ISSN: 0973-7510
E-ISSN: 2581-690X
Avian Encephalomyelitis (AE) is the disease caused by avian encephalomyelitis virus (AEV). The disease mainly affects young birds nervous system worldwide causing high morbidity and variable mortality rate in chicks and noticed egg dropping and hatchability in mature hens. Vaccination is the only way to control AEV infection since there is no treatment yet to the avian encephalomyelitis. This study aimed to use immunoinformatics approaches to predict multi epitopes vaccine from the AEV polyprotein that could elicit both B and T cells. The vaccine construct comprises 482 amino acids obtained from epitopes predicted against B and T cells by IEDB server, adjuvant, linkers and 6-His-tag. The chimeric vaccine was potentially antigenic and nonallergic and demonstrated thermostability and hydrophilicity in protparam server. The solubility of the vaccine was measured in comparison to E. coli proteins. The stability was also assessed by disulfide bonds engineering to reduce the high mobility regions in the designed vaccine. Furthermore molecular dynamics simulation further strengthen stability of the predicted vaccine. Tertiary structure of the vaccine construct after prediction, refinement was used for molecular docking with chicken alleles BF2*2101 and BF2*0401 and the docking process demonstrated favourable binding energy score of -337.47 kcal/mol and -326.87 kcal/mol, respectively. Molecular cloning demonstrated the potential clonability of the chimeric vaccine in pET28a(+) vector. This could guarantee the efficient translation and expression of the vaccine within suitable expression vector.
AEV, immunoinformatics, B cells, T cells, chimeric vaccine
Avian encephalomyelitis virus (AEV) is a positive single stranded RNA virus belonged to picornaviridea family genus tremovirus [Wei et al. 2008; Hauck et al. 2017; Senties-Cue et al. 2016; Liu et al. 2014; Marvil et al. 1999]. Avian encephalomyelitis (epidemic tremor) is a viral disease that mainly attacks the central nervous system of young chicks caused by AEV with worldwide distribution [Senties-Cue et al. 2016; Tannock and Shafren 1994]. The disease has an economic importance since it causes high morbidity and variable mortality rate in chicks and notice egg dropping in mature hen and hatchability [Tannock and Shafren 1994; Liu et al. 2014; Taylor et al. 1955]. The disease affected all avian species such as chicken, turkey, pheasant, quail and pigeons [Senties-Cue et al. 2016; Suarez 2013]. Experimental infection could be noticed in ducklings and guinea fowls [Wei et al. 2008]. The AEV affects poultry and could be transmitted horizontally (feacal and oral route) and vertically (transmitted via embryos) [Wei et al. 2008; Senties-Cue et al. 2016; Tannock and Shafren 1994; Calnek et al. 1960] causing disease characterized by neurological symptoms such as ataxia, rapid tremors of the head and neck and paralysis in young flocks, thus, the named epidemic tremor [Wei et al. 2008; Hauck et al. 2017; Markson and Blaxland 1958; Westbury and Sinkovic 1978; McNulty et al. 1990; Olitsky 1939].
The AEV genome is consist of 7.5 kilo basepair comprising a single open reading frame (ORF) encoding proteins of 2134 amino acids [Wei et al. 2008; Marvil et al. 1999; Wei et al. 2004]. This ORF consists of three parts: part1, part2 and part3 with four viral structural proteins named as vp1, vp2, vp3 and vp4. The vp2 and vp3 regions encode nonstructural proteins [Wei et al. 2008; Marvil et al. 1999]. However the vp1 region is considered as the most immunogenic part of virus and demonstrated neutralized antibodies against it [Wei et al. 2008; Muir et al. 1998].
There is no treatment yet to the avian encephalomyelitis, and the only way to control it by vaccination of flocks [Lin et al. 2018; Calnek 1998]. Flock vaccination programs designed to produce offspring with maternal antibodies that can result in well performance of the offspring and stop the transovarian transmission of disease during the stage of susceptibility (one to three weeks) post hatchability[Lin et al. 2018; Yu et al. 2015]. In china vaccination is mainly done from 14 to 16 weeks of breed age by administration of live vaccine (live field virus) into their drinking water or by using wing web inoculation via intracutaneous injection [Lin et al. 2018; Smyth et al. 1994]. This immunity protects hens during laying and their progeny through maternal antibodies [Hauck et al. 2017; Lin et al. 2018; Westbury and Sinkovic 1978]. In addition to that, in Chinese chicken industry section the inactivate vaccine was used in a wide range. However it is not effect enough like a live vaccine [Lin et al. 2018]. Study by Sarma et al., 2019 showed that live virus vaccine containing three mixture of avian encephalomyelitis (AE), fowl pox (FP), and pigeon pox (PP) viruses in a single mixture preparation demonstrated safety and provided efficacious when examined in layers. Poultry producers vaccinated their flocks with AE and FP live virus vaccines [Baxendale 1971; Garrett et al. 1985; Shafren et al. 1992; Fatunmbi and Reed 1996; Singh et al. 2000; Sarma et al. 2015]. However, although vaccination protocol was performed, immunity failure was reported due to very virulent or variant field strains of the virus were reported in the FP vaccinated flocks.
In this study we attempt to use the reverse vaccinology approach to predict multi epitopes vaccine from the polyprotein of the avian encephalomyelitis virus (AEV) that could elicit both B lymphocytes and T lymphocytes and consider as a safer vaccine candidate.
Viral proteome retrieval
Avian encephalomyelitis virus (AEV) had only one chromosome encoding for polyprotein named glycoprotein-1 or AEV polyprotein with accession number (NP_653151.1). The length of this polyprotein is 2134 amino acids. The National Center for Biotechnology Information (NCBI) at (https://www.ncbi.nlm.nih.gov/genome/browse/#!/proteins/5472/891046%7CTremovirus%20A/viral%20segment%20Unknown/) was used to retrieve the polyprotein sequence and entirely used for vaccine epitopes prediction.
Strains retrieval of AEV polyproteins
A set of six strains of avian encephalomyelitis virus polyproteins were retrieved from the NCBI at (https://www.ncbi.nlm.nih.gov/protein/?term=aev+polyprotein) on 15.4.2020 with the following accession numbers NP_653151.1; ALR74730.1; CAA12416.1; sp|Q9YLS4.1; sp|Q6R325.1 and sp|Q6WQ42.1. These proteins were further used to obtain the conserved epitopes sequences among the retrieved strains.
