ISSN: 0973-7510
E-ISSN: 2581-690X
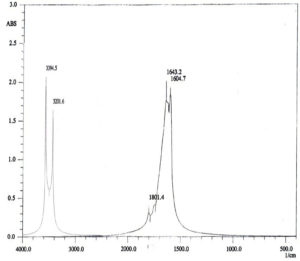
The antimicrobial agents of silver nanoparticles (AgNPs) have been applied a little while back in diverse therapeutic studies. In this analysis, AgNPs were biosynthesized using an ecologically welcomed and cost-effective simple of bio-reduction. An isolate of Aspergillus flavus KF946095 (A. flavus) was found to biosynthesize AgNPs; the size of AgNPs was (56nm) and detected by UV-Vis analysis at (400 nm). The reducing properties for biosynthesis of AgNPs are mainly due to the protein functional surface reactive groups detected by Fourier Transform Infrared spectroscopy (FTIR).Whereas, FTIR for AgNPs showed different peaks at 3994.5, 3201.6, 1801.4, 1643.2 and 1604.7 cm-1 that shared with the biosynthesize and stability of AgNPs as protein capping agents. Transmission Electron Microscope (TEM) confirmed the scattering of biosynthesized AgNPs within a sol with oval and round shapes. The antibiotic susceptibility test was studied for some pathogenic bacteria. Staphylococcus aureus DSM 1104 (S. aureus) appeared to be the more resistant strain; it resisted the action of 6 antibiotics out of 8 ones tested. MIC value of AgNPs was 20µg/mL and antibiotic ciprofloxacin was 30µg/mL. Mixture of MIC values or double MIC values distinctively inhibited the multidrug resistant (MDR) S.aureus.
Silver nanoparticles, Aspergillus flavus, FTIR, Staphylococcus aureus, antibacterial activity
The existence of antibiotic resistant strains is increased recently and their inhibition by alternative agents are necessary (Goffeau, 2008; Enan et al., 2013; Osman et al., 2016; El-Gazzar., etal.,2020 Abdel-shafi, 2020;Enan et al.,2020). S. aureus is a Gram-positive, coccid-shaped, facultative anaerobic and it forms yellow colonies on routine agar medium and forms black colonies with halo zones after its growth on its specific medium Baired Parker agar. S. aureus is non-motile, non-spore former, ferments glucose and produces lactic acid; it shows both a- and b- blood hemolysis capability and is characterized by positive coagulase reaction (Colombari et al., 2007).
Previous studies (Osman et al., 2016; Enan et al.,2020) isolated and characterized MDR S.aureus from both foods and patients. Consequently, the biocontrol of the S.aureus pathogen is of interest for detection incoming effective protocols to inhibit such pathogen in vitro. In this regard, there is an inevitable and urgent medical need for developing other novel antimicrobial mechanisms with eco-friendly properties’ (kim et al, 2008; Jo et al., 2009; Abdelshafi 2020; El-Sayed, 2020).
From the Elders, silver and its compounds have been considered to be Functional Antimicrobial active factors (Silver, 2003; El-Gazzar and rabie, 2018; Ngozi et al., 2019; El-Gazzar and Enan 2020; El-Gazzar and Ismail, 2020; Samson et al., 2020). Nano-silver has received particular attention as a possible antimicrobial agent, owing in particular to recent advances in metal nanoparticles research (Melaiye et al., 2005). Silver nanoparticles have been widely subjected due to their physicochemical properties (Lok, 2006) and are recently applied as anti-bacterial factors in food storage and health industries. The anti-microbial activity of Ag-Nps has now been well recognized and they are definite to possess anti-inflammatory, anti-viral, anti- platelet and anti-fungal efficiency (Panacek et al., 2009).
