ISSN: 0973-7510
E-ISSN: 2581-690X
The current investigation was conducted to maximise the production of the natural anticancer drug from the microbe isolated from the marine soil sample of the Coromandel Coast of the Bay of Bengal region of India. Yellow to red colour pigmented microbes separated by crowd plate method. Bacteria are producing strong colour product subjected to future study. The isolated strains were detected based on biochemical, morphological, and genetic characteristics. Pigment formation was found to be influenced strongly by conditions of the environment. The water-insoluble pigment extracted by acidified methanol and showed maximum absorbance at 535nm. A statistical screening procedure was adopted to select the optimum condition to produce the pigment. The carbon, nitrogen, medium pH, growth condition temperature and revolution of agitation were screened using the response surface methodology statistical model. The near optimum conditions for the production medium were affected by the concentration of peanut, L-proline, percentage inoculum pH and incubation time. When these conditions were employed yield increased as two-fold as the concentration of prodigiosin 789 mg/l.
Prodigiosin, Marine soil, Optimization, Microbial pigment
Insertion of biotechnology into the environment of marine which has opened new horizons for locating novel organisms by means of their trapping potential resources. However, demonstrations of independent methods that marine sediments contain a honest range of varied microorganisms1. The ocean’s rich biodiversity continues to attract more natural materials from the sea, which serve as a foundation for new medicinal agents. At present, there have been are some success stories of marine proved drugs at specific levels of segment I-III scientific clinical trial2.
New chemicals are produced by marine organisms to cope with extreme changes in salinity, pressure and temperature, then prevailing in their environment and the substances produced that are special in structural, diverse and functional characteristics. In order to remove drugs from the ocean started the efforts maintained in the late 1960s. The inquiry started in the 1970s, however. Almost 2500 new metabolites from the collection of marine organisms were identified during the period from 1977 to 1987. And studies have shown that the marine ecosystem can be a great source of new chemicals. To date, it is likely that 10,000 compounds remoted from marine species with several new compounds are nevertheless discovered once a year. Around 300 patents were granted on bioactive marine natural products merchandise among 1969 and 19993.
The utilization of marine microbes is mostly due to the primary supply of bioactive natural compounds, which is vital in understanding the supply issue in the event of prospective medications from microbes. On cultivation of micro-organism within the laboratory, however, are often challenging. It is partly because of the shortage of culture media to adequately mimic natural environments. Besides, many species aren’t viable or may require long growth periods before they seem. Earlier studies have cultured novel microbes successfully by using various methods (encapsulation procedures, casein, diffusion growth chambers, and microorganism specific agars, enriched media with the selective antibiotic)2.
One of the main sources of chemical diversity and possible viable therapeutic use of natural products. Pigments produced by organisms because the reminiscence of its secondary metabolism commonly mentioned as Biopigments. These Biopigments can obtain from two primary sources, plants and microorganism. Biopigments from the organism are preferred over those from plants due to their stability and therefore the availability of their cultivation technology throughout the year. The organism which may produce pigments in high yields may have various industrial also as pharmaceutical uses4.
Prodigiosin may be a red pigment produced by some strains of Serratia species also by other species like Pseudomonas magnisiorubra, Hahellachejuensis, Vibrio gazogenes and Vibrio psycroerythreus. Serratia species, a Gram-negative bacterium, belongs to the family Entero bacteriaceae. This bacterium grows well on standard media and produces red to dark pigment, and therefore the red colour pigment is named prodigiosin. The pigment biosynthesis may be a bifurcated process during which mono and bi pyrrole precursor are synthesised separately and assembles to make prodigiosin. Strains of Pigmented Serratia marcescens are shown to cause infections in much lower frequency than non-pigmented strains, thus reducing the danger of disease during production of pigment5-7.