Sequence alignment of the retrieved strains and epitopes conservancy
Sequence alignment of the polyprotein sequences from the retrieved strains was obtained using multiple sequence alignment (MSA) tools, Clustal W in the BioEdit program, version 7.0.9.0 [Hall 1999]. The purpose of MSA was to obtain 100% conserved epitopes that could elicit the screened B and T lymphocytes.
Phylogenetic tree construction
The six retrieved strains sequences of AEV polyproteins were subjected to phylogenetic analysis using MEGA6 software [Tamura et al. 2013]. Phylogenetic tree was built to demonstrate the common ancestor of each retrieved strain.
B-cell epitopes prediction
The propensity scale and hidden Markov models programmed software from Immune epitopes database (IEDB) analysis resource (http://toolsiedb.ofg/bcell/) were used to predict epitopes from AEV polyprotein interacting with B lymphocytes. Three tools in the IEDB analysis resources were used to analyze B cell epitopes. For the epitope to be considered as a B cell epitope it should be linear epitope, located on the surface of the antigen and antigenic eliciting immune response. Thus linear, surface accessible and antigenic epitopes were assessed by BepiPred linear epitopes prediction [Larsen et al. 2006; Ponomarenko and Bourne 2007; Haste Andersen et al. 2006], Emini surface accessibility prediction tool [Emini et al. 1985] and kolaskar and tongaonker antigenicity method [Kolaskar and Tongaonkar 1990], respectively.
T cell epitopes prediction
Since the data of epitopes binding to MHC class I and MHC class II is not yet developed for chicken in the IEDB, the human alleles were used for the prediction of the T cell epitopes.
Epitope-MHC class I binding predictions
For MHC class I the peptide binding analysis was evaluated by the IEDB MHC-I estimated tool at (http://tools.iedb.org/mhci/) using Artificial Neural Network (ANN) method. Epitopes lengths was set as 9mers and all the conserved epitopes that bound to alleles at score less than or equal to 2 percentile rank were used for further analysis [Kim et al. 2012; Nielsen et al. 2003; Lundegaard et al. 2008; Sidney et al. 2008].
Epitope-MHC class II binding predictions
Analysis of peptide binding to MHC II molecules was assessed by the IEDB MHC II prediction tool at (http://tools.iedb.org/mhcii/result/) using Neural Networks Align to identify the binding affinity and MHCII binding core epitopes. All conserved epitopes that bound to alleles at score equal or less than 100 percentile rank were selected for further analysis [Wang et al. 2008].
Determination of antigenicity, allergenicity and toxicity of the predicted epitopes
The VaxiJen v2.0 server at (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html) was used to predict the antigenic epitopes of B and T lymphocytes. The server threshold was set to the default threshold (0.4). Antigenic epitopes were further investigated for allergenicity with AllerTOP server [Dimitrov et al. 2013] and toxicity with ToxinPred server [Gupta et al. 2013].
Construction chimeric vaccine
The antigenic, nonallergic and nontoxic epitopes were used to generate the vaccine construct against AEV. Therefore the proposed B cell epitopes and epitopes with high allelic interaction against cytotoxic and helper T lymphocytes from AEV polyprotein were used to construct the chimeric vaccine. Moreover epitopes that interacted with both MHCI and MHCII alleles were used once in the structure of the chimeric vaccine as MHCI or MHCII epitopes. The GGGGS linker was used to link B cell and MHC11 epitopes. While KK linker was used to link MHC1 predicted epitopes. The 50S ribosomal protein L7/L12 of Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv, uniprot accession no P9WHE3) was used as an adjuvant on the amino terminal of the vaccine construct to enhance the immunogenicity of the chimeric vaccine. This adjuvant was linked to the epitopes via EAAAK linker. A six his-tag was added at the carboxyl terminal of the vaccine construct to ease the isolation and identification of the vaccine.
Chimeric vaccine physical and chemical properties
ProtParam (https://web.expasy.org/protparam/) is a web used to compute physical and chemical properties of a given protein sequence. The chimeric vaccine construct from the predicted epitopes was analyzed for the physical and chemical properties. The physical and chemical properties comprises the vaccine protein molecular weight (MW), theoretical isoelectric point (pl), amino acid and atomic compositions, extinction coefficient, estimated half-life, instability index, aliphatic index and grand average of hydropathicity (GRAVY).
Secondary structure prediction
The Self-optimized prediction method (SOPMA) at (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html) [Combet et al. 2000] was the tool used to analyze the number of the helix, coiled structures and beta sheets in the secondary structure of the vaccine protein construct.
Tertiary structure prediction
The chimeric vaccine sequence was submitted to PHYRE-2 protein folding recognition server (http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id=index ) [Kelley et al. 2015]. The output PDB file obtained was used in refinement and adaptation of the chimeric vaccine structure.
Tertiary structure refinement and validation
GalaxyWEB web server was used for the refinement process [Shin et al. 2014; Ko et al. 2012]. The server refines the protein structure via utilizing a refinement process that obtains repack side chains and finally achieving complete relaxation of the structure via integrating dynamic simulations [Heo et al. 2013]. The refinement was performed to ameliorate the physical quality of the structure. ProSA-web server at (https://prosa.services.came.sbg.ac.at/prosa.php ) was initially used for the model validation. This server calculates the overall quality score for a specific input protein PDB structure. Secondly the model validation was achieved via Ramachandran plot in RAMPAGE server at (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php ) [Lovell et al. 2002; Al-Hakim et al. 2015].
Solubility of the chimeric vaccine
Protein-sol (https://protein-sol.manchester.ac.uk/) is a web based algorithms for determination of protein solubility [Hebditch et al. 2017]. The server predicted the solubility of the chimeric vaccine as QuerySol scaled solubility value compared to E. coli experimental dataset that expressed as a population average solubility (PopAvrSol) with 0.45 averages. For instance if the solubility of particular protein was equal to or greater than the population average solubility of E. coli (0.45) the protein considered as soluble protein.
Stability of the vaccine construct (Disulfide bonds prediction)
Disulfide by Design 2.0 (DbD2) for disulfide engineering in proteins was applied to achieve disulfide bonds between the chimeric vaccine residues [Craig and Dombkowski 2013]. The residue pairs are evaluated for proximity and geometry composition for formation of disulfide bonds, assuming that the residue pairs were mutated to cysteines.
