ISSN: 0973-7510
E-ISSN: 2581-690X
The purpose of this study is to establish thermal processing validation of tender coconut water by using SYBR green Real time PCR assay for direct detection of the biological indicator bacteria, B. cereus, a frequent cross contaminant in packaged tender coconut water. Nucleotide sequences coding for the hemolytic enterotoxin hemolysin BL (Hbl) widely associated with B. cereus food poisoning was used as the target gene for the real time PCR assay. Specific amplification was observed with B. cereus when the specificity of the assay was examined using other closely related spore forming food spoilage Bacilli. The present real-time PCR assay provides a detection of limit (LOD) of 103 CFU/ml of tender coconut water. The thermal resistance of B. cereus (ATCC 14579) was found that D95 2.46 min and Z value 10.45°C in tender coconut water. To validate the thermal processing conditions, B. cereus was artificially contaminated into coconut water, which was thermally processed and checked with real-time PCR assay to detect the survival of B. cereus in tender coconut water bottles without pre-enrichment. The bacterial load enumerated by the real-time PCR assay correlated with the time consuming standard conventional culture method. The thermal processing of tender coconut water packed in polypropylene bottles was validated with 6D concept and minimum 18 min of process time is required to achieve sterilization value (F-value) of 15. The product was analyzed for its physical, chemical properties and commercial sterility after the validated processing conditions and products were acceptable for 6 months without preservative.
Coconut water, F value, thermal processing, PCR, real-time PCR.
Tender coconut (Cocos nucifera L.) water, a refreshing natural health drink, is the liquid endosperm inside a young coconut with various minerals such as sodium, potassium, phosphorus, chloride and magnesium, besides vitamin C and sugars (Campos et al., 1996; Nadanasabapathy and Kumar, 1999). The consumption of coconut water has increased in the last few years, mainly because of the increase in awareness among consumers regarding natural and healthy products. Preservation of tender coconut water makes it possible to get available in natural form beyond the borders of tropical countries (Jerard et al., 2008). The main concern in preserving tender coconut water is to maintain natural taste, safety and quality. (Goblin et al., 2009) The pH value above 4.5 and water activity close to 1.0 of tender coconut water provide favorable conditions for microbial growth and contamination. When the coconut water inside the nut is sterile and due to the introduction of microorganism from the processing environment contact surfaces leads the microbial load to higher (Jeff et al., 2013). After collection and cold-stored natural (unprocessed) coconut water itself Escherichia coli and Salmonella have been detected in addition to Staphylococcus aureus and Bacillus cereus populations of up to 8.0 x 104 CFU/ml and \ 1.6 x 105 CFU/ml, respectively due to cross contamination (Walter et al., 2009). Bacterial spores cause major problems in the food industry due to their ubiquitous occurrence and their intrinsic high stress resistance characteristics (Burl et al., 2011). B. cereus, a gram-positive, spore forming bacterium has long been known for its significance in food spoilage and causing severe gastroenteritis upon intake of food that usually contains more than 105 CFU/g of B. cereus cells or spores (Overcast and Atmaram, 1974; Jeff et al., 2013; Granum, 2002).
The different methods to process and preserve tender coconut water include acidification, addition of chemical preservatives followed by pasteurization and High Temperature Short Time (HTST) aseptic processing (Jayasundera et al., 2014; FAO 2007). Thermal and non-thermal treatments, sometimes combined with chemical additives have also been tried with varying degrees of success (Gobin et al., 2009). A successful processing technology with help of bio-preservatives and pasteurization have been developed to get natural sensory attributes (Srivatsa et al., 1995,1997). However, the low acid content of the tender coconut water need a proper inactivation strategies and validation of process conditions/parameters to ensure the safety and prevent spoilage. Thermal process validation study with surrogates of pathogens or indicator microorganism have been reported previously for food products (Janelle et al., 2012) wherein the indicator organisms for specific food and process validation are selected based on their ability to survive the processing conditions of the particular food matrix. Thermal death times of non-spore forming E. coli in young coconut endosperm beverage have been reported (Gabriel et al., 2009), literature is still scanty regarding thermal resistance of spore forming organisms in coconut water.
Biological validation is found to be more reliable method used to validate the thermal processing conditions and verify the microbiological food safety of products. Inoculated pack study is very common method used as biological validation of packaged foods. The National Advisory Committee on Microbiological Criteria for Foods has published the challenge studies on pathogen guidelines for conducting inhibition and inactivation of organisms in variety of foods (NACMCF, 2010). For the inoculated pack study, standard culture plating methods are used for enumeration of survival microorganism after thermal processing and there is a sustained requirement for the development of rapid methods for detection and enumeration of bacterial endospores in different foods while evaluating the efficacy of new alternative food-processing technologies. Conventional PCR method for the identification of pathogenic strains of B. cereus and the detection of the toxin gene have been used elsewhere (Ehling-Schulz et al. 2004, 2006). Real-time quantitative PCR (qPCR) represents a powerful tool that could greatly help to assess the safety and quality of foodstuffs. The purpose of this study is to develop and standardize a real-time PCR method for detection of B. cereus a frequent cross contamination indicator organisms in packaged tender coconut water during its bulk production without elaborate enrichment techniques and to establish thermal process conditions of tender coconut water packed in polypropylene bottles using real-time PCR assay.
Chemicals, reagents, culture media
All the chemicals and reagents of analytical grade were obtained from Sigma-Aldrich and culture media were from Himedia, India.
Bacterial strain
Bacillus cereus ATCC 14579 was used as test strains in the present study. The bacterium was inoculated in 10 ml Brain Heart Infusion (BHI) broth and incubated overnight at 30 °C. Five-hundred-micro liter of culture broth were spread onto surface of sporulation agar media plate reported elsewhere (Rawsthorne et al., 2009). The broth culture spreaded plates were incubated at 30 °C for 3 to 5 days to get 95% confluency of spores. Spore growth on the agar surface of plates were harvested in cold, sterile distilled water and washed repeatedly (5 times) and stored at 5°C temperature.
Determination of thermal resistance
Experiments were conducted by taking 1 ml of 106 B. cereus spores suspended in phosphate buffer and tender coconut water separately in covered micro tubes to prevent moisture loss before placed in a water bath for thermal resistance study. The temperature was measured using thermocouples (E. Val flex, M/s. Ellab, Denmark) placed in the center of the additional control tube. Testing was performed at 75°C, 85°C and 95°C the tubes were removed at different intervals time. After removing from heat, tubes were placed into an ice bath for 60 s to minimize any further thermal effects. Tubes were mixed then serially diluted and survival microbial load were tested on nutrient agar at 30°C for 48 h. incubation. The colonies were counted by using a colony counter. Survival curve drawn by plotting log of survival counts vs. their corresponding heating times. D values were calculated from the slope of the regression line obtained with the values of the straight portion of the survival curves. Z values were determined from the regression line obtained by plotting log D values vs. their corresponding heating temperatures.