Prodigiosin excels out among other natural products derived from microorganisms due to its pharmacological activity8 because the pigment prodigiosin may be a promising drug due to its reported characteristics of antimalarial/antiprotozoal, five antimicrobial, algaecides, insecticide, immunosuppressive, antineoplastic, other related cytotoxic activities and its unique application as a natural based dye for olefins and textiles9-11. This chemically characterised pigment as an alkaloid that features, flat chemical structure (pyrrole,3-methoxy pyrrole,2-methyl-3-amyl pyrrole), a linear, tripyrrole with the formula of C20H25N30 and molecular mass of 324Da2,10-12. Flat structures are characterized by prodigiosin, which acts as an intercalating agent within the DNA molecule. The intercalating drugs, which primarily function topoisomerase I and II inhibitors can induce DNA damage with structural alterations. This inhibitory impact has the potential to produce DNA damage, which has been linked to cytotoxicity. Interaction between DNA and Prodigiosin can, therefore, end in genomic damage, which characterises this drug as a possible a selective anti-tumor agent13-14. Prodigiosin has recently been found useful in natural marine settings as a biological control agent against toxic algae, so large amounts are ready to fulfil potential needs. The expansion of biotechnology considerably increased the necessity to hunt to overproduce microbial strains for the development of the assembly and obtaining better yield2,10.
One of the designs for screening discovering relevant factors among a large number of potential factors is the Plackett-Burman design. In the traditional approach, each source category is screened at an arbitrarily set level of each source, one at a time, while the other category is kept constant, also at arbitrarily determined levels1,5,15-16.
Many factors have been shown to influence prodigiosin synthesis, including species type and environmental parameters such as phosphate availability, dissolved oxygen level, light, medium composition, temperature, pH, and incubation period. Low-cost substrates could help reduce the price of prodigiosin production17-20.
Biotechnological principles have driven industrial production will be highly beneficial to meet the required production of Prodigiosin and could also help in improving the quantitative and qualitative production. In vitro, Prodigiosin production is highly dependent on the type of bacterial strain employed and other environmental factors like dissolved oxygen level, phosphate availability, temperature, pH, light, media composition, and incubation time. Cost-effective production of Prodigiosin can be achieved by using inexpensive substrates. The present work focuses on screening of microbes new strains that produce more quantities of prodigiosin and to see the environmental conditions, by using analytical models, active production can be carried out.
Gathering of marine soil
Twelve soil samples have been were collected from four various locations in natural urban beach of Chennai along the Bay of Bengal, part of Indian Ocean, with Latitudes of 13.07° and Longitude of 80.28° marines crustaceous area of Kasimedu, Mahabalipuram, Pattinampakam and Kottikuppam beaches and these samples were collected after removing the approximately 3cm of the soil surface, in polythene zip lock cover then transported immediately to the laboratory and processed for bacteriological analysis21-22.
Isolation of pigment-producing microbes
Soil samples of different places 10gm weighed, and it suspended in 90ml of sterile physiological saline. Under aseptic condition by thorough mixing, the sample allowed to settle down for 30 minutes. In the clear supernatant, one ml of the same was then serially diluted, using 9ml of physiological saline and 0.1ml of prepared dilution was used as inoculation for nutrient agar medium and incubated at 37 +0.2°C for 48hrs. Resulted in pink, yellow, red coloured colonies were isolated for further study. Eight potential isolates obtained, and these used for further work. These cultures are maintained on nutrient agar slants and stored at the refrigerator23.
Isolation of the maximal prodigiosin producing strain
The pigmented bacterial strains were further grown on 100ml of nutrient broth in 250ml Erlenmeyer flasks for three days high pigment strains were selected by quantification method and used throughout of the study24.
Quantification of Pigment & Cell growth
To quantify the amount of pigment produced by the organism, one millilitre of culture broths were centrifuged at 1200Xg for 10 minutes, then the pellet was suspended and vigorously mixed in 1 ml of acidified methanol. The solution was then centrifuged at 1200Xg for 10 minutes. Resultant solution’s optical density was calculated at 535nm. Using the formula below, the total amount of prodigiosin (mg/L) was calculated.
Total Prodigiosin (mg/L) = [AXDXV1 / 70700.0] XV2
A is the absorbance of methanol measured at 535nm, V1 – Volume of methanol added, D the dilution ratio, 7.07X104 is the extinction coefficient of prodigiosin, V2-Volume of fermentative liquid. From the total amount of prodigiosin, most high quantity producing microbes selected for the further studies2.
Biomass Assay
The cell biomass determined with the help UV–VIS spectrometer at 600 nm (OD600) by measuring the absorbance of the sampling broth. The broth filtered through 0.45 mm paper, to get dry cell weight measurement, the followed by washing twice and drying at 50°C. A calibration curve was used to link the OD600 value to the cell dry weight24.
Characterisation of the soil isolate
The bacterial strains subjected to biochemical characterisation according Manual of Determinative Bacteriology (Taxonomic Scheme of Bergey’s)25.
