod | -ve | × |
| KGG17 | White | Undulate | Flat | Circular | Rod | -ve | × |
| KGG20 | Yellow | Entire | Flat | Irregular | Rod | -ve | × |
| KGG22 | White | Undulate | Flat | Circular | Rod | -ve | × |
| KGG25 | White | Entire | Flat | Circular | Rod | -ve | × |
| KGG28 | White | Undulate | Flat | Circular | Rod | -ve | × |
| KGG29 | White | Undulate | Flat | Circular | Rod | -ve | × |
| KGG35 | Brown | Erose | Flat | Circular | Rod | -ve | × |
| KGG38 | White | Undulate | Flat | Circular | Rod | -ve | × |
| KGG44 | White | Undulate | Flat | Circular | Rod | -ve | × |
| KGG50 | White | Undulate | Flat | Circular | Rod | -ve | × |
| KGG45 | White | Undulate | Flat | Circular | Rod | -ve | × |
| KGG51 | White | Entire | Flat | Circular | Rod | -ve | × |
| KGG53 | White | Undulate | Flat | Circular | Rod | -ve | × |
| KGG59 | White | Undulate | Flat | Circular | Rod | -ve | × |
| KGG61 | White | Entire | Convex | Circular | Rod | -ve | × |
| KGG62 | White | Undulate | Flat | Circular | Rod | -ve | × |
Table (3):
Morphological characterization of the bacterial isolates
|
Isolates |
Carbohydrate Fermentation | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| D (-)
Arabinose |
D(+)
Dextrose |
D(+)
Dulcitol |
D(-)
Fructose |
D(+)
Lactose |
D(-)
Mannose |
D(+)
Raffinose |
D(-)
Ribose |
D(+)
Sucrose |
D(+)
Xylose |
|
| KGG2 | + | + | – | – | + | + | – | – | – | – |
| KGG6 | – | + | – | – | – | – | – | – | – | – |
| KGG13 | – | – | – | – | – | – | – | – | – | – |
| KGG14 | – | – | – | – | – | – | – | – | – | – |
| KGG15 | – | + | – | – | – | – | – | – | – | – |
| KGG16 | – | + | – | – | – | – | – | – | – | – |
| KGG17 | – | + | – | – | – | – | – | – | – | – |
| KGG20 | + | + | – | – | + | + | – | – | – | – |
| KGG22 | – | + | – | – | – | + | – | – | – | – |
| KGG25 | – | – | – | – | – | + | – | – | – | – |
| KGG28 | – | + | – | – | – | – | – | – | – | – |
| KGG29 | – | + | – | – | – | – | – | – | – | – |
| KGG35 | – | – | – | – | – | – | – | – | – | – |
| KGG38 | – | + | – | – | – | – | – | – | – | – |
| KGG44 | – | + | – | – | – | – | – | + | – | – |
| KGG50 | – | + | – | – | – | – | – | – | – | – |
| KGG45 | – | + | – | – | – | – | – | – | – | – |
| KGG51 | – | – | + | – | – | – | + | + | – | – |
| KGG53 | – | – | + | – | – | – | + | + | – | – |
| KGG59 | – | + | + | – | – | – | – | – | – | – |
| KGG61 | – | – | – | – | – | – | – | – | – | – |
| KGG62 | – | + | + | – | – | – | – | – | – | – |
The isolates were tested against various carbohydrates and the (+) sign indicates that the isolates were able to ferment that particular carbohydrate and (-) sign indicates that the isolates were unable to ferment that particular carbohydrate.
Antibiotic resistance
Recent studies have detected antibiotic resistant bacteria and their resistance genes in the natural environment which are geo-graphically isolated from such area30-32. Interestingly from Kanchengayao glacier ice core samples we also found three class of antibiotic resistant bacteria against penicillin (Ampicillin 100%), glycopeptides (Vancomycin 86.36%) and Tetracycline 27% respectively (Table 4). This paves for an interesting study for future as these glaciers are never exposed to antibiotics and also are usually free of any anthropogenic activities.