DNA template preparation
Overnight growth of B. cereus bacterial culture in BHI broth was centrifuged at 7000xg for 10 min at 4°C. Bacterial pellet was washed once in normal saline and DNA was extracted using kit method. (DNeasy Tissue kit, Qiagen, Gmbh) with manufacturer’s instruction. The DNA was quantified by NanoDrop ND-1000 Spectrophotometer (Bio Photometer, Eppendorf, Hamburg, Germany). This DNA was stored in -20°C for further use.
SYBR green real-time PCR assay
Based on extensive literature survey, the Hbl gene which encodes for the hemolytic enterotoxin protein and highly conserved among B. cereus was chosen as the target for real time PCR assay. The forward (BCEA (F):–TACAGGGTTATTGGTTACAGCA) and reverse (BCEA (R):- ACTCTCTTACATTTGCCTTTGC) primers were designed using the Primer 3 and BLAST algorithms of Primer BLAST tool of NCBI and custom synthesized from M/s Sigma India. The primers were also evaluated for their insilico specificity by Primer BLAST and would produce a 269 bp amplicon. Employing these primers, SYBR green based real-time PCR was developed by varying critical factors like primer concentrations, annealing time and temperature. The CFX96 real-time PCR thermal cycler (Bio-Rad) and Eva Green supermix (Bio- Rad) were used for qPCR assay. The master mix consisted of 0.4µl of 10 pM primers, 10µl of SsoFast Eva Green supermix (Bio- Rad), and 5 µl template DNA in a total volume of 20 µl made up with sterile deionised water. The thermal cycling conditions comprised of initial denaturation at 95°C for 10 min, followed by 39 cycles of denaturation at 95°C for 15 s, annealing 64°C for 5 s and elongation for 2 S at 72°C. The amplification cycle was followed by a melting curve analysis in 65-95°C with increment of 0.5°C. All the samples were analyzed in triplicate and control reactions without template were included in each assay.
Specificity
The specificity of the real time PCR assay was evaluated using the DNA extracted from four different Bacillus strains viz B. cereus ATCC 14579, B. subtilis MTCC 2117, B. licheniformis NCIM 2467 and B. coagulans MTCC 7543 along with no template PCR control.
RT-PCR Efficiency and minimum detection limit studies
Efficiency of the RT-PCR assay was estimated by plotting the mean of Ct values obtained in duplicate against the B. cereus bacterial culture dilutions in freshly collected filter sterilized tender coconut water. 1mL of sample drawn from the culture dilutions with 106 to 101 bacterial cells was centrifuged and resuspended in 100µL filter sterilized tender coconut and kept in dry bath maintained at 100°C for 10 min for the bacterial lysis. The bacterial lysate was centrifuged at 8,000 x g for 10 minutes and 5 µL of the supernatant was used for the PCR assay.
Tender coconut water collection and packaging
Tender coconut water was collected from 6-7 month old nuts after washing and sanitizing the outer surface with 50 ppm chlorine water. Incision was made using sterilized SS cutter and water was collected using a sanitized pump fitted with filter into a sterile stainless steel vessel. Collected tender coconut water was aseptically filled in 200 ml capacity polypropylene bottles and the top opening hermetically sealed with sealing wade. The dimension of polypropylene bottles used for this study was 154.5 mm height, 45mm diameter and 1 mm thickness. The flow chart of the bottled tender coconut water processing is given in Fig. 4.
Thermal process calculations
B cereus is an aerobic spore forming microorganism with higher thermal resistance than B. anthracis (Thomas et al., 2005). Hence thermal processing was aimed to reduce/achieve 6D of B. cereus for safety. The D value for B. cereus in tender coconut water was used for the study and the process lethality was calculated using the following equation:
F = [10 (T-Tref)/z ] t
Where T is the temperature of the product, Tref is the sterilization/pasteurisation reference temperature that is 95°C; D value used 2.46 min., Z value used 10.45°C. Thermal processing also can be aimed to reduce/achieve 12 D reduction of B. cereus, It depended upon the initial loads of contamination estimated through plate count. Sterilisation/pasteurisation value (F-values) is calculated during processing by Ellab software.
Thermal Processing
The steam jacketed kettle temperature was maintained at 98°C for heat penetration studies and processing was carried out to achieve 6 D reduction of B. cereus based on the thermal resistance properties. The bottle was fixed with thermocouple glands through which thermocouples were inserted and the bottles were placed at different locations in the steam jacketed kettle. For validation of four bottles were artificially contaminated with 103 to 106 spores of B. cereus was processed for required F value and under process sterilization/pasteurisation value and 2 bottles were processed till 15 F value and another till 18 F value. Once bottle removed from the kettle and cooled to room temperature. Three independent batches of bottles were processed to verify the reproducibility with change in spore concentration. Thermocouple output was measured using an Ellab system and process time and sterilization/pasteurisation value (F value) was calculated using the software. Sample microbial load was estimated by plating in nutrient agar and incubation at 37°C for 48 h and incubation along with RTPCR assay.
Real-time PCR assay of artificially contaminated tender coconut water
Processed samples were incubated at 37°C for 1 h for resuscitation and 1 ml of sample drawn after incubation was centrifuged at 8,000 x g and the pellet was resuspended in 100µl phosphate buffer solution and kept it dry bath maintained at 100°C for 10 min for the bacterial lysis. The lysate was further centrifuged at 8,000 x g for 10 min and 5µl was used as template for bacterial count by real-time PCR.
Physical, Chemical properties and commercial sterility test
Physical, chemical properties were analyzed before, after processing and during storage, the Hunter colour meter was used for measuring L a b values, for total soluble solids content (Brix) with a hand refractometer and expressed as a percentage. Measurement of pH with a pH-meter (Eutech instruments), total titratable acidity by titration with 0.1 N sodium hydroxide solution (results expressed in grams citric acid per 100mL of sample) and sugars was estimated with standard method. The commercial sterility of processed tender coconut water during the storage periods was carried. (BAM, Chapter 21A, 2001).
Thermal resistance characteristics
The B. cereus spore suspensions in the micro tubes reached the required set temperature within 1 min. Neither shoulders nor tails were observed in the spore survival curves. The D-values calculated from all the three temperatures,75°C, 85°C and 95°C were within 10% standard error from duplicate experiments and survival curves with a coefficient of correlation (R2) > 0.9 were obtained. The decimal reduction time ranged from 2.46 min at 95°C to 212 min at 75°C in coconut water (Table 1). The thermal resistance for the spores observed in tender coconut water was higher than that observed with phosphate buffer. The tender coconut water is comprised of 4.5 total soluble solid that offer protection to the test organism during the heat treatment (Marshall et al., 2006). The average r2 values of the linear regression analysis were used to determine the Z-values and it was found to be 0.983. The thermal resistance of B. cereus in coconut water was D95 =2. 46, Z value 10.45°C. and thermal processing was carried out based on this value for validation.