Genetic characterisation of isolate using 16S rRNA Sequencing
16S rRNA Sequencing was performed for pigmented strains identification. RNA Extraction from pigmented strains and further analysis of the sequence of its 16s RNA was carried out using the ion employing two universal primers, 8F & 1541r26. The horizontal gel electrophoresis method was used to examine the heat cycler product. ABI PRISM BigDye TM Terminator Cycle Sequencing Kits with AmpliTaq RNA polymerase (FS enzyme) (Applied Biosystems) and ABI 3730X1 sequencer were used to sequence the amplified 16s rRNA (Applied Biosystems)23-27.
Purification and Identification of the Pigment
Earlier work with modification separated and refined the pigment further. Briefly, the culture broth mixed with acidified methanol & organic portion was separated after centrifugation at 10,000Xg for 10 minutes. The crude extract was evaporated, and the resulting concentrated extract was passed through the activated silica gel column to trap the target product within the column. The sample eluted with chloroform: methanol: ethyl ether as a ratio of 6:3:1, at a flow rate of 0.1 ml/min. The eluted fractions then analysed by scanning in the UV-visible spectrophotometer. The selected scanning range was 200-700nm. The scanned fraction which showing high peaks in 535nm were cumulated and dried. The partially purified resulting pigment was further characterised using Thin-layer Chromatography in (Hi-250 F) silica gel plate, and Fourier transforms infrared spectrophotometer in the range of 4,000-400 cm-1. The pigment was separated and blended with potassium bromide powder before being crushed into a pellet for FTIR spectroscopy at the chosen frequency28,29.
Identifying the significant variables using Plackett-Burman Design (PBD)
Plackett-Burman Design was known for spotting important variables and their interactions between variables relatively few experiments are enough as compared to the one factor at a time technique. High (+1) and Low (-1) levels were investigated for each independent variable. Glucose, Mannose, Sucrose, C-proline, Cysteine, time period, pH, inoculum size, agitation, temperature, and peanut % were the variables utilized for this investigation. These variables were used to determine the relative value of various nutrients for prodigiosin production in shaking flask culture, and the experimental design was created using Design Expert trial 9.0.4.1 software (Stat Ease Inc., USA). Each column in Table: 1 represents a different variable, and each row represents an experiment. In 250mL Erlenmeyer flasks containing 50mL medium, all tests are performed in triplicate30-32.
Table (1):
Design of variables and coding selection of Nutritional components and Environmental parameters of the medium for the production of Prodigiosin using Serratia marcescens A2.
| Coded factors | Environmental and Nutritional Factors | Level of variables in the design | |
|---|---|---|---|
| Low level (-1) | High level (+1) | ||
| X1 | A:Glucose(gm/l) | 2 | 3 |
| X2 | B:Mannose(gm/l) | 2 | 3 |
| X3 | C:Sucrose(gm/l) | 2 | 3 |
| X4 | D: L-Proline(gm/l) | 0.25 | 0.75 |
| X5 | E:Cysteine(gm/l) | 0.25 | 0.75 |
| X6 | F:Incubation Period(Hrs) | 24 | 72 |
| X7 | G:pH | 6 | 8 |
| X8 | H: Inoculum Size (%) | 5 | 10 |
| X9 | I:Agitation(rpm) | 75 | 125 |
| X10 | J:Temperature(0C) | 28 | 37 |
| X11 | K:Peanut(gm/l) | 1 | 3 |
Plackett-Burman design for statistical analysis method
Plackett-Burman’s methodology and results from the ANOVA of the program Design Expert trial version 9.0.4.1 were used to analyze the experimental data. The effect of every variable decided. Mean squares of every variable calculated. The multivariate analysis revealed that they had a substantial impact on prodigiosin production (p<0.05).