The nanoparticles syntheses are characterized by an effective and a potent role in application as novel antimicrobial factors (El-Gazzar and Enan 2020). The newly nano-products have already applied against pathogenic bacteria (Kim et al., 2008; El-Gazzar and Ismail, 2020). So, the biosynthesis and functionalization of nanoparticles create the most potent antimicrobial products. Thus, this work was aimed to biosynthesize of AgNPs by A.flavus and investigates the antimicrobial effect of AgNps alone or in combination with antibiotics to inhibit MDR S.aureus.
The fungal strain used for biosynthesis of AgNPs
The fungal strain used in this study for biosynthesis of AgNPs was Aspergillus flavus KF946095. It was isolated from soil contaminated with wastes from Photographic Industries (El-Sharkia Governorate, 80 km North Cairo, Egypt) (El-Gazzar, 2015). It was stored in glass beads and subcultred into potato dextrose broth (Oxoid) (Abdel-Salam et al., 2001).
Biosynthesis of AgNPs
A. flavus KF946095 was isolated, identified and characterized to possess the capability to biosynthesize AgNPs by its extracellular enzyme to reduce AgNO3 solution at 1mM to AgNPs according to El-Gazzar and Rabie, (2018). The color of AgNO3 at the beginning of the experiment was colorless and the developed AgNPs were visually monitored from the intensity of yellow-brown color. The concentration and physical properties of the developed AgNPs were determined (El-Gazzar et al., 2020).
Characterization of the bio synthesized AgNPs
The prepared post-biotransformation fluid of bio synthesized AgNPs was detected with (T80+UV Flash spectrophotometer PG Instrument LTD), (DeAlba-Montero et al., 2017).The diameter of particles determined by nanosizer (Malvern, UK) at agriculture research Center, Egypt (Halima and Archna, 2016). The morphology of AgNPs was subjected by Transmission electron microscope (TEM) (JEOL.JEM.1010) in National Research Center, Egypt. Silver nanoparticles size, shape and size distribution profile were determined (Jain et al., 2011). Fourier transform infrared spectroscopy (FTIR) (Thermo Nicolet model 6700 spectrum) in Micro-Analytical Center, Egypt, using KBr methods with 400–4000 cm-1 range (DeAlba-Montero et al., 2017).
Antibiotics susceptibility test of some pathogenic bacteria
Staphylococcus. aureus DSM 1104, Streptococcus. pyogenes ATCC 19615, Listeria. monocytogenes LMG10470, Escherichia. coli LMG 8223, Klebsiella. pneumonia ATCC 43816 and Pseudomonas. aeruginosa LMG 8029 were subjected.. Stock bacterial cultures were routinely were sub-inoculated in brain heart infusion broth (BHIB) (Oxoid) (Abdel-shafi et al., 2016), then kept at 4°C for uses in this investigation.
The susceptibility of Gram positive bacteria; S. aureus DSM 1104, St. pyogenes ATCC 19615, L. monocytogenes LMG10470 and Gram negative bacteria; E. coli LMG 8223, K. pneumonia ATCC 43816 and P. aeruginosa LMG 8029 to 8 antibiotics were tested by standard disc diffusion technique (Enan et al., 1996; Enan e t al., 2014; Abdel-shafi et al.,2019). The cultures were grown in nutrient broth (Oxoid) for 12 h. Inocula were subjected at 105 CFU/mL and then were plated onto Muller Hinton agar (Hi-Media, Mumbai, India). The following antibiotic discs with their concentrations determined in parenthesis were applied (All from Johnson& Johnson, Egypt. Branch, Heliopolis, Cairo, Egypt) viz. clindamycin (DA: 2 µg), ampicillin (AM: 10 µg), tetracycline (TE: 30 µg), ciprofloxacin (CIP: 5 µg), ofloxacin (OFX: 5 µg), amoxicillin (AMC: 30µg), penicillin G (P: 10 µg), methicillin (ME: 5 µg). These antibiotics were put within Muller Hinton agar dishes which were inoculated by the tested bacteria; plates were then inverted and incubated at 37°C for 24 h (Enan et al., 2018). Results were indicated by determination inhibition zones diameters (IZDs) by millimeters as described by CLSI (2008). The indicator of Multiplied antibiotics impedance was determined according to Gu et al., (2003); Enan et al.,(2014): Abdel-shafi et al., (2019).
