ISSN: 0973-7510
E-ISSN: 2581-690X
Monitoring the distribution and resistance of antibiotics to enterococcal species is critical aspect to controlling and preventing enterococcal infection. The aim of the present study is to screen the antimicrobial resistance genes within Enterococcus species isolates that collected from Taif governorate, Saudi Arabia. Out of 134 clinical samples, nineteen enterococcal isolates were identified using 16S rRNA sequence gene. Phylogenetic tree analysis using 16S rRNA gene sequence of the 19 strains divided them into 15 strains as E. faecalis and 4 strains as E. faecium. In addition, these the species of these isolates were recognized using VITEK-2 COMPACT system. The PCR technique was used to screen the multi-drug resistant genes within enterococcal isolates. The KpsII, tetL, aac(6)-Ie-aph(2)-Ia, vanA and Erm(B) genes were found in all strains. The distribute of resistance against antibiotic drugs were differs greatly between the two species, a considerably higher prevalence of resistance to penicillin, gentamicin, cefoxitin, cefotaxime, clindamycin, erthromycin and fusidic acid was identified in E. faecalis than in E. faecium, while greater spread was detected to resist to Trim/Sulf and tetracycline in E. faecalis. Finally, rep-PCR markers investigated genomic diversity of Enterococcus strains. Results of rep-PCR markers generated 142 distinct loci; 96 were polymorphic (67.6%) and 46 were monomorphic (32.4%). Number of loci for individual rep-PCR primers ranged from 9 for rep-08 to 18 for rep-02.
Enterococcus spp.; antimicrobial resistance; Antibiotic resistance genes, Saudi Arabia.
The genus Enterococcus consists of thirty-eight species. The most common identified living microorganisms usually found in the intestines of human and animal cells including E. faecalis and E. faecium1,2. E. faecalis is accountable for about 80% of human infections2. Intestinal microorganisms are opportunistic organisms that become pathogenic due to an imbalance in the immune system. Currently, Olawale et al.3 reported that intestinal tract infections can cause infections in hospitals, especially in urinary tract, surgical sites and bloodstream. Several reports pointed out that gastroenteritis is the third organism isolated between infection in hospitals of Saudi Arabia and the most isolated microorganisms common in the bloodstream4,5. Approximately 90% of intestinal endocarditis occurs due to E. faecalis, with less than 5% affected by E. faecium4. Infrequently, additional species are documented in disease conditions, including E. avium, E. casseliflavus, E. durans, E. gallinarum, E. hirae, E. mundtii and E. malodoratus6. Primarily in the last twenty years, these strains caused infections in the hospitals of Saudi Arabia. Enterococcus comprises a necessary piece of the gastrointestinal microflora of people, animals and insects. Notwithstanding their reality as commensals in the gastrointestinal tract, a few strains exist as sharp pathogens. In a previous study, vancomycin resistance to intestinal microorganisms (VRE) accounts for about 43% of all intestinal catheter isolates, an amount that rises due to unavailability of vancomycin for clinical use in USA. Two types of VRE isolates are included; E. faecium and E. faecalis. Both species were isolated to resist eight antibiotics3. Antibiotic resistance may be help in Enterococci development and consider as a threat to public health. The high distribution of Enterococcus spp. has developed as a public health threat7. Enterococcus is the most common bacterial infection in hospitals7. Enterococcus types have many virulence factors such as gastrointestinal protein and aggregation content, which increases the colonization process of epithelial lining of host cells8. The gene coding for ESP was commonly identified in medical infection isolates9. Antibiotic resistance factors are transferred by mergers, plasmids or transposons that can act as carriers. These genes are transferred to other members of the same microbial species and transmission of horizontal genes may arise through conjugation, transduction and transformation5,10.
The polymerase chain response (PCR) is a less complex method that gives results inside a short timeframe and at lower costs. By this technique, diverse primer sits, homologous, aleatory or degenerated, are utilized to intensify locales of the DNA particle. Band designs are in this way delivered, allowing the gathering of comparative strains and the separation of irrelevant ones11. Repetitive (REP) sequences are extragenic units found in various locales of the DNA of bacterial species12. The amplification of the areas between these units creates a helpful unique fingerprint to separate Enterococcus strains13.