Molecular dynamics simulation
To explore the collective motions of proteins or nucleic acids the online server iMODS (http://imods.chaconlab.org/) was used [Lopez-Blanco et al. 2014; Lopez-Blanco et al. 2011; Kovacs et al. 2004]. The server was used to analyze the stability of protein-protein complex using normal modes analysis (NMA) in internal coordinates and further effectively assess the structural dynamics of protein complex. The iMODS demonstrated the direction and magnitude of the motions in protein complex in the form of deformability, B-factors, eigenvalues, covariance, variance map in the residue index and elastic network in the atoms index.
Molecular docking of the chimeric vaccine with chicken alleles
HDOCK server (http://hdock.phys.hust.edu.cn/) that used protein-protein and protein-DNA/RNA docking was used to dock the vaccine construct with chicken alleles [Yan et al. 2017]. The vaccine construct PDB file (ligand) was submitted to the server with PDB ID of chicken alleles, MHC class 1 BF2*2101 molecule and MHC class 1 BF2*0401 molecule. These alleles were retrieved from the NCBI database with the following PDBIDs: 4D0C, CAK54661.1 for BF2*2101 molecule and 4D0C, CAK54660.1 for BF2*0401 molecule and were used as receptors in the docking process.
Codon adaptation and in silico cloning
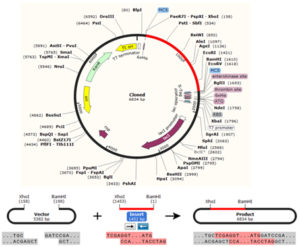
In silico cloning was performed to guarantee the expression of the vaccine construct in the selected host. The protein sequence of the chimeric vaccine was first converted into DNA sequence via Java Codon Adaptation Tool (JCAT) server (http://www.prodoric.de/JCat). The rho independent transcription termination, prokaryote ribosome binding site and cleavage site of restriction enzyme were avoided [Shey et al. 2019]. In the JACT, codon adaptation index (CAI) score is 1.0 but >0.8 is considered a good score [Morla et al. 2016]. The favourable GC content of a sequence ranged between 30–70%. The sequence of the BamHI and Xho1restriction enzymes were placed at the 5 and 3 primes of the DNA sequence, respectively. The SnapGene restriction cloning module was used to insert the DNA sequence into pET28a (+) vector between the BamHI and Xho1
Sequence alignment and epitopes conservancy
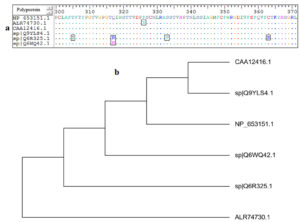
MSA of all retrieved strains of AEV polyprotein was achieved by using ClustalW embedded in Bioedit software. The software was used to obtain 100% conserved epitope from the retrieved strains. Epitopes conservancy was determined via alignment of the reference sequence and the sequences of the other retrieved strains. Fig. (1-a) provided the conserved regions by the amino acids identity among the retrieved sequences.
Fig. 1. (a): MSA was performed using Bioedit software. Dots within the sequences indicated conservancy, while letters within squares indicated non conserved regions. (b): the phylogenetic tree showed molecular divergence between the strains.
Phylogenetic tree construction
Although mutated regions were observed, the alignment provided conserved regions among the retrieved strains. The mutated regions resulted in evolutionary divergence among each retrieved strains. For instance in Fig. (1-b) the strains CAA12416.1 and sp|Q9YLS4.1 were much related to each other in their evolution. On the other hand the strain ALR74730.1 was far related to all other strains and demonstrated far molecular divergence.
B-cell Epitopes Prediction
The AEV polyprotein reference sequence was analyzed by Bepipred linear epitope prediction, Emini surface accessibility, Kolaskar and Tongaonkar antigenicity analysis tools in IEDB with thresholds of 0.06, 1.000 and 1.049 respectively (Fig. 2). Epitopes passed the three tools were considered as potential epitopes determinants of the B cell. The three tools predicted 76 linear conserved epitopes, 44 epitopes on the surface and 30 antigenic epitopes. However only 14 epitopes overlapped the three tools and were further investigated for antigenicity using Vaxijen software with default threshold (0.4), allergenicity and toxicity. Upon investigation only three epitopes were shown to be antigenic, nonallergic and nontoxic. The three epitopes, their position and their scores in in different tools were shown in Table (1).
Table (1):
The predicted B cell epitopes, their antigenicity, allergenicity and toxicity from the AEV polyprotein.
| Peptide | Start | End | Length | Emini surface accessibility 1.000 | Kolskar & Toangankar antigenicity 1.038 | Vaxijen antigenicity (0.4) | Allergenicity | Toxicity |
|---|---|---|---|---|---|---|---|---|
| LAPGKPRHTV | 542 | 551 | 10 | 1.211 | 1.052 | 0.7094 Antigenic | Non-allergen | Non-toxin |
| SHPCPKT | 1253 | 1259 | 7 | 1.381 | 1.071 | 1.0545 Antigenic | Non-allergen | Non-toxin |
| VGSKTKYH | 1664 | 1671 | 8 | 1.992 | 1.038 | 1.7085 Antigenic | Non-allergen | Non-toxin |
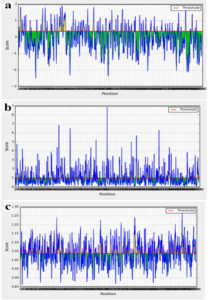
Fig. 2. A- Bepipred linear epitope prediction, B- Emini surface accessibility and C- Kolaskar and Tongaonkar antigenicity prediction. The threshold for each tool was shown by red line.
Cytotoxic T-lymphocyte epitopes prediction
Based on Artificial Neural Network (ANN) analysis tool only 22 epitopes were shown to interact with numerous MHC1alleles. All the MHC-1 predicted epitopes were further analyzed for antigenicity, allergenicity and toxicity. Ten epitopes demonstrated antigenicity and were shown to be nonallergic and nontoxic. The ten epitopes with their scores in the different tools were shown in Table (2) and were elected as cytotoxic T lymphocytes epitopes.
Table (2):
The predicted T cytotoxic cells epitopes, their antigenicity, allergenicity, toxicity from AEV polyprotein.