Table (4):
Antibiotic Assay of the bacterial isolates
| Antibiotic disc with their Concentration | ||||
|---|---|---|---|---|
| Zone of inhibition (in nm) | ||||
| Isolates | Van30 | TE30 | Amp10 | S10 |
| KGG2 | R | R | R | S |
| KGG6 | R | R | R | S |
| KGG13 | R | R | R | S |
| KGG14 | R | S | R | S |
| KGG15 | R | I | R | S |
| KGG16 | R | I | R | S |
| KGG17 | R | I | R | S |
| KGG20 | S | R | R | S |
| KGG22 | R | I | R | S |
| KGG25 | S | R | R | S |
| KGG28 | R | I | R | S |
| KGG29 | R | I | R | S |
| KGG35 | S | R | R | S |
| KGG38 | R | I | R | S |
| KGG44 | R | I | R | S |
| KGG50 | R | I | R | S |
| KGG45 | R | I | R | S |
| KGG51 | R | S | R | S |
| KGG53 | R | S | R | S |
| KGG59 | R | I | R | S |
| KGG61 | R | I | R | S |
| KGG62 | R | I | R | S |
The isolates were tested against various antibiotics and the (R) sign indicates that the isolates were resistant, (I) sign indicates that the isolates intermediate and (S) sign indicates that the isolates sensitive.