Table (1):
Thermal resistance characteristics of B.cereus ATCC 14579 spores
| Temp (0C) | Phosphate buffer
Time (Min) |
Tender coconut water
Time (Min) |
|---|---|---|
| D –value | ||
| 75 | 195.04 | 211.87 |
| 85 | 21.14 | 22.27 |
| 95 | 2.22 | 2.46 |
| Z –value | ||
| 10.28 | 10.45 | |
Optimization and efficiency of SYBR green real time PCR:
The B. cereus strains exhibit genotypic and phenotypic diversity with respect to the presence virulence genes associated with human infection. Immunoassays for detection of B. cereus enterotoxins like Hbl, Nhe (Beecher and Wong, 1994) are available; however these assays do not allow quantitative estimation of the B. cereus bacterial load that is required to validate the effectiveness of processing. Alternatively, Real Time PCR assays involving gene targets associated with food poisoning offer accurate quantitative detection of the bacterial load in processing environment and is simple and easy to perform. We chose nucleotide sequences coding for the hemolytic enterotoxin protein (Hbl) widely associated with B. cereus food poisoning as the target gene for developing Real Time PCR assay to validate the TCW processing (Birgit et al.,1999; Ten et al., 2000; Wehrle et al., 2009). The primers designed for the specific detection of B. cereus provided amplication curves with the SYBR green dye and agarose gel electrophoresis of the PCR products yielded 269 base pair amplicon (Fig. 1a). When the amplicons were assessed by melt curve analysis at 65-95°C with increment of 0.5°C, a single peak at 78.5°C was observed (Fig. 1b). No significant peaks corresponding to non-specific amplification and primer dimers were noticed. The minimum detection limit of the present real-time PCR assay was found to be 103 CFU/ml in coconut water (Fig. 2a) when culture dilutions ranging from 103 to 106 were examined using DNA extracted by thermal lysis method. The standard graph constructed by plotting log of bacterial concentrations against their mean respective Ct values showed linear correlation with R2 value of 0.968 (Fig. 2b). The detection of fluorescence signal beyond the threshold level in early cycles of the Real-Time PCR assay correlated with the higher copy numbers pertaining to lesser dilution of bacteria (103 CFU) while higher cycle numbers correlated with in bacterial dilution. Amplification curves were observed in only in