Epitope |
Start |
End |
Percentile rank |
Interacted alleles |
Vxijen antigenicity (0.4) |
Allergenicity |
Toxicity |
|---|---|---|---|---|---|---|---|
FSMDFLDLV |
993 |
1001 |
0.05 |
7 |
1.5832 antigenic |
Non-allergen |
Non-Toxin |
FTDTSSKTV |
735 |
743 |
1.2 |
5 |
0.4488 antigenic |
Non-allergen |
Non-Toxin |
HTVDHMDLY |
549 |
557 |
0.26 |
6 |
0.7126 antigenic |
Non-allergen |
Non-Toxin |
IVYFVPEGV |
625 |
633 |
1.9 |
4 |
0.5752 antigenic |
Non-allergen |
Non-Toxin |
KFMGRAHYL |
558 |
566 |
0.3 |
5 |
0.6343 antigenic |
Non-allergen |
Non-Toxin |
MTFAGKTNV |
614 |
622 |
2 |
4 |
0.9433 antigenic |
Non-allergen |
Non-Toxin |
SSFTDTSSK |
733 |
741 |
0.26 |
4 |
0.5549 antigenic |
Non-allergen |
Non-Toxin |
TPFGAVKAM |
518 |
526 |
0.31 |
4 |
0.6289 antigenic |
Non-allergen |
Non-Toxin |
TVYSMLAWK |
2058 |
2066 |
0.05 |
4 |
0.9225 antigenic |
Non-allergen |
Non-Toxin |
YATGKLVCY |
434 |
442 |
0.94 |
4 |
0.9918 antigenic |
Non-allergen |
Non-Toxin |
Helper T lymphocytes epitopes prediction
Based on NN-align analysis method, 77 epitopes were predicted interacting with MHC-II alleles. Among them, only 14 epitopes demonstrated antigenicity and were shown to be nonallergic and nontoxic. Thus these epitopes were elected as helper T cell epitopes and were shown in Table (3).
Table (3):
The predicted T helper cells epitopes, their antigenicity, allergenicity, toxicity from AEV polyprotein.
Epitope |
Start |
End |
Percentile rank |
Interacted alleles |
Vaxijen antigenicity (0.4) |
Allergenicity |
Toxicity |
|---|---|---|---|---|---|---|---|
FCFWRGDIV |
348 |
356 |
15.91 |
93 |
0.4891 antigenic |
non-allergen |
Non-Toxin |
FSMDFLDLV |
993 |
1001 |
1.26 |
107 |
1.5832 antigenic |
non-allergen |
Non-Toxin |
IVYFVPEGV |
625 |
633 |
3.89 |
86 |
0.5752 antigenic |
non-allergen |
Non-Toxin |
LDHERFTLV |
1553 |
1561 |
4.52 |
73 |
0.8378 antigenic |
non-allergen |
Non-Toxin |
LHMNRSIQV |
1794 |
1802 |
50.43 |
124 |
0.5397 antigenic |
non-allergen |
Non-Toxin |
LIAALVPYD |
143 |
151 |
37.56 |
88 |
0.4585 antigenic |
non-allergen |
Non-Toxin |
LKYHSYARF |
117 |
125 |
9.22 |
71 |
0.4211 antigenic |
non-allergen |
Non-Toxin |
LRLNMANLE |
1227 |
1235 |
43.52 |
75 |
1.2891 antigenic |
non-allergen |
Non-Toxin |
LYIERAGTI |
1488 |
1496 |
35.92 |
88 |
0.6164 antigenic |
non-allergen |
Non-Toxin |
VLGAITTQI |
691 |
699 |
26.69 |
98 |
0.7395 antigenic |
non-allergen |
Non-Toxin |
YARFGLDVI |
122 |
130 |
17.02 |
83 |
0.7700 antigenic |
non-allergen |
Non-Toxin |
YHGIKDADV |
1476 |
1484 |
30.33 |
62 |
0.6852 antigenic |
non-allergen |
Non-Toxin |
YKTSVGAIR |
650 |
658 |
31.59 |
73 |
0.7440 antigenic |
non-allergen |
Non-Toxin |
YLQVTYYIS |
708 |
716 |
0.57 |
104 |
0.7252 antigenic |
non-allergen |
Non-Toxin |
Construction of muli-epitopes vaccine
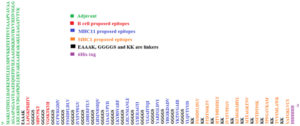
The chimeric vaccine includes the B and T cell predicted epitopes. Three epitopes were proposed as B cell epitope, ten epitopes as cytotoxic T cell and fourteen epitopes as helper T cell. The chimeric vaccine composed of 482 amino acids after addition of the adjuvants, linkers and 6-His-tag (Fig. 3). The chimeric vaccine demonstrated antigenicity in Vaxigen server with score of 0.5857 and was nonallergen in the Allertop server.
Fig. 3. The sequence of the predicted multi-epitopes vaccine. The sequence composed of 482 amino acids.
Physical and chemical properties of the vaccine construct
The MW of the chimeric vaccine was 49.80192 KDa with pI value of 9.37. The total number of negatively (Asp+Glu) and positively (Arg+Lys) charged residues was 45 and 61 respectively. The Extinction coefficient was 37945 indicating all pairs of Cys residues form cysteines. The estimated half-life was 30 hours (mammalian reticulocytes, in vitro), >20 hours (yeast, in vivo) and >10 hours (Escherichia coli, in vivo). The instability index (II) was 28.38 demonstrating the stability of the chimeric vaccine. Aliphatic index was 75.29 and the GRAVY was -0.127 indicating the hydrophilicity of the chimeric vaccine.
Chimeric vaccine secondary structure prediction
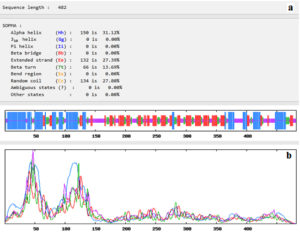
Fig. (4) showed that the 482 amino acids of the predicted vaccine showed that 150 aa (31.12%) involved in formation of alpha helices, 132 aa (27.39%) were extended strands, 66 aa (13.69%) were beta turns while 134 aa (27.80%) were random coils with no unambiguous or any other states.
Fig. 4. Secondary structure prediction plot of the vaccine construct. Alpha helices were shown in blue colour, while extended strands and beta turns were shown by red and green colours, respectively. (a) The visualization of the prediction and (b) the score curves for each predicted state.
Tertiary structure prediction
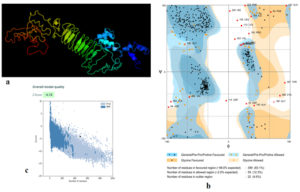
Fig. (5) provided the 3D structure of the chimeric vaccine predicted by PHYRE2 server. The model was further assessed by Ramachandran plot after refinement and demonstrated that 399 residues were in the favoured region (83.1%), 59 residues were in the allowed region (12.3%) and 22 residues were in the outlier region (4.6%). Moreover proSA server Z-score of the chimeric vaccine was -4.18 which represents the good quality of the model.