Isolation of pigment-producing microbe
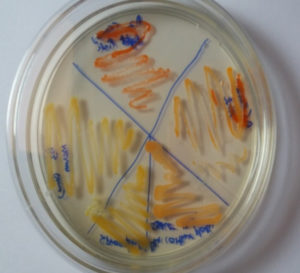
Marine microbial populations secrete a wide range of products that are dependent on their habitat and ecological activities. Microorganisms in the maritime environment are compared to their distinct living surroundings and their genetic traits under harsh chemical conditions, such as high salinity, high pressure, low temperature, and particular illumination conditions15. Pigments produced by bacteria possess enormous efficiency as medicinally essential products. Pigment producing microbes were obtained from crustacean soil of different localities on east coast region of Bay of Bengal (Marina beach) like Kasimedu, Mahabalipuram, Pattinampakam and Kottikuppam beaches near to the fish material wastes were dumped. Organisms which produced pink to orange colour colonies were isolated due to the prodigiosin usually appear these colour range and named as A1 to A10. These strains were maintained on nutrient agar medium for further studies. From these three strains got from the source area of Nellikupam and five more from Kasimedu area. Remaining two strains of microbe separated from Mahabalipuram. (Fig. 1) These microbes were grown in nutrient broth, and the amount of pigment produced by strain scanned through UV-spectrum of 200-700nm. The quantification study indicated that the sample A2 produced relatively more amount of Prodigiosin and this bacterium adapted for the further study.
Purification and Identification of Pigment
The pigment exhibits different λ max in acidic and alkaline environments. In acidic conditions λ max was absorbed in 535nm as red, were as in salty environment orange-yellow band at 465nm was absorbed. Protonation of the pyrrole group in the prodigiosin structure causes this. The ring becomes non-aromatic when the pyrrole group protonates on one of the carbon atoms in the second position in the ring rather than the nitrogen atom. The colour of prodigiosin fades as a result of this extension. The OH- group deprotonated the amine group in the structure, generating the anion, at alkaline pH20. The Rf value of obtained pigment was found to be 0.86 in Thin-layer Chromatography (TLC) on standard silica gel plates with chloroform: methanol: ethyl ether (6:3:1) as eluents. Spectrophotometric analysis of pigment showed the single peak at 536nm. In FTIR, the dye showed the strong band at 3,378cm-1 for O-H stretch, 2,950cm-1 of C-H and C=O stretch. The peak at 1,097 and 1,197cm-1 showing C-O stretch and C-N bend (amines).
Characterisation of the soil isolate
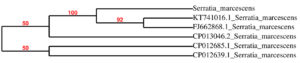
Isolate A2 was chosen for further research since it is the most efficient by using quantification test in terms of pigment production. (Fig. 2) The biochemical and morphological obtained in this study lead to the taxonomic conclusion that the red pigment-producing bacteria isolated is Serratia species. By using 16S rRNA gene sequencing, this isolate was identified as Serratia marcescens and designated as S.marcescens A2 (Fig. 3). For multiple sequence alignments, the program MUSCLE 3.7 was utilized. For phylogenetic analysis, the program PhyML 3.0 aLRT was utilized, and the substitution model was HKY85. Tree rendering was done with the Tree Dyn 198.3 program. The results were in agreement with the investigation by Y.Aparna et al., research on isolated microbes of Serratia sps through Genetical approach18.
Fig. 3. Neighbour-joining phylogenetic representation of the strains A2 and their closest relatives based on 16S rRNA gene sequences. Dendrogram constructed by maximum-likelihood method using PhyML 3.0 aLRT.
Nutritional and environmental parameter statistical screening by S.marcescens using Plackett-Burman Design (PBD)
To produce the maximum pigment the phenomenon of the analysed by Plackett Burman design, where we evaluate the effect of eleven different culture elements on the production of pigment by the isolate including environmental and nutritional conditions. (Table 2) Estimated efforts (t values and p values) on pigment production and biomass creation for each independent variable. Hence prodigiosin is a secondary metabolite, and the medium and environmental conditions influence the growth as well as the pigment production. It determined the adequacy of the model. Statistically important effects were exhibited by the variables and screened using ANOVA. Factors with a p-value of less than 0.05 are known to have a significant impact on production.
Table (2):
Plackett-Burman experimental design for Prodigiosin and Biomass production by Serratia isolate A2.