Tolerance to different; Temperature, NaCl and pH
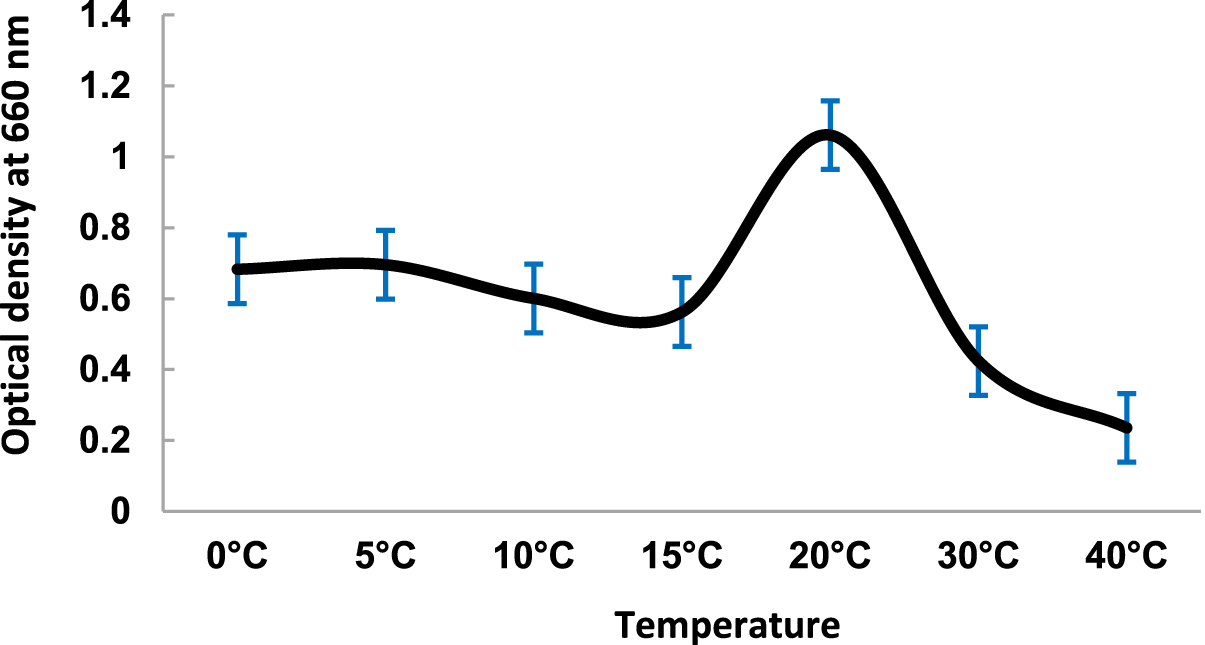
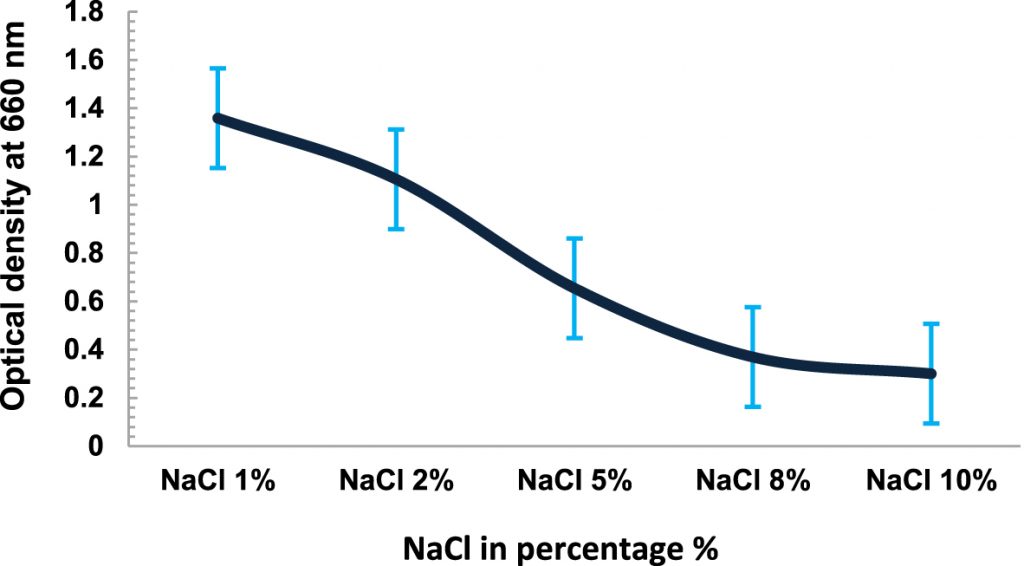
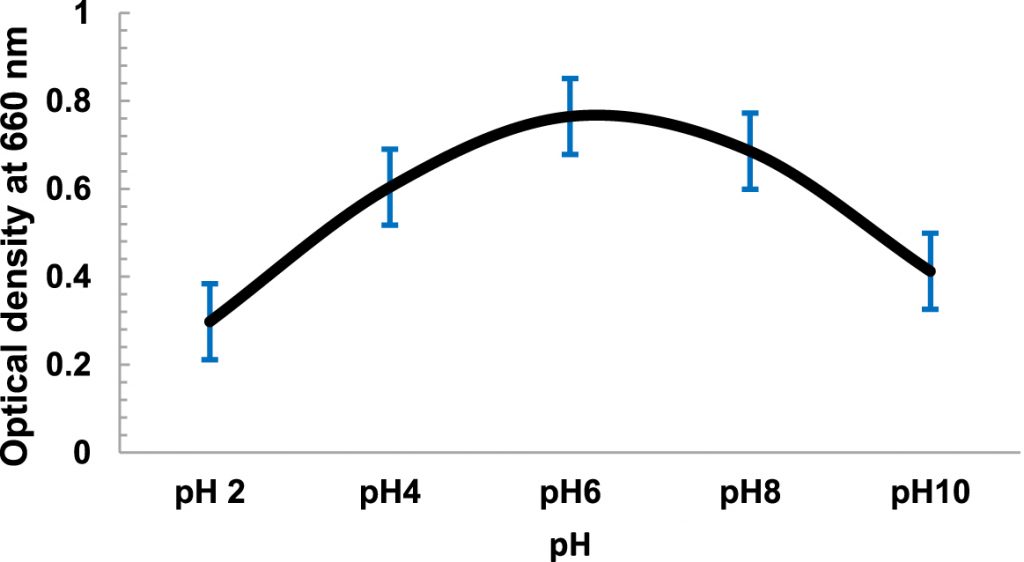
The temperature dependent growth profile of the isolates showed that the majority of the isolates had optimum growth at 20°C but all could tolerate temperature >25°C£ 40°C (Fig. 3). It suggested that these isolates are psychro-tolerant, rather than psychrophilic in nature. The salinity tolerance was also tested and it was found that majority of the isolates grew well at minimum concentrations of NaCl at 15°C (Fig. 4). pH dependent growth profile suggested that nine isolates showed good growth at pH5 and pH6, followed by three isolates at pH8 and two isolates at pH2 (Fig. 5). From our study we found that 11 bacterial isolates were acidophilic in nature, while others preferred neutral pH or slightly alkaline pH.