The purpose of the present research was to screen the distribution of the antimicrobial resistance genes within Enterococcus spp in patients from King Faisal Hospital in Taif governorate, Saudi Arabia. Moreover, the genetic diversity between the Enterococcus spp. strains were investigated using rep-PCR markers.
Molecular identification of Enterococcus strains
Out of 134 bacterial isolates, nineteen multiple-drug resistant enterococcal strains were obtained from King Faisal Hospital in Taif governorate, Saudi Arabia. Isolates were recognized by high-throughput using a VITEK-MS (bioMיrieux, France) system according to manufacturer’s protocol. The DNA extraction kit (Gena Bioscience, Germany) was used to isolate genomic DNA from all 19 isolates accordance the manufacturer’s directions. For each isolate and as per previously described methods, one fragment of 16S rRNA gene (about 1465 bp) was amplified from each stain as previously reported14. The specific band was purified from the gel using QIAGEN purification kit (QIAGEN, USA). The purified band was sequenced via DNA Analyzer 3146 (Applied Biosystems, USA). To perform the BLAST searched, NCBI service was used (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). The phylogenetic tree was constructed using neighbor joining algorithm with Kimura 2 parameter distances in MEGA 7.1 software. The bar indicates the Juke-Cantor evolutionary distance.
Antibiotic Susceptibility Testing
All nineteen Enterococcus strains were screened for antimicrobial susceptibility using an automated VITEK-2 (bioMיrieux) system. In this protocol, broth microdilution minimum inhibitory concentration (MIC) method for susceptibility testing and accomplish repetitive turbidimetric screen of bacterial growth through an abbreviated incubation period. MIC results were taken based on the Clinical and Laboratory Standards Institute guidelines15. The criteria of Hassan et al. were used to defined MDR isolates16. The susceptibility of Enterococcus isolates to 16 antibiotics including vancomycin, gentamycin, cefoxitin, efotaxime, ampicillin, daptomycin, trime/sulf, teicoplanin, vancomycin, clindamycin, erythromycin, fusidic acid, linezolid, nitrofurantoin, ciprofloxacin, moxifloxacin and tetracycline was determined.
Detection of Antimicrobial Resistance Genes
Five virulence genes [KpsII, tetL, aac(6)-Ie-aph(2)-Ia, vanA, and Erm(B)] were detected in all Enterococcus strains in multi-Drug resistant according to Hassan et al.14. The resistance genes against antibiotic were detected in all 19 strains as previously reported4. The antibiotic resistance genes were isolated from the 19 strains by PCR with primers synthesized by the macrogen Co., Ltd. (Seoul, Korea) (Table 1). The PCR reaction for each gene was accomplished as previously described4.
Table (1):
PCR primers for detection of some antibiotic resistance and virulence genes in Enterococcus spp. isolates.
Genes |
Primer sequence (5′→3′) |
Size (bp) |
|---|---|---|
Tuf |
TACTGACAAACCATTCATGATG AACTTCGTCACCAACGCGAAC |
112 |
KpsII |
GCGCATTTGCTGATACTGTTG
CATCCAGACGATAAGCATGAGCA |
272 |
Dd1-E |
CAAACTGTTGGCATTCCACAA TGGATTTCCTTTCCAGTC ACTTC |
550 |
Dd2-E |
GAAGAGCTGCTGCAAAATGCTTTAGC GCGCGCTTCAATTCCTTGT |
941 |
Van-A |
GTAGGCTGCGATATTCAAAGC CGATTCAATTGCGTAGTCCAA |
231 |
Erm(B) |
CATTTAACGACGAAACTGGC GGAACATCTGTGGTATGGCG |
405 |
aac(6)-Ie-aph(2)-Ia |
CAGAGCCTTGGGAAGATGAAG CCTCGTGTAATTCATGTTCTGGC |
348 |
tet (L) |
GTMGTTGCGCGCTATATTCC GTGAAMGRWAGCCACCTAA |
696 |
Notes: M = A or C; R = A or G; W = A or Tuf gene specific for Enterococcus; KpsII gene specific for capsule formation in Enterococcus species; Dd1-E gene specific for Enterococcus faecalis; Dd2-E gene specific for Enterococcus faecium; Van-A gene specific for vancomycin resistance; erm(B) gene specific for erythromycin resistance; aac(6)-Ie-aph(2)-Ia gene specific for gentamycin resistance; and tet(L) gene specific for tetracycline.
