ISSN: 0973-7510
E-ISSN: 2581-690X
Mutation in katG gene of Mycobacterium tuberculosis encoding catalase-peroxidase that damage its enzyme activities is well associated with isoniazid (INH) resistance. The katG gene from INH resistant strain of M. tuberculosis clinical isolate L19 has been observed in previous study, carrying mutations of G234A and C625T, and changed the arginine residue at position 209 with cysteine in its KatG protein. However the activities of the mutant protein has been not known yet. Expression of the katG gene L19 that was done in Escherisicia coli BL21(DE3) using pCold II-DNA generated KatG protein with 80 kDa in SDS PAGE electroforegram. The mutant protein of KatG L19 decreased 43% of catalase activity and 11% of peroxidase activity against to KatG wild type (H37RV). The model structure of the mutant KatG protein had deviation structure toward KatG wt as 0,13 for number of Root Mean Square Deviations (RMSD). The mutant KatG (Arg209Cys) losed two hydrogen interactions and a van der Waals bond which present in KatG wild type. In the KatG wt protein, the both hydrogen bonds was formed between the Arg209 residu to Glu201, while the van der Waals bond occured by lingking of Arg209 residu to Glu217. Disruption in the some chemical interactions might trigger the decline in catalase-peroxidase activities of mutant KatG L19 and further it bring out the INH resistance in the clinical isolate L19.
katG, catalase-peroxdase, isoniazid resistance, M. tuberculosis.
Mycobacterium tuberculosis, the causing agent of tuberculosis (TB) disease, recently many strains have been found resistant to the TB drugs. This events caused the TB cases more difficult to resolve. Multi-drug resistant tuberculosis (MDR-TB) is TB resistant to at least two potent anti-TB drugs, such as isoniazid and rifampicin together, or with resistance to first-line anti-TB drugs, namely, pyrazinamide, ethambutol and streptomysin.1 Indonesia posed the third ranks for TB cases in the world after India and China, having the total of TB burden are around 395,000 per one thousand populations in 2015 and 126,000 of them are deaths. Of TB cases, as many as 32,000 are cases of MDR-TB1,2. A better understanding in the antituberculous drug resistance is needed to make easy in the TB therapy.
The mechanism of drug resistance in Mycobacterium tuberculosis can occur in several ways, (1) a decrease in the effectiveness of the drug due to over production of its target protein, (2) drug efflux that cause the drug could not reach to its target, and (3) alteration of in the drug target protein due to mutations of their genes. Most research showed that the main cause of M. tuberculosis resistance to antibiotics were due to mutations of the genes targeted by anti-tuberculosis drugs, such as gene rpoB for rifampin, katG gene for isoniazid, pncA gene for pyrazinamide, and embB gene ethambutol.3,4.
Isoniazid (hidrazid acid 4-piridinkarboksilat, INH) is a mainstay chemical agent used for the treatment of TB, since first introduced in 1951 to the present. This is caused by it having a high bactericidal effect and relatively cheap. Isoniazid is a prodrug that is converted to an active intermediates (Isonicotinoyl acyl radical) by the catalase-peroxidase enzyme encoded by the katG gene of M. tuberculosis. Isonicotinoyl acyl radical which formed, then reacts with NADH to form INH-NADH complex in the enzyme active site of enoly ACP reductase (InhA) and 2-ketoacyl ACP synthase (KasA), then impede the activity of two enzymes in the biosynthesis of mycolic acid, the main component in the cell wall of mycobacteria5,6,7. Disruption in the biosynthesis of mycolic acid can cause the mycobakteri cells death. The analysis of of catalase-peroxidase activities is required to determine the involvement of the enzyme in the emerging of isoniazid resistance in M. tuberculosis. Disruption of catalase-peroxidase function in isoniazid activating can cause INH resistance to M. tuberculosis.7,8.
Purkan (2011) have identified a katG gene of INH resistant M tuberculosis clinical isolates (L19). The clinical isolate which had resistant to INH at a level of 0.2 mg / mL carried double mutations of G234A and C625T in katG gene. The katG mutations changed the amino acid of arginine at position 209 to Cysteine in KatG protein. Although katG mutation in the clinical isolate L19 has been connected with INH resistance, but there is no explanation for the catalytic activity of the KatG protein. It is important to clarify, because mutational event can be linked with the emergence of other traith in the cells that are not associated with INH resistance phenotype. The katG gene of M. tuberculosis clinical isolate L19 has been cloned in Escherichia coli using pCold II-DNA vector. To explain in detail the causes of INH resistance in clinical isolates of L19, since it was determined the catalase-peroxidase activity of the mutant protein of KatG (Arg209Cys), then followed by modelling of its structure.
