ISSN: 0973-7510
E-ISSN: 2581-690X
The microbial community in oral cavity associated with oral infection and disease, variety of microbes have been recorded in oral cavity with different genetic properties, the present study aims to study some oral cavity isolates using classical methods and PCR for molecular detection O-antigen genes, results show that there were four isolates, three were G- E. coli; Klebsilla pneumonia and Cardiobacterium hominis while the fourth species was Staphylococcus aureus, it had different response to antibiotics E. coli and Klebsiella pneumonia were more resistance. The plasmid profile didn’t show any plasmids as well as O- antigen genes it was negative for all isolates. The present study concluded that oral cavity have variety species with different genomic properties, it need more investigation about other genes and bioactivity.
Oral cavity, O-antigen, plasmid profile
The oral and dental infection cause by population of Microbial like periodontal diseases, pericoronitis, gingivitis, endodontitis, peri-implantitis, and post extraction infections each of these have distinct clinical and microbial features, The r microbial flora in the oral cavity contains about 1010 of bacterial species , more than 500 species yet to now recognized as normal inhabitants in oral cavity, However, only 150 microbial species have been isolated and cultured from root canals (Marina and Romana, 2007)
There are many species related with oral infections recoded by different studies like Campylobacter rectus, Prevotella intermedia, Bacteroides forsythias, Actinobacillus actinomycetem-comitans, Porphyromonas gingivalis Eubacterium species, Fusobacterium nucleatum also Eikenella corrodens, and Pepto-streptococcus micros. Treponema pallidum-related spirochetes have been associated with acute necrotizing ulcerative gingivitis, Also Porphyromonas endodontalis recorded to be specifically related to endodontic infections (Paster et al., 2000; Paste et al., 2001; Aas et al., 2005;; Dewhirs et al., 2010) it was responsible of two major oral diseases; caries (tooth decay) and periodontitis (gum diseases) Dahlén et al., 1993; Marsh et al., 1994; Nishihara et al., 2000).
The species Isolation which causing infection in oral cavity and teeth canal can be collected by strictly aerobic and anaerobic techniques, also Rapid selective culture and DNA probe have been used in identification of oral species microbes, however the detection by Molecular methods of oral microbial genera or by 16S rRNA genes sequencing improved that the mouth of human provides a habitat for about 700 species of bacteria, and that between 100 and 200 different species in the healthy mouth of any individual (Paster et al., 2006). other surveys suggested that the variety of bacteria in an individual oral cavity may be slightly higher by using pyrosequencing technologies, around 500 species (Zaura et al., 2009).
Sample collection: samples collected using trans media swaps from dental canal after extraction , all patients were visited porsepa center for dentist clinic, swaps collected by specialist dr. Saadi shershap, then it transfer to lab for culture and analysis.
Culture and diagnosis: aerobic and anaerobic culture, then isolates were re-culture for purification isolates and diagnostic. Macroscopic and microscopic diagnostic : The features of colonies, culture in differential media and gam stain using to diagnostic isolates then antibiotic sensitivity was detected using disc methods to cefixime, erythromycin, gentamicin, amikacin, meropenem, Nitrofurantoin, ceftazidime, norfloxacin, tetracycline, cefotaxime, aztronam.
DNA extraction and PCR experiment: DNA was extracted and PCR performed according to PCR colony methods then the following primers were used to amplify game negative isolates O45 wzx1 (F) 3´-CCG GGT TTC GAT TTG TGA AGG TTG-5´ ,(R) 3´-CAC AAC AGC CAC TAC TAG GCA GAA-5´, and O45 wzy1 (F) 3´-GAA ATT ATG CCA TCT TGG CGA GCG-5´ , (R) 3-´CAT GTG AAG CCT GAA GGC AAA CTC-5´ (DebRoy et al.., 2004). PCR conditions and size products: PCR experiments performed as a following; pre-denaturation for 5 min at 94ÚC, then 35 cycles ( 30 s at 94ÚC, 30 s at 59ÚC, 30 s at 72ÚC, and finally 10 min at 72ÚC). Electrophoresis was implemented using 1% agarose 0.5 X for 60 min , 70 V and 20 mA all testes had control represented by E coli HB101 [pomega ]
The results of bacterial culture show four species of bacteria three of its was gram GV- included E. coli; Klebsilla pneumonia and Cardiobacterium hominis , the fourth species was Staphylococcus aureaus.