Antibacterial assay of AgNPs
Brain Heart infusion agar plates (DifcoTM, Maryland, USA) were prepared and seeded with log phase cells (105 CFU/mL) of S. aureus DSM 1104 as it was the most resistant organism to the antibiotics tested, then the sterile singly AgNPs were applied to discs (6mm diameter) which were added onto the above plates compared with controls discs that saturated with sterile distilled water. All samples were incubated at 37°C for 24–48 h. IZDs was determined (Aziz et al., 2014; El-Gazzar and Enan, 2020).
Determination of minimum inhibitory concentration (MIC) of AgNPs
The antibacterial activity of AgNPs was studied by different concentration of AgNPs discs and placed onto Muller Hinton agar (DifcoTM, Maryland, USA) that seeded previously with activity growing cells of S. aureus (105 CFU/mL). MIC was visually measured trough the minimum concentricity of AgNPs which suppress bacterial development (Abdelshafi et al., 2019 a, b, c, d).
The efficiency of Ciprofloxacin and AgNPs incorporation against bacteria
The antibiotic Ciprofloxacin that inhibited the S. aureus DSM 1104 was combined with MIC concentration of AgNPs. Sterile filter paper discs were impregnated either by the antibiotic only or by the AgNPs only and also by combinations them and tested for their antimicrobial activity against S. aureus as described above. Mixtures of AgNPs with Ciprofloxacin were prepared as follows: (30 µg/mL Ciprofloxacin + 20 µg/mL AgNPs) and (60 µg/mL Ciprofloxacin + 40 µg/mL AgNPs (Osman et al., 2016)
The preliminary investigation showed that the A.flavus KF946095 changed the color of medium into yellowish brown indicating on conversion of AgNO3 into AgNPs as revealed from the necessary analysis used in further experiments.
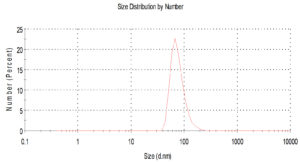
Fig.2. Histogram shows particle size distribution analyzed by Dynamic light scattering system (DLS) Zeta sizes; an average particle size based on intensity distribution for AgNPs at 56 nm.
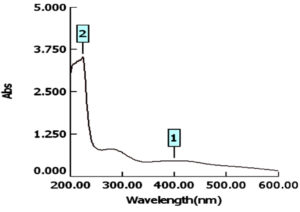
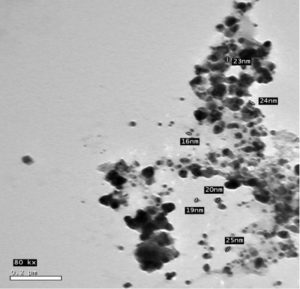
For characterization whether the yellowish brown fluids contain AgNPs or no; this suspension was subjected to spectral scan UV-visible which had maximum absorption at 400 nm corresponding to surface Plasmon resonance indicating the formation of AgNPs (Fig. 1). The diameter of nano-product showed an average particle size at 56 nm that based on intensity distribution (Fig. 2). Moreover, TEM measurements showed well distribution of AgNPs without any agglomeration at 6.49 nm with oval and round-outline (Fig. 3). The results of FTIR for AgNPs included different peaks at 1643.2 cm-1, 1604.7cm-1 and 1801.4 cm-1 (Fig. 4). The band at 1643.2 cm-1 and 1604.7 cm-1 classified into the amide bands. Whereas, the band at 1643.2 cm-1 arises due to carbonyl stretch, unsaturated aldhyde, ketone and alkene group. While the band at 1604.7 cm-1 occurs because of N H stretching vibrations in the protein’s amide linkages. At1801.4 cm-1 is assigned to c=c stretch alkenyl stretch from the proteins in the solution which is a useful tool for quantifying secondary structure in metal nanoparticles protein interaction. In addition, it shows the presence of 3201.6 and 3994.5 cm−1 bands. The band 3201.6 cm−1 is characteristic of amide II band also alchole / phenol O=H stretch. At 3994.5 cm−1, the band reflected the N=H stretch at the amide.