Samples
The Escherichia coli BL21 (DE3) carrying of pCold II-katGwt and pCold II-katG L19 recombinant respectively was obtained from the laboratory of biochemistry, Faculty of Sciences and Technology, Airlangga University. The isolates were resulted from the previous research.
Culturing of Eschericia coli
The recombinant of E. coli was cultured in liquid and solid Luria Bertani (LB) media containing 100 mg/mLof ampicillin. The liquid LB media had compositions of 0,5% (w/v) yeast extract. 1% (w/v) tryptone and 1% (w/v) NaCl. The solid media had the same compositions with the liquid media, but it cantained 2% (w/v) agar.[9]
Expression of katG gene and protein isolation
The E. coli BL21 (DE3) carrying of (pCold II-katG) recombinant plasmid was grown in 50 mL liquid LB media containing ampicillin 100 mg / mL, then incubated with shaking at 150 rpm in the temperature of 37 °C. The obtained cultures was immediatly treathed with cool shock at a temperature of 15oC for 30 minutes without shaking. The culture cells then was added with 0.1 mM IPTG and re-incubated with shaking at 150 rpm in the temperature of 15oC for 24 hours. The culture cells was separated by centrifugation at 5.000g in the temperature 4°C for 10 minutes. After the cell pellet was washed with lysis buffer (50 mM Tris-Cl pH 7,4; 200 mM NaCl), they was centrifugated at 5.000g in the temperature 40C for 10 minutes. The obtained cell pellet was resuspended in 7-10 mL of 0.02 M phosphate buffer pH 7, then lysed with the sonicator for 10 minutes on the frequency scale 4. The sonication process was carried out in cold conditions (incubated in ice). The obtained cell suspension then was centrifuged at 10.000g in the temperature of 40C for 20 minutes. The supernatant containing of KatG protein was stored at -20oC10,11
Protein Purification
The purification of KatG recombinant protein was performed by affinity chromatography using HP HisTrap column containing Ni-sepahrose matrix. The protein sample was firsly adjuted in 0.02M phosphate buffer pH 7.4 containing 25-50 mM NaCl and 10 mM imidazole. The matrix in the HiTrap HP column was balanced with binding buffer (50 mM NaH2PO4 pH 7.4, 25 mM NaCl, 10 mM imidazole). The crude extract protein which has undergone initial preparation was filled in the chromatographic column slowly. All the liquid coming out of the column was collected. The column was washed with binding buffer as much as 5 times of the column volume. In the final step, the target protein was eluted with elution buffer (50 mM NaH2PO4 pH 7.4, 25-50 mM NaCl) containing 50-200 mM imidazole. Each fraction was accommodated per 1 mL, then the presence of katG protein in the fraction was detected by polyacrylamide gel electrophoresis10,11
SDS-PAGE Electrophoresis
The Extract protein was analyzed by SDS PAGE using 12% (w/v) acrylamide for separating gel and 4% (w/v) acrilamide for stacking gel. The running process was done in 120 mV for 1,5 hours. This method was adopted from Sambrook et al, 19899
Assay of Enzyme Activities
Catalase activity is determined by the method of Patti and Bonnet-Maury (1953)12 based on the color formed from the reaction of titanium with H2O2 reagents in the reaction mixture containing substrate of 12.5 mM H2O2 and enzyme extract, The test is performed in 10 mM potassium phosphate buffer pH 7 with a total volume of 1 mL containing 12.5 mM substrate H2O2. The amount of remaining substrate in the mixture was determined by the addition of 2.5 ml of titanium reagent, then the absorbance of the yellow colour fromed was determined at l 410 nm. One unit of enzyme activity is defined as the amount of enzyme that catalyzes the decomposition of 1 mol of hydrogen peroxide per minute at the experimental conditions11
Peroxidase enzyme activity is determined by reaction of 100 µM O-dianisidine in the presence of 25 mM tert-butyl hydroperoxide (t-BHP) in 50 mM potassium buffer (pH 4.5) and 12.5mm H2O2 substrate13. The product of O-dianisidine quinonediimine formed was determined at » 460 nm (µ460 = 11.3mM-1 cm-1). One unit of enzyme activity is defined as the amount of enzyme that catalyzes the formation of 1 mol product per min on the experimental conditions10,11.