Fig. 5. (a) The 3D structure of the chimeric vaccine obtained by PHYRE2 server. (b) The 3D model after refinement in Galaxyrefiner server was validated by Ramachandran plot analysis that demonstrated 83.1%, 12.3% and 4.6% of protein residues in favoured, allowed, and disallowed (outlier) regions respectively. (c) ProSA server Z score was -4.18
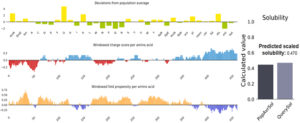
Solubility of the vaccine construct
Fig. (6) showed the solubility of the chimeric vaccine, QuerySol scaled solubility value, was 0.470 compared to the experimental dataset (PopAvrSol) of 0.45 for E. coli proteins. This result showed that the chimeric vaccine is potentially soluble.
Fig. 6. Vaccine construct solubility as obtained by protein sol server. The solubility of the vaccine construct was shown to be 0.470 compared to 0.45 of the population average solubility of E. coli.
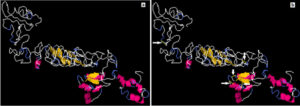
Stability (Disulfide bonds prediction) of the chimeric vaccine
A total 65 pairs of amino acids residues probably shown to be implicated in disulfide bond formation. Among them five residues were evaluated to form disulfide bond based on the chi3 residue screening (between −87 and +97), B-factor value and energy value less than 3, (Fig. 7). The five residue pairs that strongly implicated in disulfide bond formation were the following amino acids at the following positions 67LEU, 71GLY, 73LYS, 181GLY and 407PRO if were mutated to the following amino acids 98ASP, 124GLY, 76GLY, 206TYR and 410VAL respectively.
Fig. 7. Stability of the vaccine construct by disulfide bond engineering (a) the original form and (b) the mutant form. Five disulfide bonds were strongly implicated in disulfide bond formation and shown in golden sticky forms indicated by white arrows in the mutant form.
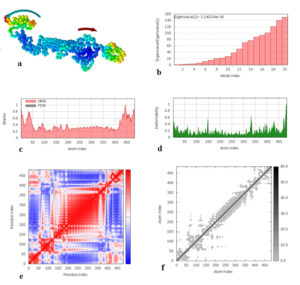
Fig. 8. Molecular dynamics simulation of vaccine protein complex. Stability of the vaccine was analyzed via (a) mobility, (b) eigenvalue, (c) B-factor, (d) deformability, (e) covariance and (f) elastic network analysis.
Molecular dynamics simulation
Molecular dynamics simulation of the vaccine protein was performed by NMA (Normal mode analysis) in the iMODS server and presented in Fig. (8). As shown in Fig. the arrows indicated the direction of the mobility of each residue in the chimeric vaccine construct (Fig. 8a). Moreover the deformability of the molecule associated with the residues individual distortion, presented by hinges in the chain (Fig. 8b). Experimental B-factor was obtained from the corresponding PDB field and obtained from the calculation of the NMA (Fig. 8c). The eigenvalue which demonstrated the stiffness of the motion was shown to be 3.145184e−06 (Fig. 8d), where the lower the eigenvalue, the easier the deformation. Covariance matrix provided the coupling between pairs of residues, i.e. whether they experience correlated (red), uncorrelated (white) or anti-correlated (blue) motions (Fig. 8e). The elastic network model defines which pairs of atoms are connected by springs in which each dot showed one spring between the corresponding pair of atoms. Dots are colored based on their stiffness, where the darker grays dots demonstrated stiffer springs and vice versa (Fig. 8f).
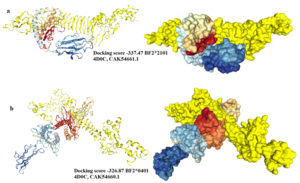
Molecular docking of chimeric vaccine with chicken alleles
The chimeric vaccine was used as a ligand and the chicken alleles (BF2*2101 & BF2*0401) as receptors. As shown in (Fig. 9a), the docking process of the vaccine construct with PDB: 4D0C, CAK54661.1 demonstrated that the binding energy score was -337.47. For PDB: 4D0C CAK54660.1 the binding energy score was -326.87 (Fig. 9b). These negatively scored values demonstrated the strong binding between the vaccine protein and the chicken alleles.
Fig. 9. Molecular docking between the vaccine protein and the chicken MHC1 alleles (a) BF2*2101 and (b) BF2*0401. The vaccine construct was shown in yellow colour and the chicken alleles in rainbow colours. A cartoon structure of the vaccine construct docked with BF2*2101 was shown in Fig. (a-left) while the ball structure in (a-right). A cartoon structure of the vaccine construct docked with BF2*0401 was shown in Fig. (b-left) while the ball structure in (b-right).
In silico cloning
The In silico cloning of the DNA sequence of the vaccine protein provided CAI-Value of 0.9742, providing higher proportion of most common abundant codons while the GC-content was 49.23928%, demonstrating favourable GC content. Fig. (10), showed that DNA sequence was cloned into pET28a (+) vector between BamH1 and Xho1restriction enzymes cutting sites.
The prevention and control of avian encephalomyelitis virus via vaccination of young chickens is of great importance [Calnek and Jehnich 1959a; Calnek and Jehnich 1959b; Schaaf 1958]. However some drawbacks were reported during the course of the vaccination process. For instance the wing-web and intramuscular vaccines was more pathogenic than is desirable although the vaccine was capable of eliciting immune response in the vaccinated birds. Furthermore the oral administration of virus provided extremely high dosage resulting in inadequate immunity of the birds [Calnek and Taylor 1960]. The vaccine failure and restrictions on the use of live or attenuated virus vaccines create the need for a safer and effective vaccine [Sarma et al. 2019; Li et al. 2014; Lo et al. 2013]. In this study reverse vaccinology approach was used to design safe multi-epitopes vaccine from avian encephalomyelitis virus polyprotein.
In this study only the 100% conserved epitopes from the AEV polyprotein were elected to interact against the B and T lymphocytes. The predicted B cell epitopes were investigated to be linear, surface accessible and antigenic using IEDB prediction tools. For T cells, large numbers of epitopes were shown to bind to different MHCI and MHCII alleles. The predicted epitopes from B and T lymphocytes showed antigenicity and were tested non allergic and nontoxic, thus, were used as a vaccine candidate. The epitopes were joined together using appropriate linkers sequences [Meza et al. 2017]. The linkers EAAAK, GGGGS and KK were introduced between the selected B and T cells epitopes to generate a sequence with minimal junctional immunogenicity [Pandey et al. 2018; Hasan et al. 2019; Ali et al. 2017; Khatoon et al. 2017; Shey et al. 2019]. An adjuvant was linked to the epitopes since adjuvants work as an immunomodulator enhancing the immunogenicity of the vaccine protein [Mohan et al. 2013; Solanki and Tiwari 2018].