The results were extended to study the antibiotic sensitivity test of different pathogenic bacteria as given in Materials and Methods. Results are given in Table (1). MAR index of the indicator bacteria tested could be arranged in the following descending manner: 75%; 62.5%; 37.5%; 25% for S. aureus; Listeria monocytogenus; Pseudomonas aeruginosa and Escherichia coli, respectively (Table, 1). Since the antibiotic ciprofloxacin was the more effective antibiotic and S.aureus bacterium was possessed the finger MAR index (Multidrug resistant), they were selected for further study.
Table (1):
Antibiotic sensitivity test of the studied bacteria based on the diameter of inhibition zone (mm).
| Bacterial strains | DA | AM | TE | CIP | OFX | AMC | P | ME | MAR index | |
|---|---|---|---|---|---|---|---|---|---|---|
| Gram positive |
Staphylococcus aureus DSM 1104 | 0 | 0 | 3 | 3 | 0 | 0 | 0 | 0 | 6 (75%) |
| Streptococcus pyogenes ATCC 19615 | 0.9 | 0 | 3 | 2.1 | 0 | 1.2 | 1.7 | 0.9 | 2 (25%) | |
| Liseria monocytogenus LMG10470 | 2.9 | 0 | 0 | 3 | 0 | 1 | 0 | 0 | 5 (62.5%) | |
| Gram negative | E.coli LMG 8223 | 0.9 | 0 | 1 | 2.1 | 0 | 1.6 | 0 | 0.9 | 3 (37.5%) |
| Pseudomonas aeruginosa LMG 8029 | 0.8 | 0 | 0 | 2.1 | 0 | 0 | 0 | 1.7 | 5 (62.5%) | |
| Klebsiella. pneumonia ATCC 43816 | 0 | 2.9 | 0 | 0 | 0 | 1.1 | 1.7 | 1.1 | 3 (37.5%) | |
| No. of strains (%) |
2(6) 33.3% |
4(6) 66.67% |
2(6) 33.3% |
1(6) 16.67% |
6(6) 100% |
2(6) 33.3% |
4(6) 66.67% |
2(6) 33.3% |
||
Multiple antibiotic resistance index was calculated by using the following formula: MAR Index = Number of antibiotics to which the isolate was resistant/Total number of antibiotics tested. Clindamycin (DA: 2 µg), Ampicillin (AM: 10 µg), Tetracycline (TE: 30 µg), Ciprofloxacin (CIP: 30 µg), Ofloxacin (OFX: 5 µg), Amoxicillin (AMC: 30µg), Penicillin G (P: 10 µg), Methaicillin (ME: 5 µg)
The experiments were investigated the possibility to inhibit the S.aureus strain that was resistant to 8 antibiotics tested by different concentrations of AgNPs Table (2). AgNPs inhibited the MDR S. aureus strain. The inhibition was dependant on concentration. Inhibitory activity against S. aureus was increased with higher concentrations of AgNPs. Minimum inhibitory concentration (MIC) of AgNPs was 20µg/mL. MIC of ciprofloxacin was 30µg/ disc.
Table (2):
Inhibitory activity of different concentrations of AgNPs and Ciprofloxacin against S. aureus.