Phylogenetic analysis of 16S rDNA sequencing of isolates
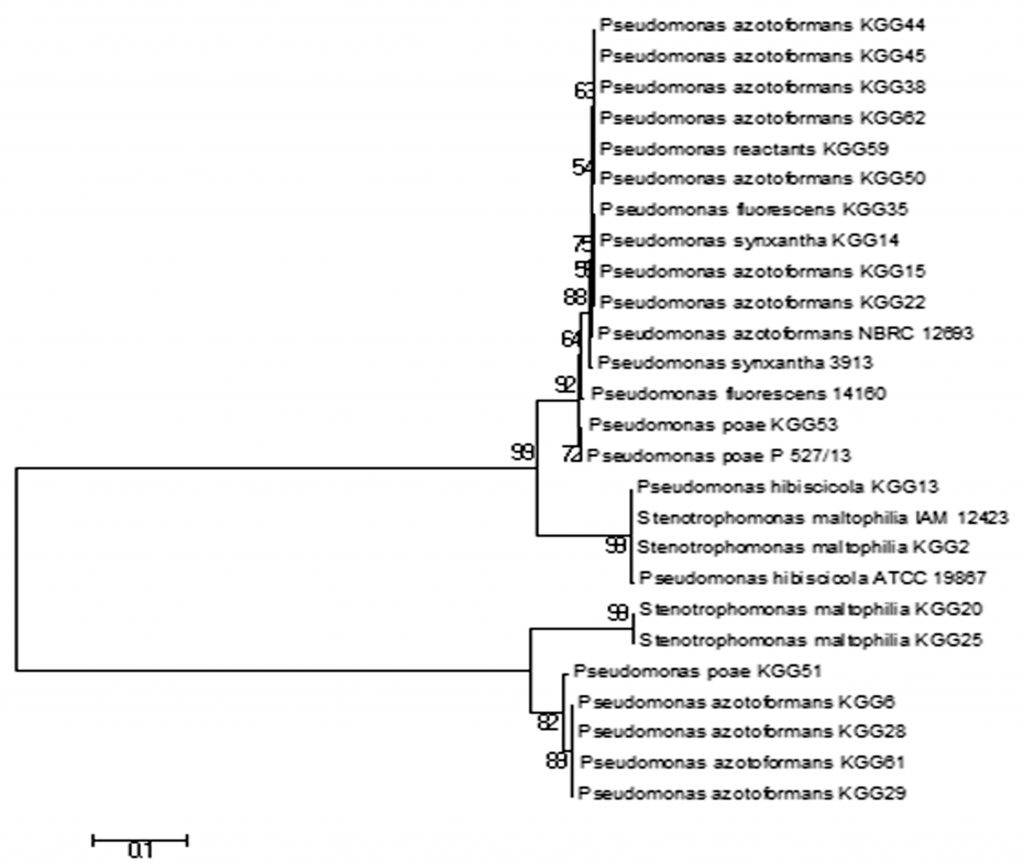
Based on the similarity criterion of at least 99% of the 16S rRNA gene sequence, it was found that all 22 bacterial isolates belonged to genus Pseudomonas and was from seven diversified Pseudomonas species. 13 isolates showed 99% homology to Pseudomonas azotoformans, two isolates had 99% homology to Pseudomonas poae, three isolates had 99% homology to Stenotrophomonas maltophilia and others were Pseudomonas fluorescens, Pseudomonas reactants, Pseudomonas hibiscicola and Pseudomonas synxantha. The accession number and identity of all the isolates are given in Table 5. The phylogenetic tree was made using MEGA7 using the Neighbor-Joining method Fig. 6.