Rep-PCR analysis
The settings of rep-PCR technique for the 19 strains of Enterococcus were standardized. Five primers (Rep-1, Rep-2, Rep-8, Rep-12 and Rep-18) were used to investigate the genetic variability between the Enterococcus strains. The primers sequence and PCR conditions were performed as previously reported17.
Enterococcus isolates identification
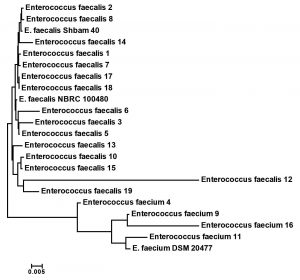
The species of the nineteen Enterococcus isolates were known by a fully automated VITEK-2 COMPACT microbiology method. Fifteen were recognized as E. faecalis and four as E. faecium. These results were confirmed by 16S rRNA sequencing. The 16S rRNA gene sequence of all 19 isolates was compared with other Enterococcus strains that placed in GenBank database. The results were somehow differed from the obtained sequence (data not shown). Ribosomal genes are greatly relevant for investigating bacterial evolution and phylogeny. When re-constructing phylogenetic relationships of microorganisms, sequencing of 16S rRNA has been generally employed. Phylogenetic tree analysis using 16S rRNA sequences of all 19 strains and other related Enterococcus species from GenBank were positioned the present Enterococcus strains into E. faecalis and E. faecium (Fig. 1).
Fig. 1. Phylogenetic relationship of the multi-Drug resistance Enterococcus isolates and related genera based on full size 16S rDNA sequences. The tree was constructed using neighbor joining algorithm with Kimura 2 parameter distances in MEGA 7.1 software. The bar indicates the Juke-Cantor evolutionary distance.
Antibiotics sensitivity test
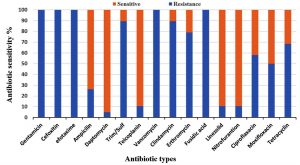
All Enterococcus strains showed resistance to the majority of selected antibiotics (Fig. 2). A significantly higher resistance to gentamicin, cefoxitin, cefotaxime, clindamycin, erythromycin and fusidic acid were found in E. faecalis than in E. faecium. On the contrary, a higher resistance to trim/sulf and tetracycline was observed in E. faecium. Furthermore, a low resistance to nitrofurantoin, linezolid and daptomycin was noticed in both species. In addition, these isolates were found to be less than 50% sensitivity to ciprofloxacin, moxifloxacin and tetracycline (Fig. 2 and 3).
Fig. 2. Antimicrobial resistance profiles of nineteen multi-Drug resistance Enterococcus strains against sixteen antibiotics.
Antimicrobial resistance genes prevalence
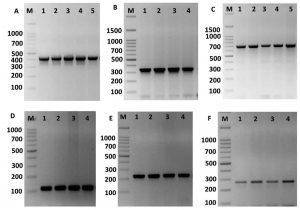
The KpsII, tetL, ermB, aac(60)-Ie-aph(200)-Ia and vanA genes were recognized in all isolates. The identification of these genes in both species were showed in table 2. The ermB gene was found in the majority of ciprofloxacin-resistant enterococci, indicating the occurrence of different systems related to the resistance of enterococci to fluoroquinolones. Similar gene were present in 79% among the erythromycin-sensitive enterococci and 10.5% among the linezolid-sensitive enterococci, demonstrating no expression of ermB gene in some enterococcus strains. Further, ermB gene occurred at a greater and significance role in fluoroquinolone-resistant enterococci compare to that in the sensitive species. Therefore, the existence of ermB gene was connected with the protection against fluoroquinolones in the Enterococcus species (Fig. 4).