Antibiotic resistance the antibiotic sensitivity of isolates were show in table (1) E coli was more resistance than other isolates it was resistance to Ceftazidime, Cefotaxime, Cefixime, Aztronam, Amikacin and Tetracycline while it more sensitive to Nitrofurantoin than other isolates. Klebsilla pneumonia isolates was resistance to seven types of antibiotics and it more sensitive to Tetracycline as well as the other two isolates, Staphylococcus aureaus was resistance to 4 types while Cardiobacterium hominis was resistance to two types only. The resistance to antibiotics types dependent on the genetic of isolates and its predisposition to acquire a new antibiotic resistance gene by conjugation, transformation and transduction way. The oral cavity was complex ecosystem that consist of hundred different taxa of microorganisms may be detected founded, some studies suggested that the incoming bacteria to oral cavity like E. coli that don’t considered as a part of the normal oral thus can able to attach to extracellular matrix or resident members , and more importantly, overcome the invasion resistance in order to integrate into the community [14], He et al., (2012) concluded that the some bacterial species in oral cavity can be integrated in this community by strategy of evasion to development oral microbial community ; which prevents detection by antagonistic oral bacteria and allows integration.
Table (1):
Antibiotic sensitivity of bacterial isolates.
Bacterial isolates |
Erythromycin 15mcg |
Ceftazidime 30mcg |
Nitrofurantoin 300mcg |
Cefotaxime 30mcg |
Cefixime 5mcg |
Norfloxacin 10mcg |
Aztronam 30mcg |
Amikacin 30mcg |
Gentamicin 10mcg |
Tetracycline 30 mcg |
Meropenem 10mcg |
|---|---|---|---|---|---|---|---|---|---|---|---|
E. coli |
17 |
0 |
35 |
0 |
0 |
23 |
0 |
0 |
16 |
0 |
20 |
Klebsilla pneumoniae |
0 |
0 |
0 |
23 |
0 |
30 |
0 |
14 |
13 |
33 |
0 |
Staphylococcus aureaus |
0 |
0 |
20 |
16 |
0 |
30 |
16 |
15 |
18 |
40 |
0 |
Cardiobacterium hominis |
25 |
10 |
30 |
0 |
0 |
27 |
20 |
27 |
23 |
44 |
60 |
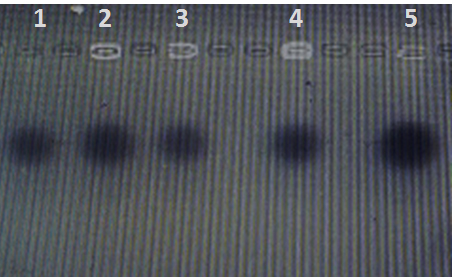
The results of DNA profile didn’t show any plasmids in all isolates (figure 1), also all isolates didn’t had o-antigen gene; the PCR product show negative results (figure 2).
Fig. 1. Plasmid profile of oral bacterial isolates lane 1 positive control E coli Hb101, lane 2-5 oral bacte-rial isolates E. coli, Klebsilla pneumonia, Staphylococcus aureaus, Cardiobacte-rium hominis respectively
Fig. 2. Negative Multiplex PCRs targeting the Owzx and Owzy genes Lane 1 M, molecular weight, lane2-8 bacterial isolates lane 9, E coli HB101. The Positive multiplex PCR showing the amplified wzx 255 bp and wzy 451 bp
The review of literature about plasmid profile in oral cavity bacterial were little, study performed by Marcia et al., (2003) about plasmid profile in oral Fusobacterium nucleatum, they found three groups of plasmid were classified according to its size. Although of absence plasmids in present isolates it were had antibiotics resistance, this can be explanation by plasmid curing in oral cavity that resulted from construction microbial biofilm initial from adherence to the oral tissues and teeth, cooperation between bacterial species, signaling between the bacteria and its role in pathogenesis, and the transfer of DNA between bacteria by different genetic transfer processes (Kolenbrander et al., 2010).