Run |
A: Glucose (gm/l) |
B: Mannose (gm/l) |
C: Sucrose (gm/l) |
D: L-Proline (gm/l) |
E: Cycteine (gm/l) |
F: Incubation Period(hrs) |
G: pH |
H:Inoculum Size (%) |
I:Agitation (rpm) |
J:Tempe- rature (0C) |
K:Peanut (%) |
Biomass (mg/L) (n=3) |
Prodigiosin mcg/L (n=3) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
1 |
2 |
2 |
2 |
1 |
0.50 |
72 |
8 |
5 |
125 |
37 |
3 |
675±09.1 |
7260±15.67 |
2 |
3 |
3 |
3 |
0.50 |
0.50 |
42 |
8 |
5 |
125 |
37 |
1 |
438±05.2 |
8450±10.89 |
3 |
3 |
3 |
2 |
1 |
1 |
72 |
6 |
5 |
75 |
37 |
1 |
625±4.53 |
7560±15.27 |
4 |
3 |
3 |
2 |
0.50 |
0.50 |
72 |
6 |
10 |
125 |
28 |
3 |
634±3.17 |
5300±35.11 |
5 |
2 |
3 |
3 |
0.50 |
1 |
72 |
8 |
5 |
75 |
28 |
3 |
597±4.23 |
6500±45.09 |
6 |
3 |
2 |
3 |
1 |
1 |
42 |
6 |
5 |
125 |
28 |
3 |
478±3.57 |
7800±20.41 |
7 |
3 |
2 |
2 |
0.50 |
1 |
42 |
8 |
10 |
75 |
37 |
3 |
467±4.63 |
6420±11.24 |
8 |
2.50 |
2.50 |
2.5 |
0.75 |
0.75 |
57 |
7 |
7.5 |
100 |
32 |
2 |
587±8.34 |
7460±20.81 |
9 |
2 |
2 |
2 |
0.50 |
0.50 |
42 |
6 |
5 |
75 |
28 |
1 |
435±5.47 |
8200±35.11 |
10 |
3 |
2 |
3 |
1 |
0.50 |
72 |
8 |
10 |
75 |
28 |
1 |
685±7.67 |
7240±20.98 |
11 |
2 |
2 |
3 |
0.50 |
1 |
72 |
6 |
10 |
125 |
37 |
1 |
643±6.89 |
5400±36.05 |
12 |
2 |
3 |
3 |
1 |
0.50 |
42 |
6 |
10 |
75 |
37 |
3 |
454±9.76 |
4900±22.67 |
13 |
2 |
3 |
2 |
1 |
1 |
42 |
8 |
10 |
125 |
28 |
1 |
412±7.54 |
6900±34.23 |
Effect on Biomass Production
The model F value of 109.79 implies the model applied for biomass production is significant. The incubation period was found to be the most significant factor in the p values of (<0.000 1). This followed by mannose (p values =0.0098) and c proline (p-value 0.1206) in biomass formation. The model was found to statically correlated with R2 value of 0.9362. The final equation on coded Model is
Amount of Biomass Formation =+545.25-18.58 * B+9.58 * D+97.92* F
B: Mannose, D: C-proline, F: Incubation period.
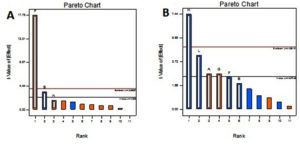
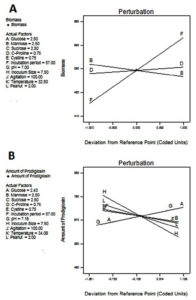
Factors of interest and slit t value factor noticed from Pareto chart were included for the design. According to the Pareto chart the factors incubation period, mannose, c-proline are above and near to the t value limit shown in Fig 4 & 5. Based on the perturbation graph incubation period showing more deviation from the reference point, mannose and C-proline given comparatively less deviation. The deviation is representing the effect on the biomass formation. From the perturbation plot, it was evident that as incubation time increases biomass also increases proportionally. Also, the dominant effect of incubation on biomass produced was also backed up by high t-value (Pareto chart) and highly significant p values.
Fig. 4. Pareto Chart showing the effect of environmental factors and media components on A) Biomass B) Prodigiosin yield.
Fig. 5. Model diagnostic plot by Perturbation graph from Plackett-Burman Design A) Biomass, B) Prodigiosin yield. In this chart A: Glucose, B: Mannose, C: Sucrose, D:L-Proline, E: Cysteine, F: Incubation Period, G: pH, H: Inoculum size, I: Agitation, J: Temperature, K: Percentage peanut.
Effect on Pigment Production
The F value applied in prodigiosin production showed significant to the model. The size of inoculum showed more significant p-value flowed by the amount of peanut and speed of agitation. P values are 0.0072, 0.0539 and 0.8862 respectively. The R2 value statistically correlated with this model. The coded equation model is as follows.
Amount of Prodigiosin = +684.58+29.9*A+9.92*D+29.75*G-79.75*H+2.75 *J-45.25* L
A-Glucose, D-L proline, G-pH, H-Inoculum size, J-Agitation, L-Peanut.