The used aged |
Concen. Of (µg/mL) |
IZD(mm) |
|---|---|---|
AgNPs |
10 |
0.00 |
20 |
12.00±0.00 |
|
30 |
15.20±0.01 |
|
40 |
18.81±0.61 |
|
50 |
34±0.11 |
|
60 |
45±0.00 |
|
70 |
46±0.45 |
|
80 |
47±0.00 |
|
90 |
48±0.42 |
|
100 |
50±0.00 |
|
Ciprofloxacin |
5 |
0.00 |
10 |
0.00 |
|
20 |
0.00 |
|
30 |
3.00±0.00 |
All values reflect the mean values of 3 replicates and standard. * Minimum inhibitory concentration (MIC) of AgNPs and Ciprofloxacin.
The experiments were further extended to check whether mixtures of both the antibiotic ciprofloxacin (the more effective antibiotic) and AgNPs will give greater inhibitory activity against the MDR S. aureus or no. Mixtures of MIC values of both the antibiotic ciprofloxacin and AgNPs showed obvious and distinctive inhibition of MDR S. aureus more than that obtained by either ciprofloxacin or AgNPs alone. Double amounts of MIC values showed distinctive inhibition of MDR S. aureus also; indicating on synergism between ciprofloxacin and AgNPs (Table,3)
Table (3):
Inhibition of S.aureus by combinations of both ciprofloxacin and AgNPs.
The used aged |
IZDs (mm) |
|---|---|
Ciprofloxacin 30µg/mL( MIC) |
3.00±0.00 |
Ciprofloxacin 60µg/mL |
9.5±0.31 |
AgNPs 20µg/mL (MIC) |
12.00±0.00 |
AgNPs 40µg/mL |
18.81±0.01 |
Ciprofloxacin 30µg/mL+20µg AgNPs (MIC values) |
28.00±0.11 |
Ciprofloxacin 60 µg/mL+ 40µg/mL AgNPs |
62.00±0.00 |
All values reflect the mean values of 3 replicates and standard.
The detection of a newly research for suppressing and blocking the various impedance microscopic organisms to antibiotic is urgent (Enan et al., 2013; Enan et al., 2014; Abdel-Shafi et al., 2020). Nanotechnology introduce effectiveness ways in inhibition of pathogenic multidrug resistant bacteria (Gu et al., 2003; El-Gazzar and Ismail 2020).The emergence of nanotechnology in the last decade offers recent biotechnology in development of drugs based nanoparticles (El-Gazzar and Enan, 2020). From the obtained data, the absorbance of AgNPs detected at 400 nm, this is similar to latter published results in this respect (El-Gazzar and Rabie, 2018; El-Gazzar and Enan, 2020).
In addition, the size of such nanoparticles was 56nm. Latter published results showed the ability of fungal strains to biosynthesize nanoparticles (El-Gazzar and Ismail, 2020). The obtained single peak indicated that the quality of the biosynthesized nanoparticles is well suitable (Nakkala et al., 2017).Also, AgNPs was scattered with oval and round shapes. Moreover, the results confirmed the presence of different molecules which are responsible on synthesis with stabilization of AgNPs. This is in confirming with previous results in this respect (El-Gazzar et al., 2020).
FTIR measurements of the prepared nanoparticles were carried out to identify the possible biomolecules presented in nanoparticles solution, for determination the stabilizing mechanism for AgNPs. The FTIR results of AgNPs detected the presence of adverse biomolecules; this is coupled with latter published work (El-Gazzar et al., 2020).Thus, the existence of diverse biomolecules with nanoparticles increases its stability and ability in metal degradation (Dhoble and Kulkarni, 2016; El-Gazzar and Rabie, 2018).
The results employed herein approved that the S. aureus was multidrug resistant bacteria, such strain showed by many authors also to resist the action of different antibiotics authors also to resist the action of different antibiotics (Osman et al., 2016). Inhibition of such MDR S. aureus by AgNPs is of interest.
Antibacterial potential of biologically synthesized silver nanoparticles was assessed. AgNPs were found to be extremely potent with inhibition of MSR S. aureus observed at 20 μg/ml concentration against all test organisms. In addition, to investigate if any synergism existed between the ciprofloxacin antibiotic and AgNPs in their antimicrobial action, mixtures containing MIC values or their doubles of them were tested. These combinations showed distinctive inhibition of MDR S. aureus than that obtained by either Ciprofloxacin or AgNPs.
