ISSN: 0973-7510
E-ISSN: 2581-690X
Hot springs are the reservoirs of novel hyperthermophilic and often mesophilic bacteria that provide information about the prevailing community structure. Here we analyzed the metagenome profile based 16S rRNA amplicon sequencing of the three different springs from Deulajhari hot spring cluster, S1, S2 and S3, having a range of temperature (43°C to 65°C), pH (7.14 – 8.10) and variation in N, P, K, TOC, Salinity, COD and TDS. These thermal spring clusters are covered with the dense vegetation of Pandanus and continuously enriched by plant leaf debris, thus resulting in a high amount of total organic carbon (TOC). The number of phyla varied among the springs: 20 in S1 (43°C), 18 in S2 (55°C) and 24 in S3 (65°C) from the 16S rRNA data. Out of the reported phyla in each spring, the most abundant were Chloroflexi, Proteobacteria, Chlorobi and Acidobacteria, which correlated with the temperature gradient. Various metabolic pathways such as ABC transporters, Two-component system, Purine metabolism were most abundantly present in the S2 sample compared to the other two. The CCA analysis revealed the correlation between physiochemical parameters and their functional annotation. The present study establishes the relation between the physiological parameters and the structural distribution of microbiota along the temperature gradient.
Deulajhari, 16S rRNA, Hot Spring, Illumina, Next-generation Sequencing
Microorganisms play a vital role in various biogeochemical cycles1 still there exists a scarcity of information about the considerate distribution of microbiota and their abundance with the variation in ecophysiological characteristics (geographic distance, temperature, pH and nutrients). 2,3 There has been an influence of the ecophysiological factors in governing the community structure. There have been various reports which link these ecophysiological parameters with the structural distribution of the microbiota.3-5 Environmental heterogeneity (pH, temperature, salinity, C/N ratio and geographic distance) strongly drives the microbial taxa-area relationship.6-8 Among the divergent environmental factors, the temperature is governing aspect that specifies the abundance and distribution of the microflora. In the previous studies, the inverse relationship between temperature and microbial diversity has been recognized,5 which stipulates the effect of temperature on the composition of the ecosystem. Bison pool hot spring study unravels the predominance of chemotrophic and phototrophic bacteria at 92°C and 56°C respectively.9 Thermophilic springs have been attracting researchers for their geological history, biogeography and physicochemical variation, as these provide a suitable habitat for thermophilic microbiota.10
Diverse studies of the thermophilic microflora in various geothermal sites have provided detailed information on the thermophilic microbial diversity and shown the path to access the bioresources that have impending relevance in biotechnological applications.11,12 Odisha, located in the eastern side of India, is bestowed with various geothermal springs, Atri, Taptapani, Tarabalo and Deulajhari, which have not been thoroughly investigated yet. Through the cultivation-based approach, few reports revealed the prevalence of thiooxydans, Chelatococcus sambhunathii, Thiomonas bhubaneswaensis and Gulbenkiania indica. 13,14 Deulajhari hot spring (Latitude 20.74 N, longitude 84.49 E) is located in the Angul district of Odisha. It has a varied temperature range (43°C to 65°C), thus providing the prospects of discovering the assorted microflora. Earlier studies over several decades on these thermophilic springs based on the cultivation-dependent approach could not present comprehensive information about the entire ecology.10,15 Metagenomics, since the last few years6,8,10 has revealed the 99% of the microflora which were earlier hard to detect through the cultivable approach. Next-generation sequencing (NGS) has enabled the study of the complete microbiome of the thermophilic springs and led to new biotechnological applications.5 Despite the diverse temperature range and varied physiochemical parameters, significant work has not been reported from the Deulajhari hot spring except a recent report on community profiling of one cluster based on the pyrosequencing approach.16
In this study, we examined the structural community of the three different springs of the Deulajhari hot spring cluster spread within a circle of 100 meters with varying temperatures. Thus, the differential microbial diversity and their relationship with the prevailing physiochemical conditions can be parceled out. Therefore, we aim to explore the abundance of the bacterial phylotypes along the 22°C temperature gradient within the Deulajhari hot spring cluster through the metagenomic 16S rRNA amplicon sequencing using the Illumina platform. This approach explains the role of temperature and the other physiochemical parameters in shaping the structural microbial ecology of the Deulajhari hot spring, which remains hidden to date.