DNA extracted from B. cereus ATCC 14579 while DNA samples from other Bacillus sp did not show any amplification indicating specificity in detection (Fig. 3 a). Melt curve analysis further confirmed obtaining the right amplicons (Fig. 3 b).
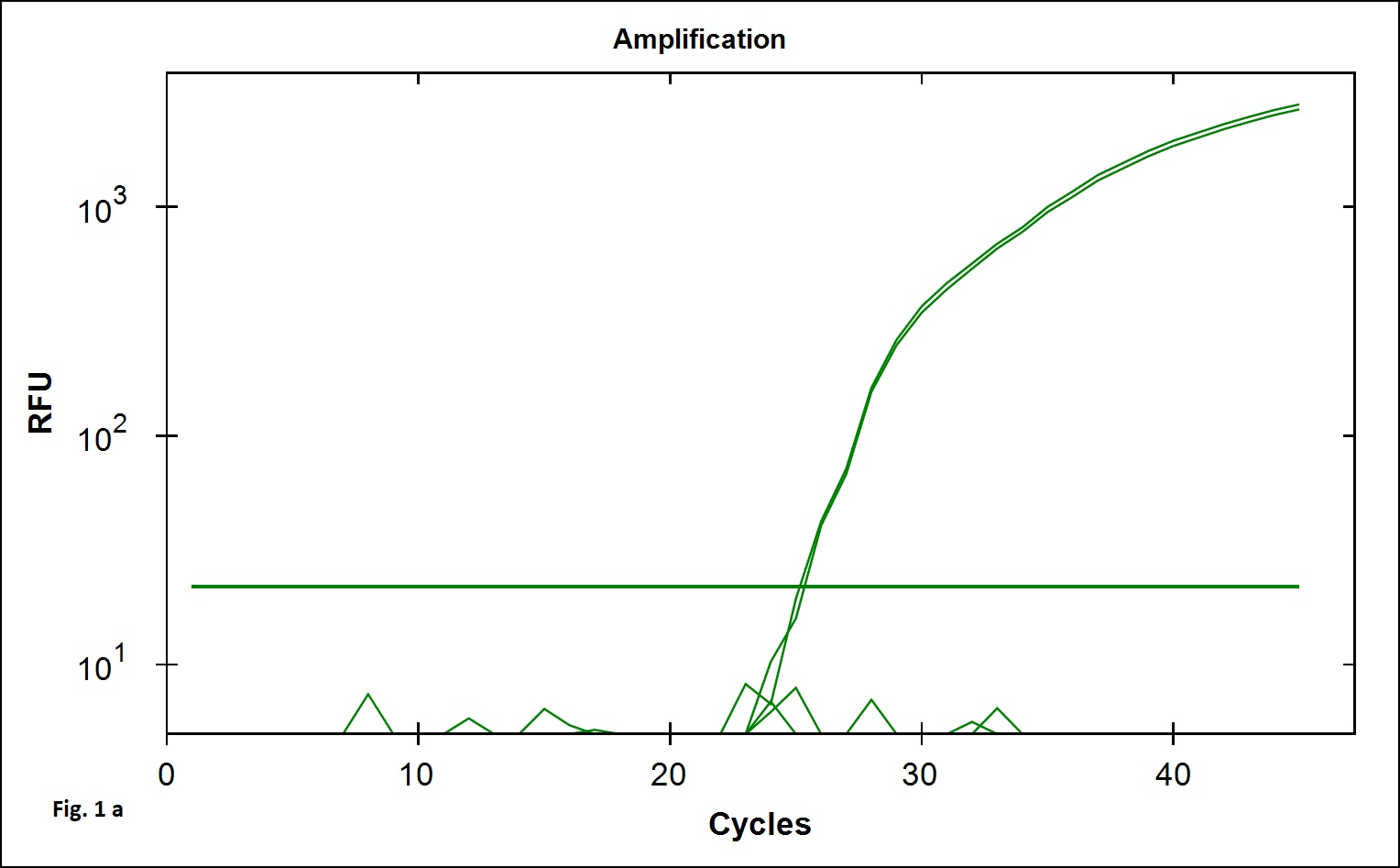
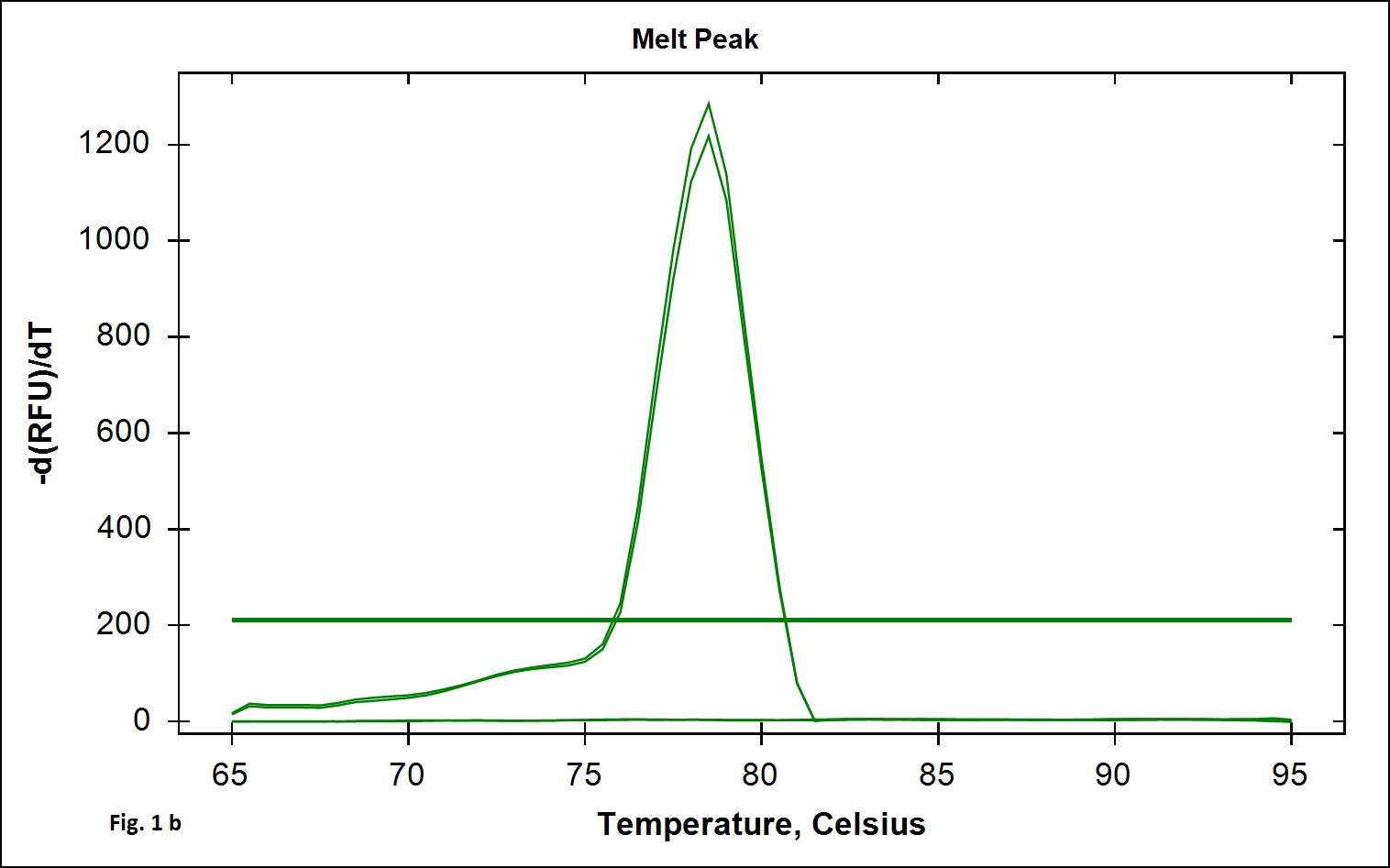 Fig. 1.SYBR green Real time PCR standardised for the detection of B. cereus hbl gene.1a. Amplification plot and 1b. melt curve analysis curve obtained with Bacillus cereus ATCC 14579 DNA.
Fig. 1.SYBR green Real time PCR standardised for the detection of B. cereus hbl gene.1a. Amplification plot and 1b. melt curve analysis curve obtained with Bacillus cereus ATCC 14579 DNA.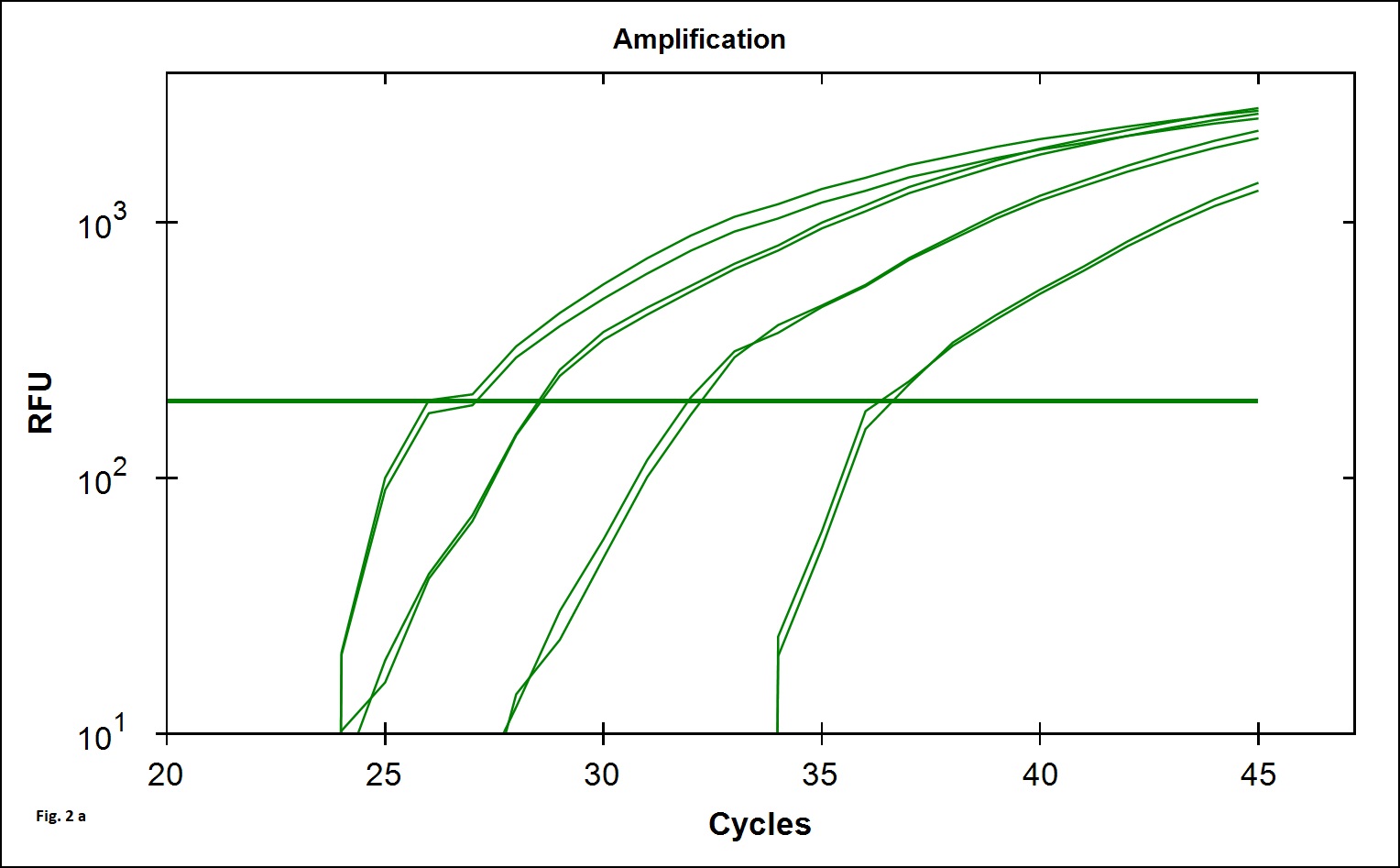
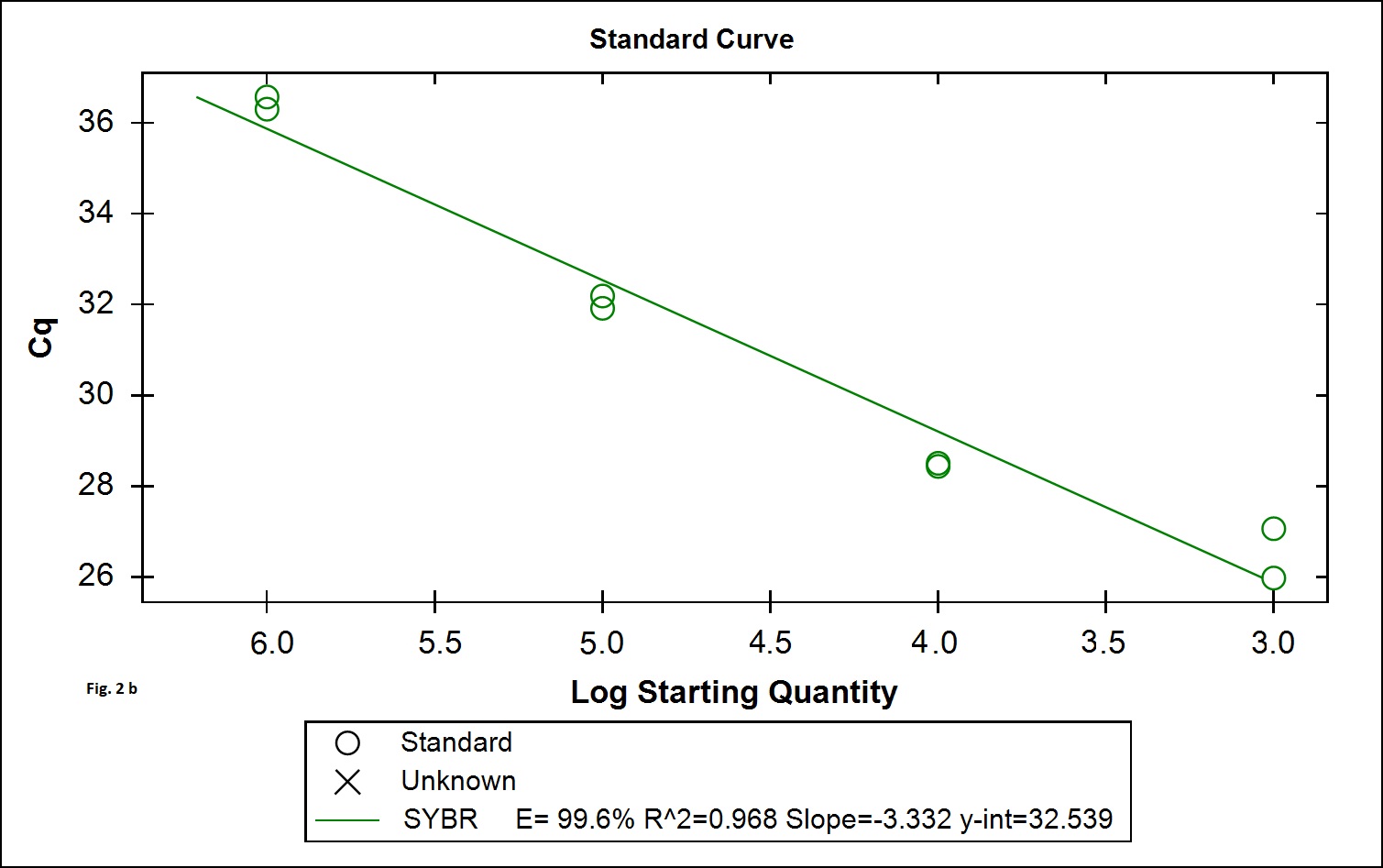 Fig. 2.Efficiency and minimum detection limit of the SYBR green Real time PCR. Fig 2a: Amplification curve obtained from 103, 104, 105, 106 bacterial cells. Fig 2b: Efficiency of the RT-PCR by plotting the mean of Ct values obtained in duplicate against the culture dilutions in freshly collected filter sterilized tender coconut water. The R2 value was found to be 0.968 and slope 3.332 at the cut off determined.