Table (2):
Antimicrobial resistance profiles of the 19 Enterococcus isolates.
| Isolates | Antibiotic resistance genes | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Tuf | KpsII | Dd1-E | Dd2-E | Van-A | Erm(B) | aac(6)-Ie-aph(2)-Ia | Tet(L) | ||
| Enterococcus faecalis (E1) | + | + | + | ــ | + | + | + | + | |
| Enterococcus faecalis (E2) | + | + | + | ــ | + | + | + | + | |
| Enterococcus faecalis (E3) | + | + | + | ــ | + | + | + | + | |
| Enterococcus faecium (E4) | + | + | ــ | + | + | + | + | + | |
| Enterococcus faecalis (E5) | + | + | + | ــ | + | + | + | + | |
| Enterococcus faecalis (E6) | + | + | + | ــ | + | + | + | + | |
| Enterococcus faecalis (E7) | + | + | + | ــ | + | + | + | + | |
| Enterococcus faecalis (E8) | + | + | + | ــ | + | + | + | + | |
| Enterococcus faecium (E9) | + | + | ــ | + | + | + | + | + | |
| Enterococcus faecalis (E10) | + | + | + | ــ | + | + | + | + | |
| Enterococcus faecium (E11) | + | + | ــ | + | + | + | + | + | |
| Enterococcus faecalis (E12) | + | + | + | ــ | + | + | + | + | |
| Enterococcus faecalis (E13) | + | + | + | ــ | + | + | + | + | |
| Enterococcus faecalis (E14) | + | + | + | ــ | + | + | + | + | |
| Enterococcus faecalis (E15) | + | + | + | ــ | + | + | + | + | |
| Enterococcus faecium (E16) | + | + | ــ | + | + | + | + | + | |
| Enterococcus faecalis (E17) | + | + | + | ــ | + | + | + | + | |
| Enterococcus faecalis (E18) | + | + | + | ــ | + | + | + | + | |
| Enterococcus faecalis (E19) | + | + | + | ــ | + | + | + | + | |
Fig. 4. Amplification of some specific genes producing in some Enterococcus isolates by single PCR. (A) ErmB gene specific for erythromycin resistance with size about of 405 bp. (B) aac(60)-Ie-aph(200)-Ia gene specific for gentamicin with size about of 348 bp. (C) TetL gene specific for tetracycline resistance with size about of 696 bp. (D) Tuf gene specific for Enterococcus spp. with size about of 112 bp. (E) VanA gene specific for vancomycin resistance with size about of 231 bp. And (F) KpsII gene specific for capsule formation in Enterococcus species with size about of 272 bp. First lane on each panel is 100 bp molecular weight markers.
Rep-PCR analysis
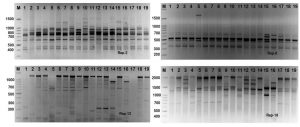
Rep-PCR markers were used to investigate the genomic diversity of Enterococcus strains. Table 3 and Figure 4 illustrates rep-PCR results. Rep-PCR markers produced 142 distinct loci; 96 bands were polymorphic (67.6%) and merely 46 bands were monomorphic (32.4%). Individual rep-PCR primers included a number of bands varying from 9 bands for rep-08 to 18 bands for rep-02. The greatest polymorphism marker was recorded for rep-08, rep-12 and rep-18. The band size of rep-18 ranged between 350 to 2100 bp (Fig. 5). The lowest polymorphism (62.5%) was detected in rep-01. Primers rep-1, rep-8, rep-12 and rep-18 produced equal mono morphism and polymorphism bands. The band size of rep-12 ranged between 100 to 1400 bp, while, the band size of rep-18 ranged between 350 to 1700 bp (Fig. 5).
Table (3):
Polymorphic bands of each genetic primers and percentage of polymorphism in nineteen Enterococcus spp. isolates based on the five rep-PCR primers.
Primers |
Total Bands |
No. of Monomorphic Bands |
No. Polymorphic Bands |
% Monomorphic bands |
% Polymorphic bands |
|---|---|---|---|---|---|
Rep-1 |
16 |
6 |
14 |
37.5 |
62.5 |
Rep-2 |
18 |
10 |
8 |
35.7 |
64.3 |
Rep-8 |
9 |
6 |
3 |
33.3 |
66.7 |
rep-12 |
16 |
6 |
10 |
33.3 |
66.7 |
rep-18 |
14 |
6 |
8 |
33.3 |
66.7 |
Total |
142 |
46 |
96 |
32.4 |
67.6 |
Fig. 5. Rep-PCR profile of 19 Enterococcus spp. isolates generated with four rep primers, rep-1, rep-2, rep-8, rep-12 and rep-18, respectively. First lane on each panel is 100 bp molecular weight markers.