Modeling of the Three Dimensional Structure of the mutan KatG Protein
Construction of the mutant katG protein structure model was performed by a server automatically modelers for protein structure (http://www.expasy.org/Swiss-Model). The structure templates used in this modeling is three-dimensional structure of KatG wild type protein (1sj2b). The model structure of the mutant katG were then minimized by Amber program 10, and aligned with the structure of the katG wild type using PyMOL 1.3 program. Deviations structure of the mutant KatG toward KatG wild type was analyzed by SuperPose program version 10 to determine the amount Root Mean Square Deviations (RMSD). Interaction between amino acid residues both mutant and wild type katG was visualized with the 1.3 PyMOL program10,11.
Expression and Perification aod mutant KatG
Catalase-peroxidase is one of the enzymes in the M. tuberculosis that is running the activation of isoniazid14,15 Mutations in katG gene encoding the catalase-peroxidase enzyme is often associated with the emergence of resistance to INH in M. tuberculosis. To associate the katG mutations in clinical isolates of M. tuberculosis L19 with the catalytic activity of KatG protein produced, it is necessary to express the gene and test its activity of catalase-peroxidase.
The expression of katG from INH sensitive M. tuberculosis (H37Rv) and from INH resistant clinical isolate (L19) were performed in the host cell of Escherichia coli BL21 (DE3) using pCold II-DNA vector, the work could generate the KatG protein 80 kDa in SDS PAGE electropherogram (Fig 1, lane 2 and 3). The 80 kDa protein band was not resulted by the control E. coli without pCold II-DNA plasmid (Fig 1, lane 1). Therefore, the expression system of pCold-II-DNA vector is controlled by the promoter cspA, a gene derived from cold shock and lac pomotor of E. coli,16 then to express the katG gene, it was done the addition of IPTG inducer and cold shock treatment to the culture of recombinant. E.coli cells. The KatG protein extract from expression result was further purified by methods Immobilized Metal Affinity Chromatography (IMAC)], using HP HisTrap column containing Ni-sepharosa matrix. This purification technique could produce the pure katG protein, as indicated by the presence of a single band of protein 80 kDa in SDS PAGE electropherogram for KatG of M. tuberculosis H37Rv (Fig 1, lane 4-5) and for KatG L19 (Fig 1, lane 6-8), The KatG protein band that obsearved in SDS PAGE electropherogram was very significantly dominant, and almost invisible presence of the other non-target protein. The purity level of katG protein sulted from the purification step was over 90%.
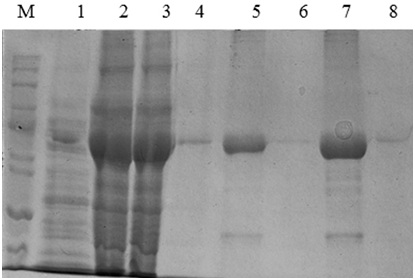
Fig 1 . The SDS PAGE Electroforegram of the KatG protein. (M), Marker protein marker; 1, crude protein obtained from E.coli BL21 (DE3); 2&3, extract protein from E.coli BL21 (DE3)[pCold-katGwt] and [pCold-katGL19]; 4&5, the fraction of KatG wt protein eluted with imidazole 100 and 150mM; 6-8, the fraction of KatG L19 protein eluted with imidazole 100 dan 150mM
Catalase-peroxidase activities of KatG protein from clinical isolate L19
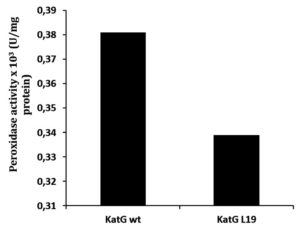
Determination of catalase-peroxidase activities for KatG protein was done by using substrate of H2O2 for catalase and o-dianisidin for peroxidase activity. The mutant of KatG L19 exhibited diminishing in catalase-peroxidase activities comparing with KatG wild type (H37RV). The mutant KatG L19 had specific activity 0,393 x 103 U/mg for catalase, and 0,69 x 103 U/mg for peroxidase activity. The catalase activity of mutant KatG L19 reduced for 43% toward KatG wt (Fig 2). Meanwhile, the mutant KatG L19 had peroxidase activity as 0,339x 103 U/mg, but the KatG wt exhibited 0,381 x 103 U/mg. The mutant KatG L19 had peroxidase activity 11% lower than KatG wt (Fig 3). The change of Arg209Cys amino acids in the KatG L19 might trigger the decreasing in the catalase-peroxsidase activities for the mutant KatG L19.