Table (5):
GenBank 16S rRNA gene sequence accession numbers
Isolates |
Closest relative species |
% Identity |
Gene Bank Accession No. |
|---|---|---|---|
KGG35 |
Pseudomonas fluorescens |
99.42 |
KY129832 |
KGG59 |
Pseudomonas reactants |
99.3 |
KY129833 |
KGG13 |
Pseudomonas hibiscicola |
99.4 |
KY129834 |
KGG2 |
Stenotrophomonas maltophilia |
99 |
KY129838 |
KGG14 |
Pseudomonas synxantha |
99 |
MH079449 |
KGG15 |
Pseudomonas azotoformans |
99 |
MH079450 |
KGG51 |
Pseudomonas poae |
99.4 |
MH079451 |
KGG62 |
Pseudomonas azotoformans |
99 |
MH079452 |
KGG6 |
Pseudomonas azotoformans |
99 |
MH157226 |
KGG20 |
Stenotrophomonas maltophilia |
99 |
MH157227 |
KGG25 |
Stenotrophomonas maltophilia |
99 |
MH157228 |
KGG28 |
Pseudomonas azotoformans |
99 |
MH157229 |
KGG38 |
Pseudomonas azotoformans |
99 |
MH157230 |
KGG44 |
Pseudomonas azotoformans |
99 |
MH157231 |
KGG45 |
Pseudomonas azotoformans |
99 |
MH157232 |
KGG53 |
Pseudomonas poae |
99.33 |
MH157233 |
KGG50 |
Pseudomonas azotoformans |
99 |
MH157234 |
KGG61 |
Pseudomonas azotoformans |
99 |
MH157235 |
KGG16 |
Pseudomonas azotoformans |
99 |
MH157236 |
KGG17 |
Pseudomonas azotoformans |
99 |
MH157237 |
KGG22 |
Pseudomonas azotoformans |
99 |
MH157238 |
KGG29 |
Pseudomonas azotoformans |
99 |
MH157239 |
Diverse form of microorganisms, including algae and fungi, has been reported on the surface of many glaciers around the world. These microorganisms make up the primary colonizer of the snow and ice, which also include cold-tolerant animals such as insects and copepods. In North East Himalayas, Kanchen-gayao glacier is located at North Sikkim, India and it is a debris-free (clear glacier) transverse valley glacier (Fig 7). This glacier originated from the southern slope of Mount Kanchengayao, treading north-south face having latitude 27°59’57.872’N and longitude 88°37’8.785’E at an altitude of 1393m. The melt water of this glacier feeds into river Thangu which is a tributary of river Lachen. Kanchengayao glacier is surrounded by a cold desert and in its lower valley small Rhododendron trees are present along with some other vegetation. Possibly, nutrient and bacteria from nearby or distantly located places gets transferred to accumulation zone of glacier through air currents33. Also, the soil bacteria might affect the distribution of Pseudomonas species on the surface of the glacier accumulation zone.