Silver was characterized by its higher efficiency versa microorganisms. Although the exact underlying mechanisms are yet to be fully elucidated, other literature has indicated that AgNPs interested by the permeability of cell membrane and in turn interferes with the bacterial respiratory chain. In addition, their connection with reactive oxygen species generation (e.g., hydrogen peroxide) and resultant oxidative stress and cell damage (Aziz et al., 2014; Loo et al., 2015).
The findings of the present study have yet again highlighted the significance of the combination between antibiotics and AgNPs as potential antimicrobial agents (Prasad et al., 2016). The extract needs to be analyzed further for isolation and identification of active principles. Biogenic silver nanoparticles are not only extremely effective antimicrobial agents, as deduced from this study, but also economically. Although the AgNPs and antibiotic displayed significant antibacterial activity, a mixture containing both in much lower concentration that have the same efficiency, through a synergistic potency (Abdel-shafi et al., 2019). Further work will be necessary to study the toxicity of AgNPs against living tissues and to study the mode of action of AgNPs alone and in combinations with antibiotics.
A. flavus KF946095 showed ability to biosynthesize AgNPs that were characterized by possible instrumental analysis herein. One strain of S. aureus DSM 1104 showed to resist the action of many antibiotics; it was inhibited by different AgNPs. The antibiotic ciprofloxacin and AgNPs in combinations showed distinctive inhibition of MDR S. aureus DSM 1104.
ACKNOWLEDGMENTS
Authors would like to thank Prof. Dr Gamal Enan Dean of Faculty of science, Zagazig University, Zagazig, Egypt for his revising the work and the isolates of bacteria.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets analyzed in the study are included in the manuscript and presented as tables and figures.
- Abdel-Salam HA, El-Khamissy T, Enan GA, Hollenberg CP. Expression of mouse anticreatine kinase (MAK33) monoclonal antibody in the yeast Hansenula polymorpha. Appl Microbiol Biotechnol. 2001;56:157-164.
Crossref - Abdel Shafi S, Osman A, Enan G, El-Nemer M, Sitohy M. Antibacterial activity of methylated egg white proteins against pathogenic G+ and G– bacteria matching antibiotics. Springer Plus. 2016;5:983.
Crossref - Abdel-Shafi S, Al-Mohammadi AR, Sitohy M, et al. Antimicrobial Activity and Chemical Constitution of the Crude, Phenolic-Rich Extracts of Hibiscus sabdariffa, Brassica oleracea and Beta vulgaris. Molecules. 2019c;24(23):4280.
Crossref - Abdel-Shafi S, Al-Mohammadi A, Hamdi S, Moustafa AH, Enan G. Biological characterization and inhibition of Streptococcus pyogenes ZUH1 causing chronic cystitis by both Crocus sativus methanol extract; bee honey singly or in combination with antibiotics: an in vitro study. Molecules. 2019b;24:2903.
Crossref - Abdel-Shafi S, Osman A, Al-Mohammadi AR, Enan G, Kamal N, Sitohy M. Biochemical, biological characteristics and antibacterial activity of glycoprotein extracted from the epidermal mucus of African catfish (Clarias gariepinus). Int J Biol Macromol. 2019d;138:773-780.
Crossref - Abdel-Shafi S, Al-Mohammadi AR, Osman A, Enan G, Abdel-Hameid S, Sitohy M. Characterization and Antibacterial Activity of 7S and 11S Globulins Isolated from Cowpea Seed Protein. Molecules. 2019;24:1082.
Crossref - Abdel-Shafi S, Al-Mohammadi AR, Almanaa TN, et al. Identification and testing antidermatophytic oxaborole-6-benzene sulphonoamide derivative (OXBS) from Streptomyces atrovirens KM192347 isolated from soil. Antibiotics. 2020;9:176.