Fig. 2.Efficiency and minimum detection limit of the SYBR green Real time PCR. Fig 2a: Amplification curve obtained from 103, 104, 105, 106 bacterial cells. Fig 2b: Efficiency of the RT-PCR by plotting the mean of Ct values obtained in duplicate against the culture dilutions in freshly collected filter sterilized tender coconut water. The R2 value was found to be 0.968 and slope 3.332 at the cut off determined.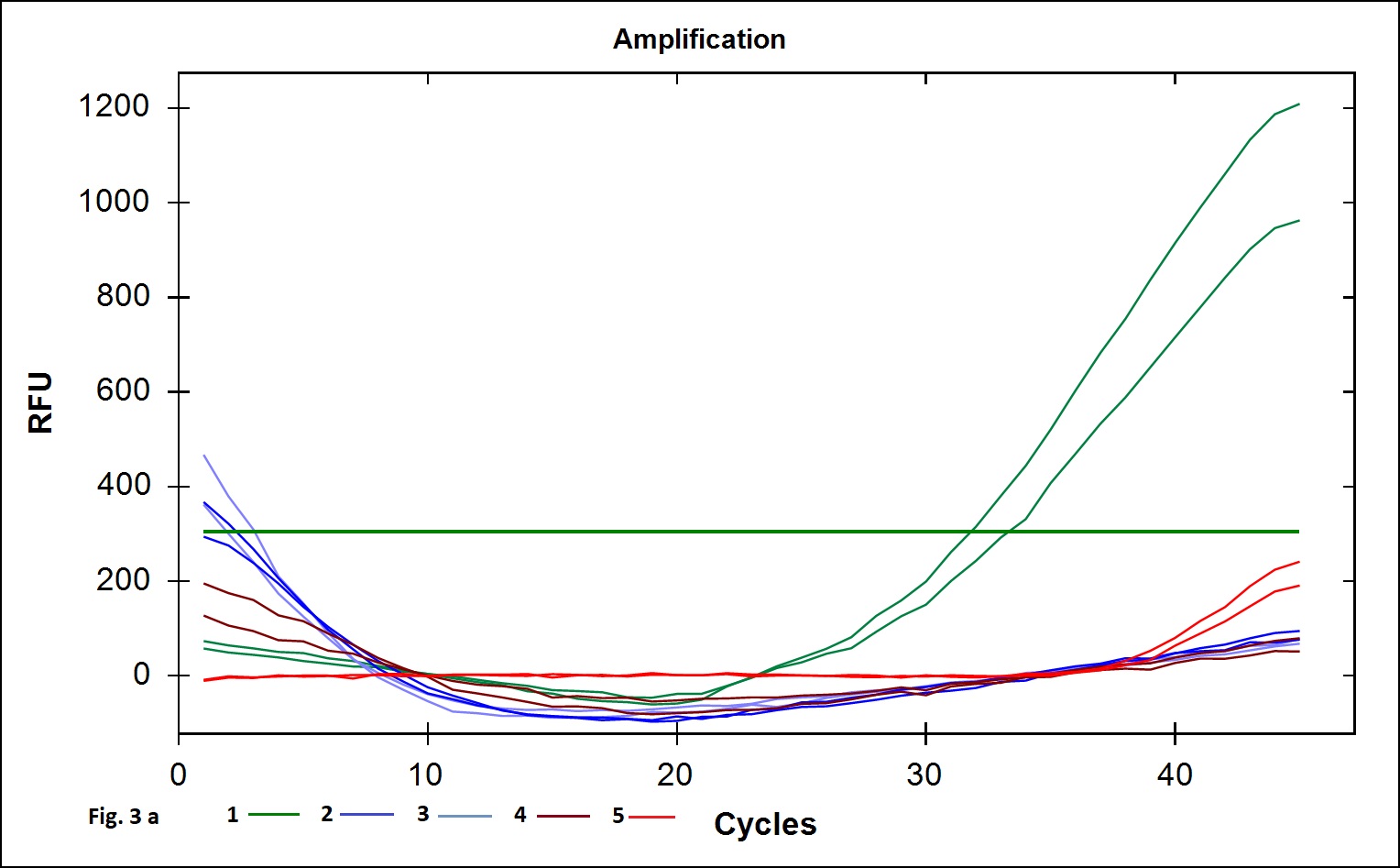
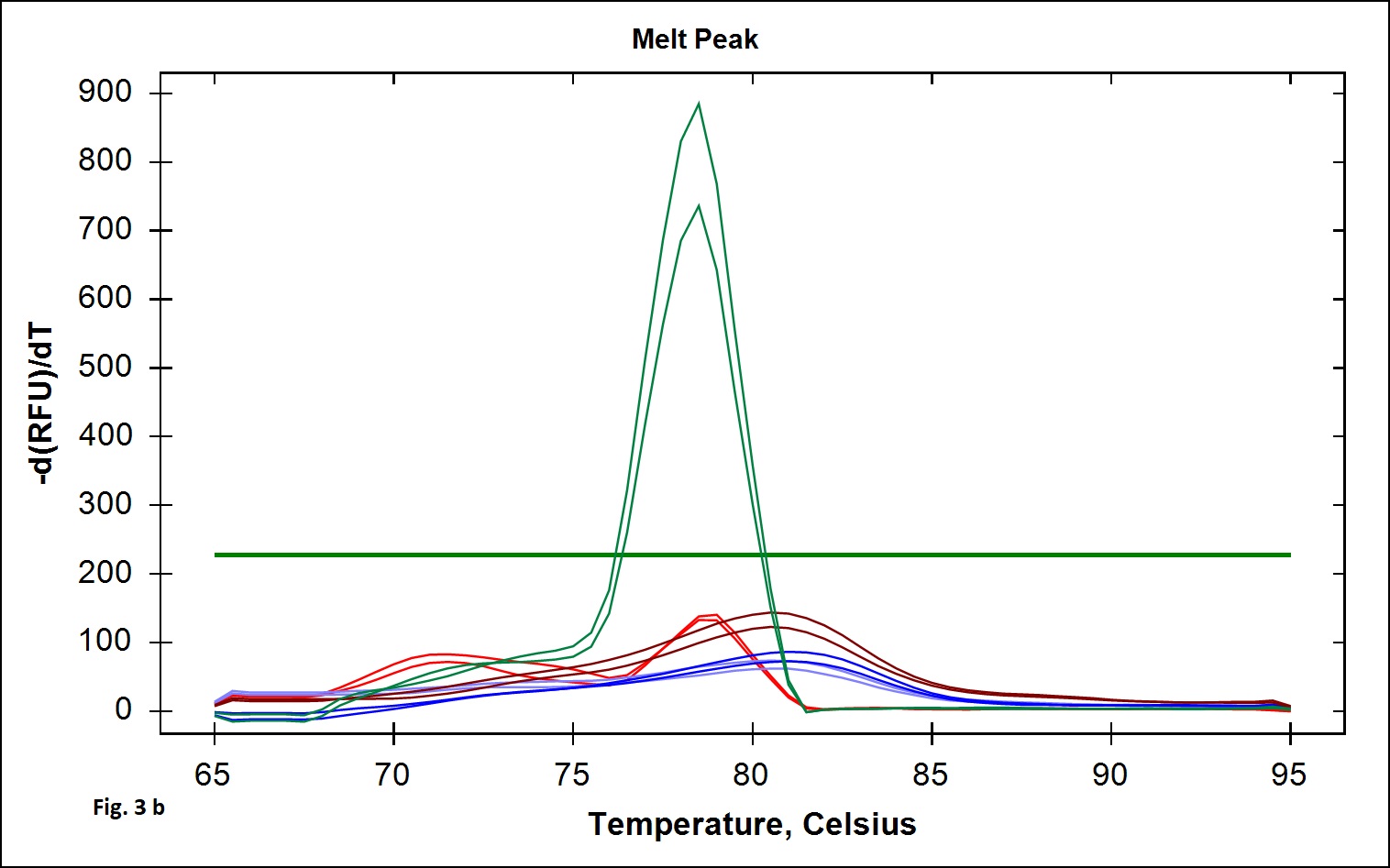 Fig. 3. The specificity analysis of the real time PCR assay. 3a: Amplifiation plot obtained when genomic DNA extracted from 1. B. cereus ATCC 14579, 2. B. subtilis MTCC 2117, 3. B. licheniformis NCIM 2467 4. B. coagulans MTCC 7543 and 5. No template PCR control. 3b: Melt curve analysis for the above samples.