Our result showed that the most dominant Pseudomonas species on Kanchen-gayao glacier were Pseudomonas azotoformans followed by Pseudomonas poae, Stenotrophomonas maltophilia, Pseudomonas fluorescens, Pseudomonas reactants, Pseudomonas hibiscicola and Pseudomonas synxantha. Diversity of Pseudomonas sp. in glaciers has been reported in various parts of the world12-14. These gram negative bacteria are abundant in environmental niches and are clinically important also. The most abundant Pseudomonas sp. in our glacier ice core samples was Pseudomonas azotoformans. This gram negative bacterium has been reported as an effective biocontrol agent against plant pathogens34-36 and it also was reported as a good biotechnological candidate which has biomineralization property to fix the concrete cracks by producing calcite37. So, it will be interesting to study this psychro-tolerant bacterium isolated from glacier in future. Pseudomonas poae is also reported to have biocontrol activity38,39. Pseudomonas fluorescens is an aerobic plant growth promoting rhizobacterium (PGPR) and has biocontrol activity40, 41. Some bacteria have the ability to synthesize nanoparticles metabolically and this has amazed the scientific community and industries at large for harnessing this unique capability to produce nanoparticles in cost effective method42,43. One of our isolates Pseudomonas hibiscicola have been reported in many literatures as a potential candidate showing this property44,45. So, it will be another interesting study in future. Stenotrophomonas maltophilia is an environmental bacterium found in various ecological niches like river, plant rhizospheres, soil etc. It is an emerging multi-drug-resistant global opportunistic pathogen and it was also present in our glacier sample.
Glacier study shall reveal in future many such diversified microorganisms and their biotechnological applications render them as suitable candidates for future industrial strains. Sikkim glacier houses plethora of microbes and it’s an important one in terms of monitoring climatic changes in the Himalayas.
This study has been funded by the Department of Science and Technology, Govt. of India (IUCCC) and Department of Biotechnology (DBT Overseas Associateship). We are grateful to Forest Department, Govt. of Sikkim for providing research permit and access to the sampling sites. Authors are thankful to Dr. Uttam Lal, Dr. Rakesh Ranjan and Dr. Smriti Basnet for their support during the field study.
The authors and planners have disclosed no potential conflicts of interest, financial or otherwise.
- Priscu J C, Christner B C, in Microbial Diversity and Bioprospecting (ed. Bull, A.) 130–145 (ASM Press, 2004).
- Miteva V, Miteva V, Psychrophiles: from biodiversity to biotechnology. Springer, New York, 2008; pp78–99
- Boetius A, Anesio A M, Deming J W, Mikucki J A, Rapp JZ, Microbial ecology of the cryosphere: sea ice and glacial habitats, Nature Rev. Microbiol, 2015; 13: 677–690. https://doi.org/10.1038/nrmicro3522
- Inouye M, Phadtare S, Cold shock response and adaptation at a near-freezing temperature in microorganisms, Sci. STKE, 2004; 9: 237-2247.
- D’Amico S, Collins T, Marx JC et al, Psychrophilic microorganisms: challenges for life, EMBO Rep., 2006; 7: 385-389.
- Siddiqui K S, Cavicchioli R, Cold-adapted enzymes, Annu Rev. Biochem, 2006; 8: 403-433.
- Lorv J S H, Rose D R, Glick B R, Bacterial ice crystal controlling proteins, Scientifica, 2014; 3: 1–20. https://doi.org/10.1155/2014/976895.
- Feller G, & Gerday C, Psychrophilic enzymes: hot topics in cold adaptation, Nat. Rev. Microbiol., 2003; 1: 200–208.
- Anesio A M, Hodson A J, Fritz A, Psenner R, Sattler B, High microbial activity on glaciers: importance to the global carbon cycle, Glob. Chang. Biol., 2009; 15: 955–960.
- Raman K V, Singh L, Dhaked R, Botechnological application of psychrophiles and their habitat to low temperature, J. Sci. Indian Res., 2000; 59: 87–101.
- Jansson J K; Tas N, The microbial ecology of permafrost, Nat. Rev. Microbiol., 2014; 12: 414–425.
- Taj M K, Ji X, Lin L, Zhang Q, Wei Y, Taj I, et al, Diversity and phylogenetic analysis of Pseudomonas strains isolated from Mingyong glacier China, Journal of Biodiversity and Environmental Sciences, 2016; 9(4): 232-236.
- Rafiq M, Hayat M, Anesio AM, Jamil SUU, Hassan N, Shah AA, et al. Recovery of metallo-tolerant and antibiotic resistant psychrophilic bacteria from Siachen glacier, Pakistan. PLoS ONE, 2017; 12(7): e0178180. https://doi.org/10.1371/journal.pone.0178180
- Mannista MK, Haggblom M, Characterization of psychrotolerant heterotrophic bacteria from Finnish Lapland, Syst Appl Microbiol., 2006, 29(3): 229–243.