Sample Collection and Analysis
Sediment and water samples were collected from the three different springs (S1, S2, and S3) of the Deulajhari hot spring cluster (Latitude 20.74 N, longitude 84.49 E) located in Odisha, India (Figure 1). The three springs (S1, S2, and S3) were selected from the widely spread Deulajhari hot spring cluster. Four sediment and water samples from each site were collected and pooled together to make it one representative sample of that particular site. The water temperature of the three springs was recorded in situ using the Enviro-safe thermometer (Sigma, USA) and the pH of both the water and sediment sample was measured using the portable pH meter (Sigma, USA). All sediment and water samples were collected in sterile plastic bags, sealed and transported on ice (4°C) to the laboratory and stored at −80°C for further investigation. Various physiochemical parameters of the sediment samples such as electro conductivity (EC), total organic carbon (TOC), nitrogen (N), phosphorus (P), potassium (K), chloride (Cl-), chemical oxygen demand (COD) and ammonia (NH3) was estimated as described by Jackson (1962) at Department of Soil science, Orissa University of Agriculture and Technology, Bhubaneswar, Odisha. Physiochemical parameters of the water such as salinity, oxidation-reduction potential (ORP), dissolved oxygen (DO), total dissolved solutes (TDS), nitrate (NO3-) and ammonia (NH3) carried out at the department of chemistry, Institute of Technical Education and Research (ITER), Siksha ‘O’ Anusandhan (Deemed to be University), Bhubaneswar, Odisha, India.
DNA Extraction from the Hot Spring Sediment
Hot spring sediment samples (5 gm) from each spring S1, S2 and S3 were mixed with 10 ml lysis buffer [100 mM Tris – HCl (pH 8.0), 10mM sodium EDTA (pH 8.0) and 100 µl of lysozyme (100mg/ml)] by horizontal shaking at 200 rpm for overnight at 37°C. First, 100 µl of proteinase K (10 mg/ml) was added after the overnight incubation and incubated for one hour (hr) with gentle end-to-end inversions every 15 min. Next, the samples were incubated at 37°C for one hr with gentle end-to-end inversions every 15 min after adding 1 ml of SDS (10% w/v) to each. After completing one hr incubation, samples were again incubated at 90°C for one hr with gentle end-to-end inversion every 15 min. The extraction was prepared with an equal volume of phenol: chloroform: isoamyl alcohol (25:24:1, v/v/v). After centrifugation at 10,000 rpm for 10 min at 25°C, the aqueous layer was collected cautiously. Precipitation of the aqueous phase was done with 0.3 M sodium acetate and two volumes of chilled ethanol at -20°C for one hr. Then the centrifugation was carried out at 10,000 rpm for 10 min at 25°C; the pellet obtained was washed with chilled 70% ethanol and resuspended in sterile deionized water.
Library Construction, Sequencing and Data Generation
Amplification of the V3-V4 region of the 16S rRNA using primers 341F, 5′-CCTACGGGAGGCAGCAG-3′ and 518R, 5′-ATTACCGCGGCTGCTGG-3′ carried out using the 50 ng of DNA (Bartram et al. 2011). PCR conditions were an initial denaturation at 98°C for 9 min, followed by 20 cycles at 95°C for 30 sec, 60°C for 30 sec, 72°C for 30 sec and ended with a final extension step at 72°C for 5 min. Min Elute gel extraction kit (Qiagen, Germany) was used to purify the PCR amplified product. The amplified V3-V4 amplicons (100 ng) as a template were cleaned using AMPure XP beads (Beckman Coulter, CA, USA) to filtred out non-specific fragments.17 The cleaned amplicons were further subjected to library preparation using NEBNext Ultra DNA Library Prep Kit for Illumin’s GAIIx sequencer (New England Biolab, UK). The prepared libraries were finally subjected to the 2×150 bp paired-end multiplex sequencing using Illumina GAIIx sequencer at Genotypic Technology Pvt. Ltd (Bangalore, India). The raw sequence reads of each sample were further quality checked using the FastQC v0.11.3.18 The adapter and low-quality ends from threads were trimmed using Trimmomatic v0.32 (https://github.com/usadellab/Trimmomatic).
Bacterial Diversity and Functional Prediction
Bacterial census data derived from next-generation sequencing of the three different springs S1, S2, S3 for 16S amplicon analysis using the QIIME2 pipeline.19 Briefly, the fastq-Join tool converted the overlapping paired-end reads into a consensus sequence of the V3-V4 region. Chimeric sequences were filtered out using the uchime-denovo with default settings. UCLUST algorithm was used for denovo OTU picking, taxonomic assignment and phylogenetic at sequence similarity of ≥97%. Finally, an average of 300 OTUs was identified at the species level. The taxonomic-to-phenotypic mapping was implemented using METAGENassist (http://www.metagenassist.ca/METAGENassist/) to predict seventeen potential energy sources at varying abundance. The functional capabilities was predicted by PICRUSt (Phylogenetic investigation of communities by reconstruction of unobserved states)20 and Tax4fun software21 based on GreenGenes v13.8 and SilvaSSU v123 databases respectively.