The Pareto chart shows inoculum size, amount of proline and agitation speed are much deviated from t value whereas glucose and pH shoes near to t values. Deviated factors influence the pigment production. The perturbation plot is evidence that inoculum size directly proportional to the pigment production. The percentage of peanut is influenced with limited extent. This shows an excess amount of peanut may not increase the pigment formation due to the availability of excess nutrients.
Depending on their role and habitat, microorganisms from marine environments secrete various components. Samples were collected from four different localities in legitimately from urban beach of Chennai along the Bay of Bengal, part of Indian Ocean. These samples illustrated from marines crustaceous area of Kasimedu, Mahabalipuram, Pattinampakam and Kottikuppam beaches. The microbes were cultured on nutrient broth and the yield of pigment production was quantified by the UV-spectrum at range of 200-700nm. The bacterial strain is having excellent pigment-producing properties adapted for further study. Based on the high yield of pigment, isolate A2 was used for further study. The taxonomical evaluation of the microbe strain was conducted and the red pigment-producing bacteria isolated were confirmed as Serratia species. Based on the software analysis MUSCLE 3.7, PhyML 3.0 aLRT (for a phylogeny), Tree Dyn 198.3 and DNA sequencing, the isolated microbe was identified as Serratia marcescens. It is named as Serratia marcescens KM. From the statistical analysis, we found that the production of pigment may highly be influenced by the size of inoculum (7.5%) along with the percentage of peanut (2%) in the medium. The mannose alters the biomass formations of microbes as carbon (2.5%) and C-proline (0.75%) as nitrogen sources. Overall, this method can be utilized to produce prodigiosin and other alkaloid hydrophobic components from bacterial cells on a big scale.
ACKNOWLEDGMENTS
The authors would like to express their gratitude to SRM College of Pharmacy, SRM Institute of Science and Technology, Kattankulathur, for providing the necessary facilities and support for this research.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
Both the authors have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
- Deepika L, Kannabiran K. Isolation and Characterization of Antagonistic Actinomycetes from Marine Soil. J Microbial Biochem Technol. 2010;2(1):1-6.
Crossref - Lin S-R, Fu Y-S, Tsai M-J, Cheng H, Weng C-F. Natural Compounds from Herbs that can Potentially Execute as Autophagy Inducers for Cancer Therapy. Int J Mol Sci. 2017;18(7):1412.
Crossref - Holland JF, Hong WK, Kufe DW, et al. Cardinal manifestations of cancer. Holland-Frei Cancer Medicine, Eds, John Wiley & Sons, Inc. 2016;3-7.
Crossref - Sundaramoorthy N, Yogesh P, Dhadapani R. Production of Prodigiosin from Serratia marcescens isolated from soil. Indian J Sci Technol. 2009;2(10):32-34.
Crossref - Mekhael R, Youse SY. The role of red pigment produced by Serratia marcescens AS antibacterial and plasmid curing agent. 2nd Kurdistan conference Biol Sci, J Duhok Univ 2009;12(1):268-274.
- Srimathi R, Priya R, Nirmala M, Malarvizhi A. Isolation, Identification, Optimization of Prodigiosin Pigment Produced by Serratia marcescens and its Applications. International Journal of Latest Engineering and Management Research. 2017;02(09):11-21.
- Shahitha S, Poornima K. Enhanced production of Prodigiosin production in Serratia marcescens. Journal of Applied Pharmaceutical Science. 2012;2(8):138-140.
Crossref - Montaner B, Perez-Tomas R. Prodigiosin-induced apoptosis in human colon cancer cells. Life Sci. 2001;68(17):2025-2036.
Crossref - Chen W-C, Yu WJ, Chang C-C, et al. Enhancing production of prodigiosin from Serratia marcescens C3 by statistical experimental design and porous carrier addition strategy. Biochem Eng J. 2013;78:93-100.
Crossref - Elkenawy NM, Yassin AS, Elhifnawy HN, Amin MA. Optimization of prodigiosin production by Serratia marcescens using crude glycerol and enhancing production using gamma radiation. Biotechnol Rep. 2017;14:47-53.
Crossref - Gargallo D, Loren J G, Guinea J, Vinas M. Glucose-6-phosphate dehydrogenase alloenzymes and their relationship to pigmentation in Serratia marcescens. Appl Environ Microbiol. 1987;53(8):1983-1986.