Physical and chemical properties of the vaccine construct were assessed via protparam server. Results showed that the computed instability index (II) classifies the protein as stable and the vaccine protein showed aliphatic side chains, indicating potential hydrophobicity. The GRAVY classified the vaccine as hydrophilic with thermal stability. Furthermore vaccine secondary and tertiary structures were evaluated since they are important in vaccine design [Meza et al. 2017]. Moreover the prediction extremely ameliorated by the refined software and demonstrated desirable characteristics on Ramachandran plot predictions. These indicated that the overall model quality of the vaccine protein was satisfactorily.
The formation of insoluble protein particles may hinder the vaccine protein native form. In addition, purification of the vaccine construct may cause structural alterations during solubilization and refolding steps [Silva et al. 2016; Dill and Shortle 1991]. Thus the study of the solubility of the vaccine construct is a cornerstone in determining the vaccine nature. It was reported that the solubility of the recombinant protein in E. coli is important for biochemical and functional analysis [Khatoon et al. 2017]. In this study the vaccine construct provided solubility index of 0.470 compared to that of E. coli (0.45) indicating the favourable solubility of the vaccine construct
Vaccines are biological products that had their own stability issues, which must be considered during development. The stability problems obstacle the efficacies of the vaccine construct, thus, it is of great importance to measure the vaccine stability. Moreover the folding stability of protein can also directly impact the availability of B-cell epitopes [Scheiblhofer et al. 2017]. For instance protein destabilization guide to improper folding of the protein tertiary structure. This effectively resulted in loss of conformational epitopes. Also this resulted in proteins with epitopes not recognized by IgE, but maintains their capacity to stimulate T-cell responses [Swoboda et al. 2007; Thalhamer et al. 2010]. In this study the stability of the chimeric vaccine was performed via disulfide bonds formation. The result showed that the stability was well indexed if five residues in the vaccine structure mutated to cysteine. Beside determination of the stability of the vaccine construct via disulfide bonds formation molecular dynamics study was performed to determine the complex stability as well. Previously macromolecules stability was linked with correlated fluctuations of atoms [Clarage et al. 1995; Caspar 1995]. Thus we performed essential dynamics in accordance to the normal modes of proteins deposited in iMODS server to further assess the complex stability of the vaccine. The analysis demonstrated that no significant distortion of the protein atoms resulting in reducing the chance of deformability with no stiffness motion of the vaccine construct thereby strengthening our prediction.
Molecular docking interaction of the vaccine construct and chicken MHC1 molecules (BF2*2101 and BF2*0401) was performed to explore the binding affinity of vaccine construct and eliciting immune response. In this study the attractive binding energy between chicken MHC1 molecules and the vaccine construct demonstrated high binding affinity. This was expressed in negative binding energy values providing strong interaction of the vaccine to MHC1 molecules, that professionally eliciting a potential protective immune response.
The clonability of the chimeric vaccine in a suitable E. coli expression vector is significant step for recombinant protein production [Chen 2012; Rosano and Ceccarelli 2014]. The protein sequence of the designed vaccine was converted to DNA sequence via reverse transcription process and adapted for E. coli strain K12 before cloning into pET28a (+) vector. The CAI value and the GC content showed successful cloning process with high-level of protein expression in the host bacteria.
Peptide–based vaccine via reverse vaccinology is becoming one of the important tools for designing vaccines. The predicted vaccine demonstrated favourable interaction against B and T-cells and induced humoral and cellular responses. The predicted vaccine showed chemical stability and solubility and demonstrated good binding with chicken alleles. Molecular cloning showed that the vaccine could be cloned and produced as a recombinant vaccine. The effectiveness and safety of the designed vaccine by this computational analysis needed to be evaluated in clinical trial experiments to confirm their efficacy in inducing protective immune response.
ACKNOWLEDGMENTS
Authors would like to thank the staff members of College of Veterinary Medicine, University of Bahri, Sudan for their cooperation and support.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
FKA: Downloaded the data from the NCBI, participated in data analysis, carried out sequence alignments, epitopes prediction from the IEDB, participated in the design of the study and drafted the manuscript.
YAA: Conceived of the study, designed the study, participated in data analysis, coordinated the study and helped draft the manuscript.
KAA: Conceived of the study, designed the study, and participated in data analysis.
MAAN: designed the study and participate in data analysis
EEME: Designed the study, participated in data analysis.
All authors gave final approval for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
The [retrieved strains and their accession numbers from the NCBI, data analysis from the IEDB server] data used to support the findings of this study are included within the article.
- Al-Hakim M, Hasan R, Ali MF, Rabbee Joy, Marufatuzzahan ZF. In-silico characterization and homology modeling of catechol 1,2 dioxygenase involved in processing of catechol- an intermediate of aromatic compound degradation pathway. Glob. J. Sci. Front. Res. G Bio-Tech Genet. 2015;15:1-13.
- Ali M, Pandey RK, Khatoon N, Narula A, Mishra A, Prajapati VK. Exploring dengue genome to construct a multi-epitope based subunit vaccine by utilizing immunoinformatics approach to battle against dengue infection. Sci Rep. 2017;7:9232.
Crossref - Baxendale W. Studies of three avian pox viruses and the development of an improved fowlpox vaccine. Vet Rec. 1971;88(1):5-10.15.
- Calnek BW. Control of avian encephalomyelitis: a histori-cal account. Avian Dis. 1998;42:632-647.
Crossref - Calnek BW, Jehnich H. Studies on avian encephalomyelitis. I The use of a serum-neutralization test in the detection of immunity levels. Avian Dis. 1959a;3(2):95-104.
Crossref - Calnek BW, Jehnich H. Studies on avian encephalomyelitis. II Immune responses to vaccination procedures. Avian Dis. 1959b;3(3):225-239.
Crossref - Calnek BW, Taylor PJ. Studies on Avian Encephalomyelitis. III Immune Response to Beta-Propiolactone Inactivated Virus. Avian Diseases. 1960;4(2):116-122.