The Biofilms consider good environment to exchange DNA because of a close proximity of cells that able DNA to trapped in extracellular matrix. Roberts et al., (2001) and Warburton et al., (2007) improved horizontal gene transfer between oral streptococci in biofilm communities. Mira (2008) clarified that the DNA transfer between some oral bacteria genera was happen conjugative transposons also t Analysis of the genomes of some oral isolates improved that past horizontal gene transfer happened in about 5% and 45% of genes in different species. DNA transferring between different strains P. gingivalis and Escherichia coli, events by conjugation; its suggested by Koehler et al., (2003). In some genera DNA can be transmitted without direct cells contact as in oral streptococci (Wang et al., 2002).
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
- Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005; 43: 5721–5732. [PubMed: 16272510]
- Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001; 183: 3770–3783. [PubMed: 11371542]
- Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontology 2000. 2006; 42: 80–87. [PubMed: 16930307]
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner ACR, et al. The human oral microbiome. JBacteriol. 2010; 192:5002–5017. [PubMed: 20656903].
- Dahlén G. Role of suspected periodontopathogens in microbiological monitoring of periodontitis. Adv Dent Res. 1993; 7: 163–174. [PubMed: 8260004]
- Marsh PD. Microbial ecology of dental plaque and its significance in health and disease. Adv DentRes. 1994; 8: 263–271. [PubMed: 7865085]
- Nishihara T, Koseki T. Microbial etiology of periodontitis. Periodontology 2000. 2004; 36:14–26.[PubMed: 15330940]
- Roberts, A. P. et al. Transfer of TN916-like elements in microcosm dental plaques. Antimicrob. Agents Chemother. 2001; 45: 2943–2946.
- Warburton, P. J., Palmer, R. M., Munson, M. A. & Wade, W. G. Demonstration of in vivo transfer of doxycycline resistance mediated by a novel transposon. J. Antimicrob. Chemother. 2007; 60: 973–980.
- Mira, A. in Molecular Oral Microbiology, (ed Rogers, A. H.) 65–85 (Caister, Norfolk, UK, 2008).
- Koehler, A. et al. Multilocus sequence analysis of Porphyromonas gingivalis indicates frequent recombination. Microbiology, 2003; 149: 2407–2415.
- Wang, B. Y., Chi, B. & Kuramitsu, H. K. Genetic exchange between Treponema denticola and Streptococcus gordonii in biofilms. Oral Microbiol. Immunol. 2002; 17: 108–112.
- Paster, B. J., Olsen, I., Aas, J. A. & Dewhirst, F. E. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol. 2000, 2006; 42: 80–87.
- Zaura, E., Keijser, B. J., Huse, S. M. & Crielaard, W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009; 9, 259.
- Kolenbrander P.,. Palmer Jr R., Periasamy S. and Jakubovics S. Oral multispecies biofilm development and the key role of cell–cell distance NATuRE REvIEWS | Microbiology, 2010; 8: 471-80.
- He, X., Hu, W., Kaplan, C. W., Guo, L., Shi, W., & Lux, R. Adherence to Streptococcifacilitates Fusobacterium nucleatum integration into an oral microbial community. Microbial Ecology, 2012; 63(3): 532–542. http://doi.org/10.1007/s00248-011-9989-2
- Marcia o. Paula, Elerson Gaetti-Jardim Jr & Mario j. Avila-campos plasmid profile in oral fusobacterium nucleatum from humans and cebus apella monkeys, rev. Inst. Med. Trop. S. Paulo, 2003; 45(1):5-9.
- Marina George, Romana Ivanaková Root Canal Microflora, Acta Medica (Hradec Králové); 2007; 50(1):7–15.
- Al-Terehi M, Al-Saadi A. H, Zaidan H.K , Alkaim Z.H.,Habeeb R. A. , Majed N. (2016) some herbal medicinal plants activity against Candida spp wich resistance to antifungal drugs. International Journal of Chem Tech Research. 2016; 9(03): pp 407-411.
- Al-Terehi1 M., Al-Saadi A, Zaidan H., Hikmat R, Haleem Z. (2015) Some plants extracts synergism effects in pathogenic bacteria. International Journal of pharm tech Research. CODEN(USA): IJPRIF, ISSN:0974-4304, 2015; 8(10), pp158-164.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.