Fig. 3. The specificity analysis of the real time PCR assay. 3a: Amplifiation plot obtained when genomic DNA extracted from 1. B. cereus ATCC 14579, 2. B. subtilis MTCC 2117, 3. B. licheniformis NCIM 2467 4. B. coagulans MTCC 7543 and 5. No template PCR control. 3b: Melt curve analysis for the above samples.Thermal Processing
The schema for the tender coconut water processing and Real time PCR based validation is provided in Fig. 4. The tender coconut water bottles were not steam exhausted or deaerated and contained 18% oxygen when examined by the head space analyzer (SYSTEC Instruments, Model No. 6600, USA) before and after processing. Higher level of O2 in the bottles do not allow the proliferation of the anaerobic spore forming bacteria Clostridium sp otherwise observed in the packaged food and hence was not considered as an indicator organism to evaluate the processing. B. cereus was chosen as indicator organisms for process validation. For determination of heat adequacy, depend upon the initial microbial load of the products is used (Fellow 1992). For 6 D reduction of B. cereus, F value calculated to 6 D reduction is 14.76 min; the target was to kill the microorganism population from 106 CFU to 100 CFU/ml. The total processing time to achieve 15 F value was 17 min at actual kettle boiling temperature of 98°C, which included 1 min come-up time (CUT) and 7 min cooling time to reach 45°C. For 12 D processing 30 min was the process time for achieving F value of 29.52 min with a total process time of 32 min at actual kettle boiling temperature of 98°C (Fig. 5). Previous studies reported the F value requirements of other spoilage bacteria indicators like Lactobacillus sp. and Leuconostoc sp. in pure citrus juice for 3D process is 11.26 min. at reference temperature 85°C. (Sukasih and Setyadjit 2008). Manually extracted unprocessed fresh coconut water quality by street vendors was studied and found 58% of pathogens. (Soares et al., 2017) Preservation of tender coconut water in pouches or bottles in bulk quantities required many types of machineries which lead to cross contamination by pathogenic microorganisms. Since spot contaminations are difficult to predict, problem awareness and appropriate control measures in the food safety management system are recommended to tackle this problem. (FAO 2007).
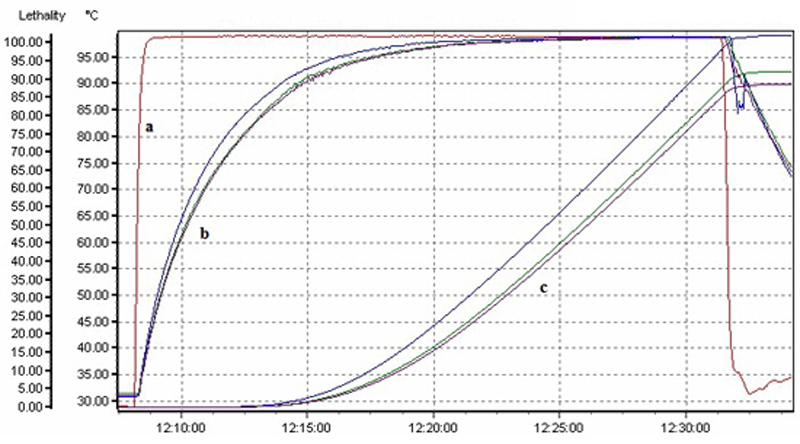 Fig. 5. Heat penetration profile and lethality value of tender coconut water. a) temperature profile from a) probe placed inside the steam jacketed kettle temperature, b) probe placed inside the polypropylene bottle bag with tender coconut water and c) cumulative lethality value of the tender coconut water calculated from the data logger to monitor the processing.
Fig. 5. Heat penetration profile and lethality value of tender coconut water. a) temperature profile from a) probe placed inside the steam jacketed kettle temperature, b) probe placed inside the polypropylene bottle bag with tender coconut water and c) cumulative lethality value of the tender coconut water calculated from the data logger to monitor the processing. Real-time PCR assay to validate the thermal processing
The real time PCR assay developed was used to quantitate the B. cereus population artificially contaminated in tender coconut water and was compared with agar plate count. Thermal processing reduced the 103 or 106 CFU/ml of artificially contaminated bacterial count based on the process F values achieved in polypropylene bottles (PP). No bacterial count was observed in PP bottles that were artificially contaminated with 103 and 106 CFU/ml with required F value of 7.38 and 14.76 respectively. The bottles inoculated with 106 CFU/ml under processed bottles with F values of 7 min showed microbial growth in plates and were also detected in RT- PCR (Table 2). The results of bacterial counts calculated from bacterial plating and RT- PCR assay correlated with each other, thus establishing the current RT- PCR assay to replace the cumbersome and time-consuming plating techniques for assessing the bacterial contamination in processed foods.
Table (2):
Microbial count and RT-PCR assay of artificially contaminated tender coconut water
| Bacterial load | Before processing | Process F value | After processing | ||
|---|---|---|---|---|---|
| Plating | RT-PCR | Plating | RT-PCR | ||
| 103 | 3.2 × 103 | 1.4 × 103 | 8 | Nil | Nil |
| 106 | 2.0 × 106 | 1.0 × 106 | 7 | 3.3 × 103 | 4.1 × 103 |
| 15 | Nil | Nil | |||
Chemical properties and commercial sterility test
Physical, chemical properties of tender coconut water are shown in Table 3. The validated process conditions did not show any significant changes in colour of the products and chemical constituents. All bottles were found to be commercially sterile during the storage periods and acceptable for 6 months of shelf life.
Table (3):
Chemical analysis of tender coconut water
| S. No | Parameters | Results** | |||||
|---|---|---|---|---|---|---|---|
| Before process | After processing | ||||||
| 1. | Colour* | L | a | b | L | a | b |
| 60.78 | -0.56 | 1.62 | 59.07 | -0.46 | 2.07 | ||
| 2. | Brix (%) | 4.5±0.5 | 4.5±0.5 | ||||
| 3. | pH | 4.9±0.2 | 5.0±0.2 | ||||
| 4. | Acidity (mg) | 44±5 | 48±5 | ||||
| 5. | Reducing sugar (%) | 2.9±0.5 | 2.9±0.5 | ||||
| 6. | Total sugar (%) | 4.9±0.5 | 5.0±0.5 | ||||
Hunter colour meter (L a b – value) , ** ± SD (n=5)
Tender coconut water is a low acid nutritional health drink and susceptible to rapid deterioration by micoroganisms. Thermal processing of packed tender coconut is a best way to preserve without preservative, a full process validation is necessary to assure the microbial safety and spoilage. We report the successful use of a real-time PCR method to estimate surviving spore populations in thermally processed tender coconut water without further elaborate enrichment. The real time PCR method of validating the processing of tender coconut water can be completed in 2 hr instead of the conventional microbiological methods that required at least 2 days.This work of process validation will probably increase the quality of young coconut water and give a chance to millions of coconut share holders to increase the value of their product. The method may also be appropriate for validation of advanced thermal and non-thermal sterilization technologies.
Acknowledgements
The authors would like to thank Director, DFRL for providing the facilities to undertake this research work.