Crossref - Calnek B, Taylor PJ, Sevoian M. Studies on avian encephalomyelitis. IV Epizootiology. Avian Diseases. 1960;4:325-347.
Crossref - Caspar DLD. Problems in simulating macromolecular movements. Structure. 1995;3(4):327-329.
Crossref - Chen R. Bacterial expression systems for recombinant protein production: E. coli and beyond. Biotechnol Adv. 2012;30(5):1102-1107.
Crossref - Clarage JB, Romo T, Andrews BK, Pettitt BM, Phillips GN. A sampling problem in molecular dynamics simulations of macromolecules. Proc Natl Acad Sci U.S.A. 1995;92(8):3288-3292.
Crossref - Combet C, Blanchet C, Geourjon C and Deleage G. NPS@. Network Protein Sequence Analysis. Trends Biochem Sci. 2000;25(3):147-150 .
Crossref - Craig DB, Dombkowski AA. Disulfide by Design 2.0: a web-based tool for disulfide engineering in proteins. BMC Bioinformatics. 2013;14:346.
Crossref - Dill KA, Shortle D. Denatured states of proteins. Annu Rev Biochem. 1991;60:795-825.
Crossref - Dimitrov I, Bangov I, Flower DR, Doytchinova IA. AllerTOP v.2- a server for in silico prediction of allergens. J Mol. Model. 2014;20(6):2278.
Crossref - Emini EA, Hughes JV, Perlow DS, Boger J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J Virol. 1985;55(3):836-839.
Crossref - Fatunmbi OO, Reed WM. Evaluation of a commercial modified live fowlpox virus vaccine for the control of “variant” fowlpox virus infections. Avian Dis. 1996;40(3):582-587.
Crossref - Garrett JK, Davis RB, Ragland WR. Correlation of serum antibody titer for avian encephalomyelitis virus (AEV) in hens with the resistance of progeny embryos to AEV. Avian Dis. 1985;29(3):878-880.
Crossref - Gupta S, Kapoor P, Chaudhary K, et al. Open source drug discovery consortium, Raghava GP. in silico approach for predicting toxicity of peptides and proteins. PLoS One. 2013;8(9):e73957.
Crossref - Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95-98. Key: citeulike: 691774.
- Hasan M, Ghosh PP, Azim KF, et al. Reverse vaccinology approach to design a novel multi-epitope subunit vaccine against avian influenza A (H7N9) virus. Microb Pathog. 2019;130:19-37.
Crossref - Andersen HP, Nielsen M, Lund O. Prediction of residues in discontinuous B-cell epitopes using protein 3D structures. Protein Sci. 2006;15(11):2558-2567.
Crossref - Hauck R, Senties-Cue CG, Wang Y, et al. Evolution of avian encephalomyelitis virus during embryo-adaptation. Vet Microbiol. 2017;204:1-7.
Crossref - Hebditch M, Carballo-Amador MA, Charonis S, Curtis R, Warwicker J. Protein-Sol: a web tool for predicting protein solubility from sequence. Bioinformatics. 2017; 33(19):3098-3100.
Crossref - Heo L, Park H, Seok C. Galaxy Refine: protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013;41(1):W384-W388.
Crossref - Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10(6):845-858.
Crossref - Khatoon N, Pandey RK, Prajapati VK. Exploring Leishmania secretory proteins to design B and T cell multi-epitope subunit vaccine using immunoinformatics approach. Sci Rep. 2017;7:8285.
Crossref - Kim Y, Ponomarenko J, Zhu Z, et al. Immune epitope database analysis resource. Nucleic Acids Res. 2012;40(W1):W525-W530.
Crossref - Ko J, Park H, Heo L, Seok C. Galaxy WEB server for protein structure prediction and refinement. Nucleic Acids Res. 2012;40(W1):W294-W297.
Crossref - Kolaskar AS, Tongaonkar PC. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990;276(1-2):172-174.
Crossref - Kovacs J, Chacon P, Abagyan R. Predictions of Protein Flexibility: First Order Measures. Proteins: Structure, Function, and Bioinformatics. 2004;56(4):661-668.
Crossref - Larsen JE, Lund O, Nielsen M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006;2:2.
Crossref - Li W, Joshi MD, Singhania S, Ramsey KH, Murthy AK. Peptide vaccine: progress and challenges. Vaccines. 2014;2(3):515-536.
Crossref - Lin W, Lu P, Li A, et al. Assessing the efficacy of a live vaccine against avian encephalomyelitis virus. Arch Virol. 2018;163:2395-2404.
Crossref - Liu Q, Yang Z, Hao H, et al. Development of a SYBR Green real-time RT-PCR assay for the detection of avian encephalomyelitis virus. J Virol Methods. 2014;206:46-50.
Crossref - Lo YT, Pai TW, Wu WK, Chang HT. Prediction of conformational epitopes with the use of a knowledge-based energy function and geometrically related neighboring residue characteristics. BMC Bioinformatics. 2013;14(Suppl 4): S3.
Crossref - Lopez-Blanco JR, Aliaga JI, Quintana-Orti ES and Chacon P. iMODS: Internal coordinates normal mode analysis server. Nucleic Acids Res. 2014;42(1):W271-W276.
Crossref - Lopez-Blanco JR, Garzon JI, Chacon P. iMods: multipurpose normal mode analysis in internal coordinates. Bioinformatics. 2011;27 (20):2843-2850.
Crossref - Lovell SC, Davis IW, Arendall WB, et al. Structure validation by Calpha geometry: ϕ, ψ and Cβ deviation, Protein. 2003;50:437-450.
Crossref - Lundegaard C, Lamberth K, Harndahl M, et al. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8-11. Nucleic Acids Res. 2008;36:W509-W12.
Crossref - Markson LM, Blaxland JD. Infectious avian encephalo-myelitis. Veterinary Record. 1958;20:12081213.
- Marvil P, Knowles NJ, Mockett APA, Britton P, Brown TDK, Cavanagh D. Avian encephalomyelitis virus is a picorna-virus and is most closely related to hepatitis A virus. J Gen Virol. 1999;80(3):653-662.
Crossref - McNulty MS, Connor TJ, McNeilly F, McFerran JB. Biological characterization of avian enteroviruses and enterovirus-likeviruses. Avian Pathol. 1990;19:75-78.
Crossref - Meza B, Ascencio F, Sierra-Beltran A P, Torres J, Angulo C. A novel design of a multi-antigenic, multistage and multi-epitope vaccine against Helicobacter pylori: An in silico approach. Infect Genet Evol. 2017;49:309-317.