- Spiers A J, Buckling A, Rainey P B, The causes of Pseudomonas diversity, Microbiology, 2000; 146: 2345-2350.
- Sherpa M T, Najar I N, Das S, Thakur N, Bacterial diversity in an Alpine debris-free and debris-cover accumulation zone glacier ice, North Sikkim, India, Indian J Microbiol., 2018; 58(4): 470-478.
- Liu Y, Yao T, Jiao N, Kang S H, Culturable bacteria in glacial meltwater at 6350 m on the East Rongbuk Glacier, Mount Everest, Extremophiles, 2009; 13: 89–99. https://doi.org/10.1007/s00792-008-0200-8
- Hong S, Zhang H, Yang W, Bacteria recovered from a high-altitude, tropical glacier in Venezuelan Andes, World J. Microbiol. Biotechnol., 2010, 3: 931–941. https://doi.org/10.1007/s11274-013-1511-1
- Shen L, Yao T, Xu B, Wang H, Jiao N, Kang S, Variation of culturable bacteria along depth in the East Rongbuk ice core, Mt. Everest, Geosci. Front., 2012; 3: 327–334. https://doi.org/10.1016/j.gsf.2011.12.013
- Xiang S, Yao T, An L, Xu B, 16S rRNA sequences and differences in bacteria isolated from the Muztag Ata Glacier at increasing depths, Appl Environ Microbiol., 2005; 71: 4619–4627. https://doi.org/10.1007/BF02931042
- Xiang S R, Yao T D, An L Z, Xu BQ, Li Z, Wu G J, Bacterial diversity in Malan ice core from the Tibetan Plateau, Folia Microbiology, 2004; 49: 269–275. https://doi.org/10.1007/BF02931042
- Zhang X, Diversity of 16S rDNA and environmental factor influencing microorganisms in Malan ice core, Chin. Sci. Bull., 2003; 48: 1146–1152. https://doi.org/10.1360/02wd0409
- Piper AM, A graphical procedure in the geochemical interpretation of water analyses Transactions, American Geophysical Union, 1994; 25: 914-928.
- Zhou M Y, Chen X L, Zhao H L, Dang H Y, Luan X W, et al, Diversity of both the cultivable protease-producing bacteria and their extracellular proteases in the sediments of the South China Sea, Microb. Ecol., 2009; 58: 582–590. https://doi.org/10.1360/19301066
- Lu M, Fang Y, Li H, Liu W S, Isolation of a novel cold adapted amylase-producing bacterium and study of its enzyme production conditions, Ann. Microbiol., 2010; 60: 557–563. https://doi.org/10.1007/s13213-010-0090-8
- Aneja R K, Experiments in Microbiology, Plant Pathology and Biotechnology, New age International (P) Ltd, New Delhi, India, 2003.
- Metsalu T, Vilo J, Clustvis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap, Nucleic Acids Res., 2015; 43: 566–570
- Tamura K, Dudley J, Nei M, Kumar S, Molecular evolutionary genetics analysis (MEGA) software version 4.0, Mol. Biol. Evol., 2007; 24: 1580–1596. https://doi.org/10.1093/molbev/msm092
- Saitou N, The neighbor-joining method: a new method for reconstructing phylogenetic trees, Molecular Biology and Evolution, 1987; 4: 406-425.
- Clemente J C, Pehrsson E C, J. Blaser M, Sandhu K, Gao Z, Wang B, Magris M, Hidalgo G, Contrera M, The microbiome of uncontacted Amerindians. Microb. Ecol., 2015; 4: 1–12.
- Goethem1 M W, Van Pierneef R, Bezuidt O K I, Peer Y, Van De Cowan D A, et al. A reservoir of ‘historical’ antibiotic resistance genes in remote pristine Antarctic soils. Microbiome., 2018; 6: 1–12.