Crossref - Aziz N, Fatma T, Varma A, Prasad R. Biogenic synthesis of silver nanoparticles using Scenedesmus abundans andevaluation of their antibacterial activity. J Nanopart. 2014.
Crossref - Colombari V, Mayer MD, Laicini ZM, et al. Food borne outbreak caused by Staphylococcus aureus: phenotypic and genotypic characterization of strains of food and human sources. J Food Prot. 2007;70(2):489-493.
Crossref - CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Eighteenth Informational Supplement. Wayne, Pennsylvania, USA. 2008.
- DeAlba-Montero I, Guajardo-Pacheco J, Morales-Sanchez E, et al. Antimicrobial Properties of Copper Nanoparticles and Amino Acid Chelated Copper Nanoparticles Produced by Using a Soya Extract. Bioinorg Chemical Appl. 2017;2017:1064918.
Crossref - Dhoble SM, Kulkarni NS. Biosynthesis and characterization of different metal na noparticles by using fungi. Scholars Acad J Biosci. 2016;4 (11):1022-1031
- El-Gazzar N, Almaary Kh, Ismail A, Polizzi G. Influence of Funneliformis mosseae enhanced with titanium dioxide nanoparticles (TiO2NPs) on Phaseolus vulgaris L. under salinity stress. PLoS ONE. 2020;15(8):e0235355.
Crossref - El-Gazzar N, Ismail AM. The potential use of Titanium, Silver and Selenium nanoparticles in controlling leaf blight of tomato caused by Alternaria alternata. Bocatalysis and Agricultural Biotechnology. 2020;27:101708.
Crossref - El-Gazzar NS, Enan G. Advances in phage inspired nanoscience based therapy. Nanobioscience, Chapter 10, pp: 237-257 eds. Saxena SK, Khurana SP. Springer Nature Singapore. 2020; Pt. Ltd.
Crossref - El-Gazzar NS, Rabie GH. Application of Silver Nanoparticles on Cephalosporium maydis in vitro and in vivo. Egypt J Microbiol. 2018;53:69- 81.
- El-Gazzar NS. Continuous production of nanometal by some fungi isolated from heavy metal polluted habitats. PhD. Thesis, Faculty of Science, Zagazig University, Zagazig, Egypt. 2015.
- El-Sayed A, Enan G, Al-Mohammadi A-R, Moustafa A, El-Gazzar N. Detection, purification, elucidation of chemical structure and antiproliferative activity of taxol produced by Penicillium chrysogenum. Molecules. 2020;25(20):4822.
Crossref - Enan G, Osman ME, Abdel-Haliem MEF, Abdel-Ghany S. Advances in microbial and nucleic acids biotechnology. Biomed Res Int. 2018;2018:3102374.
Crossref - Enan G, Abdel Shafi S, Ouda SM, Negm S. Novel antibacterial activity of Lactococcus lactis subspecies lactis Z11 isolated from Zabady. International Journal of Biomedical Science. 2013;9(3):174-180.
- Enan G, Abdel-Haliem MEF, Tartour E. Evaluation of the antimicrobial activity, starter capability and technological properties of some probiotic bacteria isolated from some Egyptian Pickles. Life Sci J. 2014;11:976-985.
- Enan G, El-Essawy AA, Uyttendael M, Debevere J. Antibacterial activity of Lactobacillus planetarium UG1 isolated from dry sausage: characterization, production and bactericidal action of plantaricin UG1. Int J Food Microbiol. 1996;30(3):189-215.
Crossref - Enan G, Al-Mohammadi A-R, Mahgoub S, et al. Inhibition of Staphylococcus aureus LC554891 by Moringa oleifera seed extract either singly or in combination with antibiotic. Molecules. 2020;25(19):4583;
Crossref - Gu H, Ho P, Tong E, Wang L, Xu B. Presenting vancomycin on nanoparticles to enhance antimicrobial activities. Nano Lett. 2003;3(9):1261-1263.