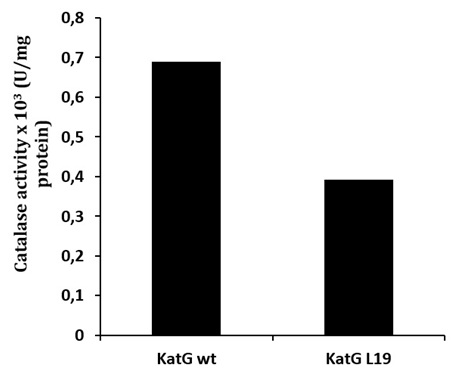
Fig. 2. The catalase activity of KatG. The mutant KatG L19 had catalase activity 43% lower than the KatG wt.
Fig. 3. The peroxidase activity of KatG. The mutant KatG L19 had catalase activity 11% lower than the KatG wt
A decrease in catalase-peroxidase activities in some variant katG of M. tuberculosis strains resistance to INH was also reported by other researchers. The mutant KatG (S315T) which was found in clinical isolates of M. tuberculosis isoniazid resistance, decreased 50% catalase-peroxidase activity of KatG wt17,18 Meanwhile, the mutant KatG (Asn238Ser) decreased the catalytic efficiency as 41% for catalase and 52% for peroxidase respectively11 Wei (2003) reported that the change of catalase-peroxidase activity of the variants katG directly correlated with activity activates INH17 To know the basis of the change in catalase-peroxidase activity in KatG L19, it was created the structure modeling of the mutant protein.
The Structure Model of Mutan KatG L19 Protein
The model structure of mutant katG L19 (Arg209Cys) was constructed using three-dimensional protein structure template katG wt (1sj2b) of M. tubeculisis (H37Rv). The structure of the mutant katG L19 resulted from modeling method indicated similar folds with KatG wt structure (1sj2b) (Fig 4, A-B). Superposition of two structures KatG showed that all residues between mutant and wild type katG impinge upon each other well (Fig 4C). Superposition of C± skeleton between mutant KatG L19 to KatG wt indicated root mean square deviations (RMSD) of 0:13 (Table 1), which meaned that there was a change in the structure of mutant katG L19 compared with the structure of katG wt. The other effect from the mutations of Arg209Cys in the KatG L9 was the change on the total charge in protein molecule, i.e -21 in the KatG wt and -22 in the KatG L19 (Table 1).
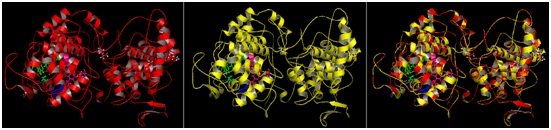 Fig 4. The allignment product of structure model of mutant KatG L19 toward KatG wt. The KatG wild type structure (A); the model structure of mutant KatG L19 (B); the superposition result of the KatG L19 toward KatG wt (C)
Fig 4. The allignment product of structure model of mutant KatG L19 toward KatG wt. The KatG wild type structure (A); the model structure of mutant KatG L19 (B); the superposition result of the KatG L19 toward KatG wt (C)
Table (1):
The model strukture RMSD of mutant KatG (Arg209Cys) toward KatG wild type
KatG |
Total muatan |
RMSD (C±) |
|---|---|---|
KatG wild type (wt) |
-21 |
0 |
KatG (R209C) (L19) |
-22 |
0.13 |
The interpretation of the model structure for KatG L19 show Arg209Cys amino acid modifications lead to changes in interactions at multiple amino acid residues around the substrate binding and catalytic side of KatG (Fig 5, A). In the structure of KatG protein, the amino acid residues Arg209 located in around of substrate binding side that composed by amino acid residues Asn137, Val230, Asn231, Pro232 and Ser315. Three of the five amino acid residues for substrate binding, namely Val230, Asn231, and Pro232 is located very close to Arg209 (Fig 5, A). The Arg209 residue has an important role to support the stability of the environment in which the substrate is bound and catalyzed. In the structure of KatG wt, amino acid residues Arg209 appears to form two hydrogen bonds with residues Glu201 and a van der Waal interaction with Glu217 (Fig 5, A). Replacement of Arg209 Cys in the mutant KatG L19 eliminated these types of interaction which present in KatG wt (Fig 5, B). The loss of two hydrogen interactions and a van der Waals interaction might cause disruption in the function of catalase-peroxidase KatG L19, in turn it decreased the enzym activity. Reduction in the catalase-peroxidase activities might further trigger the emergence of INH resistance in clinical isolates L19.