Statistical Analysis
The non-parametric richness estimates, Dominance (D), Chao1, Shannon–Weaver diversity indices (H) and Simpson index was determined by PAleontological STatistics (PAST) v3.02.22 The multivariate Canonical Component Analysis (CCA) method in PAST was used to elucidate the relationship between physiochemical parameters and taxonomic abundance as well as predicted functional capabilities. Further, to assess the community richness of the samples, the rarefaction curve was constructed using the same software.
Differential Physiochemical Characteristics of Springs of Deulajhari
Deulajhari hot spring cluster being the hub of variant springs with variable thermal conditions harbors the diverse microbial community (Figure 1). The pH of the three springs, S1, S2 and S3, was in the range of 7.14-8.10. These springs have a variant temperature range, highest in S3 (65°C) followed by S2 (55°C) and S1 (43°C), thus presenting the temperature gradient of 22°C among the Deulajhari hot spring cluster (Figure 1). The sediment analysis of the three different springs, S1, S2 and S3 showed variation in several parameters viz; electroconductivity, total organic carbon (TOC), nitrogen, phosphorus, potassium, chloride, chemical oxygen demand (COD), and ammonia. While the aqueous concentration of the phosphate, fluoride, dissolved oxygen (DO), total dissolved solutes (TDS), nitrate and ammonia also varied in the three springs of the Deulajhari hot spring cluster (Table 1). On the whole, the spring S3 had the highest concentration of ORP (194.6 m/V), temperature (65°C) and potassium (0.692 mg/l), while the spring S2 have the highest concentration of ammonia (568.1 mg/l), chloride (0.229 mg/l) and COD (29440 mg/l), whereas the spring S1 showed the dominance in the concentration of nitrogen (187.5 mg/l), TOC (3.80 H), nitrate (312 mg/l), pH (7.95), TDS (581 mg/l) and fluoride (4.06 mg/l) (Supplementary Table S1). Thus, due to the possible difference in the physiochemical parameters, these three springs of the Deulajhari cluster finds the probability of diverse ecology.
Table (1):
Physiochemical parameters of springs sediment samples.
Physiochemical parameters |
S1 |
S2 |
S3 |
|---|---|---|---|
EC (dSm-1) |
0.019 |
0.019 |
0.019 |
TOC (%) |
3.80 |
0.97 |
3.76 |
Temperature (°C) |
43.00 |
55.00 |
65.00 |
pH (1:2) |
7.83 |
7.14 |
8.06 |
Nitrogen (N) (Kg ha-1) |
187.50 |
112.50 |
175.00 |
Phosphorus (P) (Kg ha-1) |
77.80 |
57.50 |
40.00 |
Potassium (K) (Kg ha-1) |
209.70 |
153.20 |
301.00 |
COD (mg/L) |
1980.00 |
29440.00 |
23400.00 |
Ammonia |
539.70 |
568.10 |
539.00 |
Figure 1. Photographs of the A. Thermal Spring S3, B. Thermal Spring S2, C. Spring S1 and D. Location of the Deulajhari hot spring cluster.
Microbial Community Structure and Diversity Study
The three springs S1, S2 and S3 of the Deulajhari cluster exhibit a diverse microbial structure. A rarefaction curve of all species revealed as high as 90% sequence coverage (Supplementary Figure S1A). A total number of 262656, 471835 and 574661 filtered reads were obtained in S1, S2 and S3 springs, respectively. However, 99% (99.76% in S1, 98.38% in S2 and 99.12% in S3) reads were taxonomically classified as Bacteria. Archaea is reportedly negligible (0.24% in S1, 1.62% in S2 and 0.88% in S3), and the Eukarya was almost nil in all three springs.