Conflict of Interest
The authors declares that there is no conflict of interest.
Authors’ Contributions
JC designed, performed the study and wrote the manuscript; SRU provided technical support on real time PCR work; JKJ designed the work, guided on experimental work and reviewed the manuscript.
Funding
None.
Data Availability
The data that support the findings of this study are available with the first author Jayaprahash C.
Ethics Statement
No animal or human subjects was involved in the work related to this manuscript.
-
- BAM: Examination of Canned Foods, Bacteriological Analytical Manual, 2001. USFDA. 8th Edition, Chapter 21A.
- Beecher DJ, Wong AC. Identification and analysis of the antigens detected by two commercial Bacillus cereus diarrheal enterotoxin immunoassay kits. Appl Environ Microbiol, 1994; 60(12): 4614-6.
- Birgit MP, Richard D, Birgit N, Erwin, M, Siegfried, S. The Hemolytic Enterotoxin HBL Is Broadly Distributed among Species of the Bacillus cereusGroup. Appl Environ Microbiol, 1999; 65(12): 5436–5442.
- Brul S, van Beilen J, Caspers M, O’Brien A, de Koster C, Oomes S, Smelt J, Kort R, Ter Beek A. Challenges and advances in systems biology analysis of Bacillus spore physiology; molecular differences between an extreme heat resistant spore forming Bacillus subtilis food isolate and a laboratory strain. Food Microbiol, 2011; 28: 221-227.
Crossref - Campos CF, Souza PEA, Coelho JV, Glףria MBA. Chemical composition, enzyme activity and effect of enzyme inactivation of flavor quality of green coconut water. J Food Process Pres, 1996; 20(6):487-500.
Crossref - Ehling-Schulz M, Fricker M, Scherer S. Identification of emetic toxin producing Bacillus cereus strains by a novel molecular assay. FEMS Microbiol Lett, 2004; 232: 189–195.
Crossref - Ehling-Schulz M, Guinebretiere, MH, Monthan A, Berge O, Fricker, M, Svensson B. Toxin gene profiling of enterotoxic and emetic Bacillus cereus. FEMS Microbiol Lett, 2006; 260: 232–240.
Crossref - FAO. Training guide How to Bottle Coconut Water. Agricultural and food engineering, training and resource materials, 2007, http://www.fao.org/3/a1418e/a1418e.pdf Accessed 6 Aug, 2019.
- Fellow PJ. Food Processing Technology. 1992; pp252, 2nd ed. CRC Press, New York.
- Gabriel AA, Cruz KGS, Guzman JADD. Thermal death times of Escherichia coli in young coconut endosperm beverage, J. Food Process Preserv, 2009; 33: 136–144.
Crossref - Gobin A, Falade KO, Akingbala JO. Effect of packaging on physical, chemical and sensory attributes of coconut water during storage, J Food Agric Environ, 2009; 7(1): 62 -65
- Granum PE. Bacillus cereus and food poisoning. In Applications and Systematics of Bacillus and Relatives ed. Berkeley R, Heyndrickx M, Logan N. de Vos P. 2002; pp. 37–46. Malden, USA: Blackwell Science Ltd.
- Janelle L, Brown, Nai TD,Belinda C. Clostridium sporogenes PA 3679 and Its Uses in the Derivation of Thermal Processing Schedules for Low-Acid Shelf-Stable Foods and as a Research Model for Proteolytic Clostridium botulinum. J Food Protect, 2012; 75(4): 779–792.
Crossref - Jayasundera M, Dharmasena A. Preservation of tender coconut water of Sri Lankan tall coconut variety, Annals Food Sci Technol, 2014; 15(1): 111-114
- Jerard BA, Damodaran V, Jaisankar I, Velmurugan A, Swarnam, TP. Coconut Biodiversity – Nature’s Gift to the Tropical Islands. Biodiversity and Climate Change Adaptation in Tropical Islands, 2008; pp145–185, Ademic press.
Crossref - Marshall, MR., Marcy JE, Braddock, RJ. Effect of Total Solids Level on Heat Inactivation of Pectinesterase in Orange Juice. J Food Sci, 2006; 50(1): 220-222.
Crossref - Nadanasabapathy S., Kumar R. Physico-chemical constituents of tender coconut (Cocos nucifera) water. Indian J Agr Sci, 1999; 69: 750-751.
- NACMCF – National advisory committee on microbiological criteria for foods Parameters for Determining Inoculated Pack/Challenge Study Protocols. J Food Protect, 2010; 73(1): 140-202.
Crossref - Overcast WW, Atmaram K. The role of Bacillus cereus in sweet curdling of fluid milk. J Milk Food Technol, 1974; 37: 233–236. https://doi.org/10.4315/0022-2747-37.5.233
- Rawsthorne H, Dock CN, Jaykus LA. PCR-based method using propidium monoazide to distinguish viable from nonviable Bacillus subtilis spores. Appl Environ Microbiol, 2009;75(9):2936-9.
Crossref - Srivatsa AN, Sankaran R. Preservation of tender coconut water in polymeric pouches and metal cans, Indian Coconut J., 1995; 26: (1/2) 13.
- Srivatsa AN, Sabapathy SN, Nataraju S, Visweswaraiah G, Manja, KS. Mohan MS, Aravindakshan, M, Process for preservation and packaging of tender coconut water, Indian patent No. 186975, 1997.
- Soares K, Morais D, Gois V, Silva J, Costa A, Silva L. Quality of unprocessed cooled fresh coconut water manually extracted by street vendors. Arquivos do Instituto Biologico. 2017; 84.
Crossref - Sukasih E, Setyadjit. Study on heat resistance and heat adequacy to inactivate microorganism, population in pasteurized pure citrus juices cv. Siam. Indones J Agric, 2008; 1(1): 22-27
- Thomas JM, Rebecca D, Tara DS, Marcelo B, Schaffner W. Thermal Resistance of Spores from Virulent strains of Bacillus anthracis and Potential Surrogates. J Food Protect, 2005; 68(11): 2362–2366.
Crossref - Tsen HY, Chen ML, Hsieh YM, Sheu SJ, Chen YL. Bacillus cereus group strains, their hemolysin BL activity, and their detection in foods using a 16S RNA and hemolysin BL gene-targeted multiplex polymerase chain reaction system. J Food Protect, 2000; 63(11): 1496-502.
Crossref - Wehrle E, Moravek M, Dietrich R, B rk C, Didier A, Martlbauer E. Comparison of multiplex PCR, enzyme immunoassay and cell culture methods for the detection of enterotoxinogenic Bacillus cereus. J Microbiol Methods, 2009; 78(3): 265-70.
Crossref - Walter EHM, Nascimento MS, Kuaye AY. Efficacy of sodium hypochlorite and peracetic acid in sanitizing green coconuts. Lett Appl Microbiol, 2009; 49: 366–371.
Crossref
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.