Crossref - Castilla LC, Aguilar O. Effect of mineral supplementation and type of starch on the production of prodigiosin from a culture of Serratia marcescens BS303. Rev Med Ing Quim. 2015;14(3):641-652.
- Chang CC, Chen WC, Ho TF, Wu HS, Wei YH. Development of natural anti-tumor drugs by microorganisms. J Biosci Bioeng. 2011;111(5):501-511.
Crossref - Han SB , Kim HM, Kim YH, et al. T-cell specific immunosuppression by prodigiosin isolated from Serratia marcescens. Int J Immunopharmacol. 1998;20(1-3):1-13.
Crossref - Abdelhafez AA, Husseiny SM, Ali AA-A, Sanad HM. Optimization of carotene production from agro-industrial by products by Serratia marcescens ATCC 27117 using Plackett-Burman design and central composite design. Annals of Agricultural Sciences. 2016;61(1):87-96.
Crossref - Kaur B, Garg N, Sachdev A. Optimization of Bacteriocin production in Pediococcus acidilactici BA28 using response surface methodology. Asian J Pharm Clin Res. 2013;6(1):192-195.
- Williams RP, Gott CL, Qadri SM. Induction of pigmentation in non proliferating cells of Serratia marcescens by addition of single amino acids. J Bacteriol, 1971;106(2):444-448.
Crossref - Wang SL, Wang CY,Yen YH, Liang TW, Chen SY, Chen CH. Enhanced production of insecticidal prodigiosin from Serratia marcescens TKU011 in media containing squid pen. Process Biochem. 2012;47(11):1684-1690.
Crossref - De Araujo HW, Fukushima K, Takaki GM. Prodigiosin production by Serratia marcescens UCP 1549 using renewable-resources as a low cost substrate. Molecules. 2010;15(10):6931-6940.
Crossref - Siva R, Subha K, Bhakta D, Ghosh AR, Babu S. Characterization and enhanced production of prodigiosin from the spoiled coconut. Appl Biochem Biotechnol. 2012;166(1):187-196.
Crossref - Prasad MP. Optimization of media parameters for pigment production in bacteria from effluent water sample. Biolife. 2015;3(2):428-433.
Crossref - Giri AV, Anandkumar N, Muthukumaran G, Pennathur G. A novel medium for the enhanced cell growth and production of prodigiosin from Serratia marcescens isolated from soil. BMC Microbiol. 2004;4:11.
Crossref - Elkenawy NM, Yassin AS, Elhifnawy HN, Amin MA. Optimization of Prodigiosin by Serratia marcescens using crude glycerol and enhancing production using gamma radiation. Biotech Reports. 2014;14:47-53.
Crossref - Gulani C, Bhattacharya S, Das A. Assessment of process parameters influencing the enhanced production of prodigiosin from Serratia marcescens and evaluation of its antimicrobial, antioxidant and dyeing potentials. Mal J Microbiol. 2012;8(2):116-122.
Crossref - Breed RS, Murray EGD, Smith NR. Baltimore: Bergey’s Manual of Determinative Bacteriology.7th ed. Williams and Wilkins. 1962.
- Dereeper A, Guignon V, Blanc G, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465-W469.
Crossref - Shaikh Z. Biosynthesis of Prodigiosin and its applications. IOSR-JPBS. 2016;11(6):1-28.
- Linawati H, Anwar H, Rita RC. The influence of environmental conditions on the production of pigment by Serratia marcescens. Biotechnol Bioprocess Eng. 2002;7:100-104.
Crossref - Kavitha R, Aiswarya S, Ranthnavali CMG. Anticancer activity of red pigment from Serratia marcescens in Human cervix carcinoma. Int J ChemTech Res. 2010;2(1):784-787.
- Kamble KD, Hiwarale VD. Prodigiosin production from Serratia marcescens strains obtained from farm soil. International Journal of Environmental Sciences. 2012;3(1):631-637.
- Durga R, Rajagopal K. Statistical optimization of medium components by Plackett Burman design and response surface methodology for enhanced antioxidant activity by Xylaria feejeensis HMJAU22039. Int J Pharm Pharm Sci. 2016;8(11):159-164.
Crossref - ZZ Lv, XX Liu, SQ Sun, YJ Wang. Effect of surfactants on production of prodigiosins by S. marcescens. Chinese Journal of Pharmaceutical Biotechnology. 2012 ;19:324-327.
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.