- Pawlowski AC, Wang W, Koteva K, Barton H A, McArthur A G, Wright G D, A diverse intrinsic antibiotic resistome from a cave bacterium. Nat. Commun., 2016; 7: 1–10.
- Yung P T, Shafaat H S, Connon S A, Ponce A, Quantification of viable endospores from a Greenland ice core, FEMS Microb. Ecol., 2007; 59: 300–336. https://doi.org/10.1111/j.1574-6941. 2006.00218
- Fang Y, Wu L, Chen G, Feng G, Complete genome sequence of Pseudomonas azotoformans S4, a potential biocontrol bacterium, Journal of Biotechnology, 2016; 227: 25- 26 doi: 10.1016/j.jbiotec.2016.04.020
- Sang M K, Kim E N, Han G D, Kwac k M S, Jeun Y C, et al, Priming-mediated systemic resistance in cucumber induced by Pseudomonas azotoformans GC-B19 and Paenibacillus elgii MM-B22 against Colletotrichumorbiculare, Phyto-pathology, 2014; 104: 834-842.
- Anzai Y, Kim H, Park J Y, Wakabayashi H, Oyaizu H, Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence, International Journal of Systematic and Evolutionary Microbiology, 2000; 50(4): 1563–1589.
- Nonakaran S H, Pazhouhandeh M, Kevyani A, Abdollahipour F Z, Shirzad A, Isolation and identification of Pseudomonas azotoformans for induced calcite precipitation, World Journal of Microbiology and Biotechnology, 2015; 31: 1993-2001. doi 10.1007/s11274-015-1948-5.
- Zachow C, Jahanshah G, de Bruijn I, Song C, Ianni F, Pataj Z et al, The novel lipopeptide Poaeamide of the endophyte Pseudomonas poae RE*1-1-14 is involved in pathogen suppression and root colonization, Molecular Plant-Microbe Intera-ctions, 2015; 28(7): 800-810. doi 10.1094/MPMI-12-14-0406-R
- Behrendt U, Ulrich A, Schumann P, Fluorescent pseudomonads associated with the phyllosphere of grasses; Pseudomonas trivialis sp. nov., Pseudomonas poae sp. nov. and Pseudomonas congelans sp. nov., International Journal of Systematic and Evolutionary Microbiology, 2003; 53: 1461–1469. doi 10.1099/ijs.0.02567-0.
- David B V, Chandrasehar G, Selvam P N, Pseudomonas fluorescens: A Plant-Growth- Promoting Rhizobacterium (PGPR) with potential role in biocontrol of pests of crops, In: Crop improvement through microbial Biotechnology, New and future developments in microbial biotechnology and bioengineering, 2018: 221-243. doi 10.1016/B978-0-444-63987-5.00010-4
- Ganeshan G, Kumar A M, Pseudomonas fluorescens, a potential bacterial antagonist to control plant diseases, Journal of Plant Interactions, 2007; 1(3): 123-134. doi: 10.1080/17429140600907043.
- Horikoshi S, Serpone N, Introduction to nanoparticles. In: Microwaves in Nanoparticle Synthesis, Wiley-VCH Verlag GmbH and Co. KGaA, 2013.
- Babu M M G, Sridhar J, Gunasekaran P, Global transcriptome analysis of Bacillus cereus ATCC 14579 in response to silver nitrate stress, Journal of Nanobiotechnology, 2011; 9(1): 1-12.
- Punjabi K, Mehta S, Yedurkar S, Jain R, Mukherjee S, et al, Extracellular synthesis of silver nanoparticle by Pseudomonas hibiscicola – Mechanistic approach, Advances in Nano Research, 2018; 6(1): 81-92 doi: https://doi.org/10.12989/anr.2018.6.1.081
- Punjabi K, Yedurkar S, Doshi S, Deshapnde S, Vaidya S, Biosynthesis of silver nanoparticles by Pseudomonas spp. isolated from effluent of an electroplating industry, IET Nanobiotechnology, 2017; 11(5): 584-590.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.