Crossref - Mohan T, Verma P, Rao DN. Novel adjuvants and delivery vehicles for vaccines development: a road ahead, Indian. J Med Res. 2013;138(5):779.
- Morla S, Makhija A, Kumar S. Synonymous codon usage pattern in glycoprotein gene of rabies virus. Gene. 2016;584(1):1-6.
Crossref - Muir P, Kammerer U, Korn K, et al. Molecular typing of enteroviruses: current status and future requirements. Clin Microbiol. 1998;11(1):202-227.
Crossref - Nielsen M, Lundegaard C, Worning P, et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12(5):1007-1017.
Crossref - Olitsky PK. Experimental studies on the virus of infectious avian encephalomyelitis. J Exp Med. 1939;70(6):565-582.
Crossref - Pandey RK, Ojha R, Aathmanathan VS, Krishnan M, Prajapati VK. Immunoinformatics approaches to design a novel multi-epitope subunit vaccine against HIV infection. Vaccine. 2018;36(17):2262-2272.
Crossref - Ponomarenko JV, Bourne PE. Antibody-protein interactions: benchmark datasets and prediction tools evaluation. BMC Struct Biol. 2007;7:64.
Crossref - Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol. 2014;5:172.
Crossref - Sarma G, Kersting BA, Spina G. Vaccination of 1-day-old turkey poults with fowlpox vaccine by subcutaneous route. Avian Dis. 2015;59(3):419-421.
Crossref - Sarma G, Kersting BA, Spina G. Field safety and efficacy of a unique live virus vaccine for controlling avian encephalomyelitis and fowlpox in poultry. Vet World. 2019;12(8):1291-1298.
Crossref - Schaaf K. Immunization for the control of avian encephalomyelitis. Avian Dis. 1958;2:279-289.
Crossref - Scheiblhofer S, Laimer J, Machado Y, Weiss R, Thalhamer J. Influence of protein fold stability on immunogenicity and its implications for vaccine design. Expert Rev Vaccines. 2017;2017;16(5):479-489.
- Senties-Cue CG, Gallardo RA, Reimers N, Bickford AA, Charlton BR, Shivaprasad HL. Avian Encephalomyelitis in Layer Pullets Associated with Vaccination. Avian Diseases. 2016;60(2):511-515.
Crossref - Shafren DR, Tanmock GA, Groves PJ. Antibody responses to avian encephalomyelitis virus vaccines when administered by different routes. Aust Vet J. 1992;69(11): 272-275.
Crossref - Shey RA, Ghogomu SM, Esoh KK, et al. In-silico design of a multi-epitope vaccine candidate against onchocerciasis and related filarial diseases. Sci Rep. 2019;13;9(1):4409.
Crossref - Shin WH, Lee GR, Heo L, Lee H, Seok C. Prediction of Protein Structure and Interaction by GALAXY protein modeling programs. Bio Design. 2014;2(1):1-11.
- Sidney J, E Assarsson C, Moore, Ngo S, Pinilla C, et al. Quantitative peptide binding motifs for 19 human and mouse MHC class I molecules derived using positional scanning combinatorial peptide libraries. Immunome Res. 2008;4:2.
Crossref - Silva HC Junior, Pestana CP, Galler R, Medeiros MA. Solubility as a limiting factor for expression of hepatitis A virus proteins in insect cell-baculovirus system. Mem Inst Oswaldo Cruz. 2016;111(8):535-538.
Crossref - Singh P, Kim TJ, Tripathy DN. Re-emerging fowlpox: Evaluation of isolates from vaccinated flocks. Avian Pathol. 2000;29(5):449-455.
Crossref - Smyth JA, McNeilly F, Reilly GA, McKillop ER, Cassidy JP. Avian encephalomyelitis following oral vaccination. Avian Pathol. 1994;23(3):435-445.
Crossref - Solanki V, Tiwari V. Subtractive proteomics to identify novel drug targets and reverse vaccinology for the development of chimeric vaccine against Acinetobacter baumannii. Scientific Report. 2018;8(1):9044.
Crossref - Suarez DL. Avian encephalomyelitis. 2013:486-510. In: Diseases of Poultry, 13th ed. (Swayne DE, Glisson JR, McDougald LR, Nolan LK, Suarez DL, Nair V. eds.), Wiley-Blackwell, Ames.
- Swoboda I, Bugajska-Schretter A, Linhart B, et al. A recombinant hypoallergenic parvalbumin mutant for immunotherapy of IgE-mediated fish allergy. J Immunol. 2007;178(10):6290-6296.
Crossref - Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725-2729.
Crossref - Tannock GA, Shafren DR. Avian encephalomyelitis: A review. Avian Pathology. 1994;23(4):603-620.
Crossref - Taylor LW, Lowry DC, Ragg LG. Effects of an outbreak of avian encephalomyelitis (epidemictremor) in a breeding flock. Poultry Science. 1955;34(5):1036-1045.
Crossref - Thalhamer T, Dobias H, Stepanoska T, et al. Designing hypoallergenic derivatives for allergy treatment by means of in silico mutation and screening. J Allergy Clin Immunol. 2010;125(4):926-934.E10.
Crossref - Wang P, Sidney J, Dow C, Mothe B, Sette A. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol. 2008;4(4):e1000048.
Crossref - Wei L, Chee LL, Wei T, et al. The VP1 protein of avian encephalomyelitis virus is a major host-protective immunogen that serves as diagnostic potential. J Virol Methods. 2008;149(1):56-62.
Crossref - Wei L, Liu J, Yao WG, Zhang FL, Zhou J. Determination of the whole genome of avian encephalomyelitis virus isolates in China. Chinese Journal of Virology. 2004;20:230236.
- Wei L, Zhou J, Wang J, Shi L, Liu J. Development of a non-radioactive digoxigen in cDNA probe for the detection of avian encephalomyelitis virus. Avian Pathol. 2008;37(2):187-191.
Crossref - Westbury HA, Sinkovic B. The pathogenesis of infectious avian encephalomyelitis 1. The effect of the age of the chicken and the route of administration of the virus. Aust Vet J. 1978;54(2):68-71.
Crossref - Yan Y, Tao H, He J et al. The HDOCK server for integrated protein-protein docking. Nat Protoc. 2020;15:1829-1852.
Crossref - Yu XH, Zhao J, Qin XH, Zhang GZ. Serological evidence of avian encephalomyelitis virus infection associated with vertical transmission in chicks. Biologicals. 2015;43(6):512-514.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.