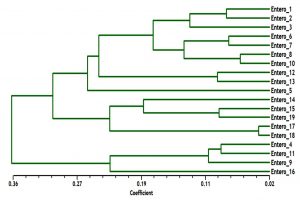
The phylogenetic analysis tree using Neighbor Joint method according to the bands that produced from rep-PCR markers was shown in Fig. 6. A low genetic distance, ranged from 0.02 to 0.36, among Enterococcus isolates was determined based on Jaccard’s similarity coefficients (Fig. 6). The Enterococcus strains were gathered into two groups with around 66% genetic similarity. The first group was included E. faecium samples (Entero-4, 9, 11 and 16), with similarity 76%. While, the second main group included E. faecalis samples, that divided into two clusters. The first cluster contained Entero-14, 15, 17, 18 and 19 isolates, and the second one contained Entero-1, 2, 3, 5, 6, 7, 8, 10, 12 and 13. These results suggested that mostly genetic distance between native Enterococcus strains was quite low among each species, while, was high between the two species (Fig. 6).
Out of 134 clinical isolates, 19 antiretroviral strains, 15 as E. faecalis and 4 as E. faecium, were obtained from King Faisal Hospital in Taif, Saudi Arabia, within four months. The antimicrobial resistance of Enterococcus spp. was recognized to be common in gastrointestinal tracts2,14,18. The frequency of both species in 206 strains of intestinal tract infections that obtained from clinical specimens at Riyadh hospitals, KSA, were 69.2% for E. faecalis and 11.3% for E. faecium19. Also, the different microorganism that isolated from intestinal patients of the University Hospital in Taif, Saudi Arabia were documented as E. faecium with 34% and E. faecalis with 33%4. In agreement to these reports, the main two species, E. faecium and E. faecalis, in our clinical specimens were found to be a natural resistance to gentamicin and aminoglycosides. Although bacteria found sensitive to these drugs in laboratory trials, effectiveness was initiate to be unsatisfactory in clinical practice20,21. Enterococcus species has been reported as widely multiple-antimicrobial resistance4,22. In the present study, a greater level of resistance to gentamicin, cefoxetin, cevotaxim, clindamycin, erythromycin and fucidic acid was detected in E. faecalis than E. faecium, while a greater resistance occurrence towards Trim/Sulf was detected in E. faecium. The resistance of E. faecalis to tetracycline was greater than E. faecium. Furthermore, a low predominance of protection against linezolid and nitrofurantoin was identified in both species. In this way, linezolid, vancomycin, and nitrofurantion are as of now generally utilized for viable treatment of intestinal fistula infection23,24. To achieve an understanding of the change in antimicrobial resistance in gastrointestinal tracts in different hospitals, a comparison of our study with other clinical intestinal strains that have resistance towards antimicrobial agents and collected from January 2014 to December 2015 was made4. There was a significant increase percentage of Enterococcus strains, where E. faecium was remain prevalent, but the component rate enlarged. Persistent antibiotic pressure is believed to lead to secondary resistance to gastrointestinal microbes4,25. Here, ermB gene was found in all intestinal cysteine resistance to ciprofloxacin. Moreover, the existence of the ermB gene was highly significant in strain of enterococci that showed resistance to fluoroquinolone than those sensitive strains. Currently, many amino-glycoside adjusting enzymes have been recognized that have tow functions of 6′-aminoglycoside acetyltransferase (AAC(6′)) and 2″-aminoglycoside phosphotransferase (APH(2″)). These enzymes usually encoded by aac(6′)/aph(2″) gene. This gene typically removes the synergistic consequence between glycopeptide or penicillin antibiotics and aminoglycosides5. The incidence of aac(60)-Ie-aph(200)-Ia gene was detected in all strains. Here, the occurrence of tetracycline-resistant genes remained in E. faecium with 10.5% and in E. faecalis with 89.5%. Consequently, protection of intestinal tract species against tetracyclines is believed to be as a result of the existence of a gene that encodes the relating enzymes. One of the mechanisms that responsible for the resistance of macrolides in intestinal microorganisms is the alteration of erythromycin target site by ermB and mef genes in the genome of enterococci microorganisms26. In this study, vanA gene was identified in all 19 strains. Vancomycin-resistant Enterococcus species have been able to transfer vanA gene to Staphylococcus aureus, leading to the appearance of vancomycin-resistant S. aureus bacteria, leading to further difficulty in the clinical treatment of intestinal infections4,26. As a result, care should be taken when vancomycin is used as a drug for treatment of gastrointestinal infection and management of viomycin-resistant enterococci need to be enhanced23.