Crossref - Halima R, Archna. A review on green synthsis of silver nanoparticle, characterization and optimization parameters. Int J Res Eng Technol. 2016.
- Jain N, Bhargava A, Majumdar S, Tarafdar JC, Panwar J, Extracellular biosynthesis and characterization of silver nanoparticles using Aspergillus flavus NJP08. A mechanism perspective. Nanoscale. 2011;3(2):635-641.
Crossref - Jo YK, Kim BH, Jung G. Antifungal activity of silver ions and nanoparticles on phytopathogenic fungi. Plant Dis. 2009;93(10):1037-1043.
Crossref - Kim KJ, Sung WS, Moon SK, Choi JS, Kim JG, Lee DG. Antifungal effect of silver nanoparticles on dermatophytes. J Microbiol Biotechnol. 2008;18:1482-1484.
- Loo SL, Krantz WB, Fane AG, Hu X, Lim TT. Effect of synthesisroutes on the properties and bactericidal activity of cryogelsincorporated with silver nanoparticles. RSC Adv. 2015;55:44626-44635.
Crossref - Melaiye A, Sun Z, Hindi K, et al. Silver (I)-imidazole cyclophane gem-diol complexes encapsulated by electrospun tecophilic nanofibers: Formation of nanosilver particles and antimicrobial activity. J Am Chem Soc. 2005;127(7):2285-2291.
Crossref - Nakkala JR, Mata R, Sadras SR. Green synthesized Nano silver: Synthesis, physicochemical profiling, antibacterial, anticancer activities and biological in vivo toxicity. J Colloid Interface Sci. 2017;499:33-45.
Crossref - Ngozi M, Samson OA, Awais A, et al. Biosynthesis of iron oxide nanoparticles via a composite of Psidium guavaja-Moringa oleifera and their antibacterial and photocatalytic study. Journal of Photochemistry and Photobiology B: Biology, 2019;199:111601.
Crossref - Osman A, El-Daidamony G, Sitohy M, Khalifa M, Enan G. Soybean glycinin basic subunit inhibits methicillin resistant-vancomycin intermediate Staphylococcus aureus (MRSA-VISA) in vitro. Int J Appl Res Nat Prod. 2016;9:17-26.
- Panacek A, Kolar M, Vecerova R, et al. Antifungal activity of silver nanoparticles against Candida spp. Biomaterials. 2009;30(31):6333-6340.
Crossref - Prasad R, Pandey R, Barman I. Engineering tailored nanoparticles with microbes: Quo vadis? Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2016;8:316-330.
Crossref - Pimia RP, Nohynek L, Meier C, et al. Antimicrobial properties of phenolic compounds from berries. J Appl Microbiol. 2001;90(4):494-507.
Crossref - Pyla R, Kim TJ, Silva JL, Jung YS. Enhanced antimicrobial activityof starchbased film impregnated with thermally processed tannicacid, a strong antioxidant. Int J Food Microbiol. 2010;137(2-3):154-160.
Crossref - Shaligram NS, Bule M, Bhambure R, et al. Biosynthesis of silver nanoparticles using aqueous extract from the compactin producing fungal strain. Process Biochemistry. 2009;44(8):939-943.
Crossref - Samson OA, Ngozi MM, Hisham A, Ishaq A, Botha S, Fabian I. Bio-inspired encapsulation and functionalization of iron oxide nanoparticles for biomedical applications. Eur Polym J. 2020:122:109371.
Crossref - Silver S. Bacterial silver resistance: molecular biology and uses and misuses of silver compounds. FEMS Microbiol Rev. 2003;27(2-3):341-353.
Crossref - Zain ME, Awaad AS, Al-Othman MR, Alafeefy AM, El-Meligy RM. Biological activity of fungal secondary metabolites. Int J Chem App Biol Sci. 2014;1:14.
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.