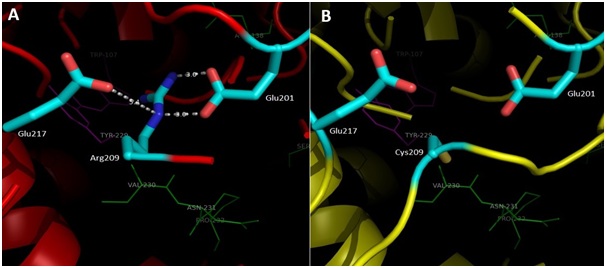
Fig 5. Illustration of the effect for Arg209Cys substitution in mutant KatG L19. Superposition result of mutant KatG L19 structure (yellow) toward KatG wt (red). The green residues like Val230, Asn231 and Pro232 constituted amino acid residues for binding sites, while the magenta residues like Trp107 and Tyr229 rep-resented as active sites in KatG. The residu Arg209 in KatG wt make two hidrogen interactions with Glu201 and van der Waal interaction with Glu217 (A). Replacement of Arg209Cys in KatG L19 eliminated the interactions (B).
Thw KatG failure in running the catalytic activity was also demonstrated in the mutant katG (Ser315Thr).[13,19,20,21] Modification of the amino acid serine at position 315 into threonine emerge an impact on changing the conformation of the inlet substrate, thus the mutant KatG (Ser315Thr) could not bind well its substrate13,21 Modification of Ser315Thr shifted the substrate channel from 6 A in the KatG wt to 4.7 A in the mutant KatG (Ser315Thr)20 As a result of this structural change, the mutant KatG (Ser315Thr) had dropped catalase-peroxidase activity around 50%19 In future, the crystal structure of the mutant KatG L19 is importantly developed to detect the real changes in its protein structure.
ACKNOWLEDGMENTS
The research was supported by DIPA DITLITABMAS Project fiscal year 2015 in accordance with the Decree of the Rector of Airlangga University, Number: 519 / UN3 /2015. We would also thank to Go Bambang Sugiarto, Ph.D, California University, Davis for critically reading the manuscript
- Anonymous. Tuberculosis. https://www.expat.or.id/medical/tuberculosis. Retrieved 2016-11-25
- Anonymous, Indonesia Tuberculosis Profile. www.who.int/tb/data. Retrieved 2016-11-25
- Shi, R., Koji, O., Hiroyuki, Y., Taiga, T., Isamu, S. Temperature-Mediated Heteroduplex Analysis for The Detection of Drug-Resistant Gene Mutations in Clinical Isolates of Mycobacterium tuberculosis by Denaturing HPLC, SURVEYOR Nuclease, Microbes and Infection., 2006; 8:128–135
- Parka, T.Y.K., Sonya, S., Sungweon R., Sang N.C., Won-Jung K., Kwond, O.J., Young S.S., Woo, J.L., dan Gill, H.B. Comparison of Drug Resistance Genotypes between Beijing and Non-Beijing Family Strains of Mycobacterium tuberculosis in Korea., Journal of Microbiological Methods., 2005; 63:165– 172
- Yu S., Chouchane, M., Magliozzo, R.S. Characterization of the W321F mutant of Mycobacterium tuberculosis catalase–peroxidase KatG, Protein Science., 2002; 11: 58–64
- Atalay, F.M.D., Nejat, A., Dilek, E.T., Derya, A., P1nar, E., Yurdanur, E.A.N. Catalase-Peroxidase Gene (KatG) Deletion in Isoniazid Resistant Strains of Mycobacterium Tuberculosis., T Klin J Med Sci., 2004; 24: 243-246.