Molecular procedures used to identify bacterial strains vary as far as unfair power, reproducibility, standardization, cost, simplicity of advancement and interpretation27. REP-PCR were established and demonstrated great outcomes when utilized to type Enterococcus strains13. There are a few other PCR-based strategies effectively applied for enterococcal separation, for example RAPD-PCR28, ITS-PCR29, tDNA-PCR30, AFLP31, and PCR-RFLP32. Additionally, multilocus arrangement examination (MLSA) is likewise a procedure utilized for recognizable proof of enterococci by utilizing partial sequences of RNA polymerase alpha subunit, phenylalanyl-tRNA synthase and the alpha subunit of ATP synthase33,34. In any case, PCR results are progressively hard to examine, since the presence of different powerless loci in the PCR profiles makes it hard to decipher the results. Rep-PCR with oligonucleotide primers used for amplified genomic DNA from enterococci and demonstrated acceptable results when used to identify E. faecium clinical isolates35. Here, the Rep-PCR method approved to be a satisfied technique to discriminate Enterococcus spp., however, additional studies should be assumed to evaluate strains obtained from other hospitals.
Enterococci have been the major pathogenic microorganisms that make clinical contaminations because of various antimicrobial resistance, and the clinical enterococcal diseases prevalently happen in the urinary framework. Antimicrobial sensitivity changes in various Enterococcus species, and the resistance of enterococci to antimicrobial antibiotics is essentially inferable from the development of antimicrobial resistance genes. The screening of antimicrobial resistance of Enterococcus species would give a manual for the fitting determination of antimicrobial agents and avert the existence of increasingly antimicrobial-resistant enterococcal strains. The present results propose a moderately high predominance of antibiotic resistance in enterococci strains, especially to gentamicin, vancomycin, cefoxitin, and efotaxime. It is of worry that the scope of antibiotics to which resistance has been gained after some time is generally wide, to the degree that it presently incorporates new developing antibiotics utilized for the treatment of enterococcal contamination.
ACKNOWLEDGMENTS
We eagerly thank all the patients who were sampled in this study. We thank the nursing and medical staff at the participating hospital for their assistance.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
WFA and EMF designed the study. AS, MA, HH and KA contributed to the collected and analysis of the clinical samples. MAF performed the molecular analysis. WFA and EMF contributed in the literature research and aided in draft the manuscript. WFA, MA and HH contributed to data analysis and manuscript with the final version. All authors read and accepted the final manuscript.
FUNDING
None.
ETHICS STATEMENT
This study was accepted by the ethics council of obstetrics hospital in King Faisal complex, Taif, Saudi Arabia.
AVAILABILITY OF DATA
All datasets obtained or studied during this study are incorporated in the manuscript.
- Gilmore, M.S, P.S. Coburn, S.R. Nallapareddy, and B.E. Murray. “Enterococcus virulence,” in The Enterococci: Pathogenesis, Molecular Biology, and Antibiotic Resistance, M. S. Gilmore, D. B. Clewell, P. Courvalin, G. M. Dunny, B. E. Murray, and L. B. Rice, Eds., 2002; pp. 325.
Crossref - John, V.U. and J. Carvalho. Enterococcus: review of its physiology, pathogenesis, diseases and the challenges it poses for clinical microbiology. J. Front. Biol., 2011; 6: 357.