- Cade, C.H., Adrienne C., Dlouhy., Katalin, F., Medzihradszky., Saida, P.S.C., Ghiladi, R.A. Isoniazid-resistance conferring mutations in Mycobacterium tuberculosis KatG: Catalase, peroxidase, and INH-NADH adduct formation activities, Protein Science., 2005; 19: 458—474.
- Ando, H., Yuji, K., Toshinori, S., Emiko, T., Seiya, K., Toru, M., dan Teruo, K. Identification of katG Mutations Associated with High-Level Isoniazid Resistance in Mycobacterium tuberculosis, Antimicrobial Agents And Chemotherapy., 2010; 54(5): 1793–1799.
- Sambrook, J. F., Maniatis, T. Molecular Cloning Laboratory Manual, 2nd edn. USA: Cold Spring Harbour Laboratory Press, 1989; pp 87-111
- Purkan., Ihsanawati., Syah YM., Retnoningrum D., Noer A., Shigeoka S., Natalia D. Novel mutations in katG gene of a clinical isolate of isoniazidresistant Mycobacterium tuberculosis. Biologia., 2012; 67(1): 41-47.
- Purkan, Ihsanawati, Natalia D, Syah YM, Retnoningrum DS, Kusuma HS. Mutation of katG in a clinical isolate of Mycobacterium tuberculosis: effects on catalase-peroxidase for isoniazid activation. Ukr Biochem J., 2016; 88 (N5): 71-81.
- Patti, F., Bonet-Maury, P. Methode Colorimetrique pour le Dosage de la Catalase. Bull. Soc. Chim. Biol., 1953; 35: 1177-80
- Wengenack, N.L., Brian, D.L., Preston, J.H., James, R.U., Gudrun, S.L.R., Leslie, H., Glenn, D.R., Franklin, R.C., Patrick, J.B., Kenton, R.R., John, J.B., Frank. R. Purification and Characterization of Mycobacterium tuberculosis KatG, KatG(S315T), and Mycobacterium bovis KatG(R463L), Protein Expression and Purification., 2004; 24: 232-243
- Bertrand, T., Eady, A.J.N., Jones, N.J., Jesmin, Nagy, M.J., Gregoire, J.N., Rave, L.E., Brown, A.K. Crystal Structure of Mycobacterium tuberculosis Catalase-Peroxidase., J. Biol. Chem., 2004; 279(37): 38991-38999.
- Wilming, M., dan Johnsson, K. Inter- and Intramolecular Domain Interactions of The Catalase-Peroxidase KatG from M. tuberculosis, FEBS Letters, 2001; 509: 272-276
- Anonymous. Cold Shock Expression System pCold™ DNA; TAKARA BIO Inc. http://www.takara-bio.com. Retrieved: 2015-2-12
- Wei, J., Benfang, L., James M.M., Shiao-Chun T.C. Isoniazid Activation Defects in Recombinant Mycobacterium tuberculosis Catalase-Peroxidase (KatG) Mutants Evident in InhA Inhibitor Production Antimicrobial Agents And Chemotherapy., 2003; 47(2): 670–675
- Devito, J.A., Morris, S. Exploring the Structure and Function of the Mycobacterial KatG Protein Using trans-Dominant Mutants., Antimicrobial Agents And Chemotherapy., 2003; 47(1): 188–195
- Kapetanaki, S.M., Salem, C., Shengwei Y., Richard S.M., Johannes, P.M.S. Resonance Raman spectroscopy of Compound II and its decay in Mycobacterium tuberculosis catalase-peroxidase KatG and its isoniazid resistant mutant S315T, Journal of Inorganic Biochemistry., 2005; 99: 1401–1406
- Zhao, X., Yu, H., Yu, S., Wang, F., Sacchettini, J.C., Magliozzo, R.S. Hydrogen peroxide-mediated isoniazid activation catalyzed by Mycobacterium tuberculosis catalase-peroxidase (KatG) and its S315T mutant, Biochemistry Journal., 2006; 45: 4131–4140.
- Kapetanaki, S.M., Xiangbo, Z., Shengwei Y., Richard, S. M., Johannes, P.M.S. Modification of The Active Site of Mycobacterium tuberculosis KatG After Disruption of the Met–Tyr–Trp Cross-Linked Adduct, Journal of Inorganic Biochemistry., 2007; 101: 422–433.
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.