Crossref - Olawale, K.O., S.O. Fadiora, and S.S. Taiwo. Prevalence of Hospital-Acquired Enterococci Infections in Two Primary-Care Hospitals in Osogbo, Southwestern Nigeria. Afr. J. Infect. Dis., 2011; 5(2): 40-46.
Crossref - Hassan, M.M. and E.B. Belal. Antibiotic resistance and virulence genes in enterococcus strains isolated from different hospitals in Saudi Arabia. Biotechnol. Biotechnol. Equip., 2016; 30.
Crossref - Al-Bloushy, A.A. and A. Elbehiry. History, Virulence Genes, Identification and Antimicrobial Resistance of Enterococcus faecalis Isolated from Diabetic Foot Patients. Int. J. Curr. Microbiol. App. Sci., 2018; 7(01): 2136-2154.
Crossref - Narayanaswamy A., K. Rajalakshmi, and M. Varadharajan. Speciation and antimicrobial susceptibility pattern of enterococci from a tertiary health care center of south India. J. Pharm. Res., 2011; 4(4): 989-90.
- Anvarinejad, M., G. Pouladfar, A. Japoni, S. Bolandparvaz, Z. Satiary, P. Abbasi, and J. Mardaneh. Isolation and Antibiotic Susceptibility of the Microorganisms Isolated from Diabetic Foot Infections in Nemazee Hospital, Southern Iran. J. of Pathogens., 2015; ID 3287967.
Crossref - Soheili, S., S. Ghafourian, Z. Sekawi, V. Neela, N. Sadeghifard, R. Ramli, and R.A. Hamat. Wide Distribution of Virulence Genes among Enterococcus faecium and Enterococcus faecalis Clinical Isolates. Sci. World J., 2014; ID 623174:6.
Crossref - Giridhara Upadhyaya, P. M., B.L. Umapathy, and K.L. Ravikumar. Comparative Study for the Presence of Enterococcal Virulence Factors Gelatinase, Hemolysin and Biofilm Among Clinical and Commensal Isolates of Enterococcus faecalis. J. Lab. Physicians., 2010; 2(2): 100-104.
Crossref - Vogan, A.A. and P.G. Higgs. The advantages and disadvantages of horizontal gene transfer and the emergence of the first species. Biology Direct., 2011; 6: 1.
Crossref - Tyrell G.J., R. N. Bethune, B. Willey, and D. E. Low. DE. Species identification of enterococci via intergenic ribosomal PCR. J. Clin. Microb., 1997; 35: 1054- 1060.
- Bachellier S., D. Perrin, M. Hofnung, and E. Gilson. Bacterial interspersed mosaic elements (BIMEs) are present in the genome of Klebsiella. Mol. Microb., 1993; 7: 537-544.
Crossref - Mercier E., E. Jumas-Bilak, A. Allardet-Servent, D. O’Callaghan, and M. Ramuz. Polymorphism in Brucella strains detected by studying distribution of two short repetitive DNA elements. J. Clin. Microb., 1996; 34: 1299-1302.
- Hassan, M.M. and A.I. Ismail. Isolation and molecular characterization of some pathogenic mobile phone bacteria. Int. J. Biochem. Biotechnol., 2014; 3: 516-522.
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement. Wayne: Clinical and Laboratory Standards Institute; 2014.
- Hassan, M.M., A. Gaber, A.O. Attia and A.R. Baiuomy. Molecular Characterization of Antibiotic Resistance Genes in Pathogenic Bacteria Isolated from Patients in Taif Hospitals, KSA. Ame. J. Phytom. Clin. Thera., 2014; 2: 939-951.
- Gaber, A., M.M. Hassan, S.E. Dessoky, and O. Attia. In vitro Antimicrobial Comparison of Taif and Egyptian Pomegranate Peels and Seeds Extracts. J. App. Biol. Biotech., 2015; 3(02): 012-017.
- Klare, I., C. Konstabel, D. Badst bner, G. Werner, and W. Witte. Occurrence and spread of antibiotic resistances in Enterococcus faecium. Int. J. Food. Microbiol., 2003; 88: 269-290.
Crossref - Salem-Bekhit, M.M., I.M. Moussa, M.M. Muharram, F.K. Alanazy, and H.M. Hefni. Prevalence and antimicrobial resistance pattern of multidrug-resistant enterococci isolated from clinical specimens. Indian J. Med. Microbiol., 2012; 30: 44-51.
Crossref - Sood, S., M. Malhotra, B.K. Das, and A. Kapil. Enterococcal infections & antimicrobial resistance. Indian J. Med. Res., 2008; 128: 111-121.
- Hollenbeck, B.L. and L.B. Rice. Intrinsic and acquired resistance mechanisms in enterococcus. Virulence., 2012; 3(5): 421-433.
Crossref - Arias, C.A. and B.E. Murray. The rise of the Enterococcus: Beyond vancomycin resistance. Nat. Rev. Microbiol., 2012; 10: 266-278.
Crossref - Arias, C.A., G.A. Contreras, and B.E. Murray. Management of multidrug-resistant enterococcal infections. Clin. Microbiol. Infect., 2010; 16: 555-562.
Crossref - Hammerum, A.M., C.H. Lester, and O.E. Heuer. Antimicrobial-resistant enterococci in animals and meat: A human health hazard? Foodborne. Pathog. Dis., 2010; 7: 1137-1146.
Crossref - Xi, R., S.F. Tian, Y.Z. Chu, and B.Y. Chen. Linezolid induces resistance in Enterococcus in vitro and the mechanism. Chin. J. Infect. Chemother., 2011; 11: 22-26.
- Portillo, A., F. Ruiz-Larrea, M. Zarazaga, A. Alonso, J.L. Martinez, and C. Torres. Macrolide resistance genes in Enterococcus spp. Antimicrob Agents Chemother., 2000; 44(4): 967-71.
Crossref - Versalovich J., T. Koeuth, and J.R. Lupski. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res., 1991; 19: 6823-6831.
Crossref - Yousif, N.M., P. Dawyndt, H. Abriouel, A. Wijaya, U. Schillinger, M. Vancanneyt, J. et al. Molecular characterization, technological properties and safety aspects of enterococci from ‘Hussuwa’, an African fermented sorghum product. J. Appl. Microbiol., 2005; 98: 216228.
Crossref - Tyrrell, G.J., R.N. Bethune, B. Willey, and D.E. Low. Species identification of enterococci via intergenic ribosomal PCR. J. Clin. Microbiol., 1997; 35: 10541060.
- Devriese, L.A., M. Vancanneyt, P. Descheemaeker, M. Baele, H.W. Van Landuyt, B. Gordts, et al. Differentiation and identification of Enterococcus durans, E. hirae and E. villorum. J. Appl. Microbiol., 2002; 92: 821827.
Crossref - Vancanneyt, M., A. Lombardi, C. Andrighetto, E. Knijff, S. Torriani, K.J. Bjorkroth, et al. Intraspecies genomic groups in Enterococcus faecium and their correlation with origin and pathogenicity. Appl. Environ. Microbiol., 2002; 68: 13811391.
Crossref - Lu, J.J., C.L. Perng, S.Y. Lee, and C.C. Wan. Use of PCR with universal primers and restriction endonuclease digestions for detection and identification of common bacterial pathogens in cerebrospinal fluid. J. Clin. Microbiol., 2000; 38: 20762080.
- Naser, S., F.L. Thompson, B. Hoste, D. Gevers, P. Dawyndt, M. Vancanneyt, and J. Swings. Application of multilocus sequence analysis (MLSA) for rapid identification of Enterococcus species based on rpoA and pheS genes. Microbiology, 2005; 151: 21412150.
Crossref - Naser, S., F.L. Thompson, B. Hoste, D. Gevers, K. Vandemeulebroecke, I. Cleenwerck, C.C. Thompson, M. Vancanneyt, and J. Swings. Phylogeny and identification of enterococci by atpA gene sequence analysis. J. Clin. Microbiol., 2005; 43: 22242230.
Crossref - J. Bedendo and A.C.C. Pignatari. Typing of Enterococcus faecium by polymerase chain reaction and pulsed field gel electrophoresis. Braz. J. Med. Biol. Res., 2000; 33: 1269-1274.
Crossref
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.