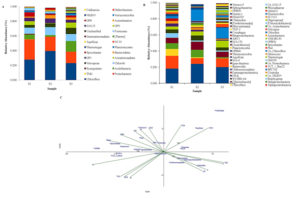
In all the three springs, S1, S2 and S3 of the Deulajhari cluster total of 20, 18 and 24 phyla were identified (Figure 2A). Under the domain bacteria, phyla Chloroflexi (0.22 in S1, 0.39 in S2 and 0.23 in S3) and Proteobacteria 0.27 in S1, 0.19 in S2 and 0.15 in S3) were abundantly found in all the three springs although the other phyla were also found specifically Chlorobi (highest in S2 i.e. 0.10 followed by 0.09 in S3 and 008 in S1), Nitrospirae (0.085 in S3, 0.050 in S2 and 0.037 in S1), Acidobacteria (0.145 in S3, 0.022 in S2 and 0.065 in S3), Armatimonadetes (0.071in S3, 0.050 in S1 and almost negligible in S2 i.e. 0.005), OP1 belonging to the Candidates division (0.062 in S2, 0.048 in S1 and 0.038 in S3), Bacteriodetes (0.046 in S2, 0.031 in S3 and 0.015 in S1) and Firmicutes (0.032 in S1, 0.022 in S2 and 0.011 in S3), and Spirochetes (0.059 in S1, 0.028 in S3 and 0.019 in S2) while several other minor phyla (whose abundance <0.001% ) were Actinobacteria, Aquificiae and Planctomycetes (Supplementary Table S2).
At the class level, 43 OTUs in S1, 34 OTUs in S2 and 48 OTUs in S3 of the Deulajhari cluster were identified (Supplementary Table S3) various classes were reported, which were common in all the three springs, although there existed variation in their abundance viz. Dehalococcoidetes (0.182 in S1, 0.241 in S2 and 0.2 in S3), Alphaproteobacteria (0.158 in S1, 0.071 in S2 and 0.108 in S3) Deltaproteobacteria (0.070 in S1, 0.092 in S2 and 0.013 in S3), Chloroflexi (0.086 in S1, 0.011 in S2 and 0.001 in S3), Spirochaetes (0.003 in S1,0.004 in S2 and 0.014 in S3), Brachyspirae (0.057 in S1, 0.014 in S2 & S3), Clostridia (0.032 in S1, 0.021 in S2 and 0.010 in S3), BPC102 (0.028 in S1, 0.133 in S3 and 0.00 in S2) and Bacteriodia (0.014 in S1, 0.024 in S2 and 0.015 in S3) (Figure 2B, Supplementary Table S3). The Venn diagram at the class level depicted that 28 (50.9%) OTUs are found common in all samples, whereas 12, 1, and 7 OTUs are uniquely present in S1, S2 and S3 samples (Supplementary Figure S1B).
Figure 2. A. Phylum level abundance of the three springs of the Deulajhari hot spring cluster metagenome, B. Class level abundance of the three springs of the Deulajhari hot spring cluster metagenome and C. Trioplot of the relative abundance of different phylotypes (≥1%) for the canonical correspondence analysis along with the fourteen different physiochemical parameters of three springs of the Deulajhari hot spring.
Although all three springs of the Deulajhari cluster shares several taxonomic groups both at the phylum and the class level, there is a significant difference in overall phylotypic composition and diversity (Table 2). Furthermore, the microbial diversity of the three springs at different taxa levels are found to be diverse irrespective of the phylum level. The above significant diversity in the phylotypic composition may be due to the three spring’s differential physiochemical variables within one geographical location. Thus, the Illumina sequencing gave us a brief outlook over the taxonomic composition of these three diverse springs, S1, S2 and S3, which shows some taxonomic diversity concerning the temperature and other physiochemical variations. In addition, this study observed that the reservoirs of diverse novel phylotypes persist in these springs of Deulajhari hot spring, which are yet to be explored by the scientific community.
Table (2):
Alpha diversity indices calculated using different metrices.
Indices |
S1 |
S2 |
S3 |
|---|---|---|---|
Taxa_S |
12 |
11 |
16 |
Dominance |
0.1868 |
0.2349 |
0.131 |
Simpson |
0.8132 |
0.7651 |
0.869 |
Shannon |
1.998 |
1.823 |
2.299 |
Chao |
12 |
11 |
16 |
Effect of Environmental Variables in the Composition of Microbial Community Structure
Environmental factors play a leading role in governing the microbial structure of hot springs. The three springs of the Deulajhari cluster exhibit the differential temperature viz. 43°C to 65°C, thus developing the temperature gradient of 22°C. The differential environmental gradients are responsible for the divergent, phenotypic characteristics.23 From the sequencing analysis, it revealed that the taxonomic richness of the microbiota in spring S1 (43°C) was more (20 phyla and 40 class) as compared to the remaining two springs S2 (55°C) with 18 phyla and 32 class and spring S3 (65°C) with 24 phyla and 46 class. Thus, this observation confirms that with the increase in the habitat temperature, the current microbiota’s taxonomic richness and structural diversity decreases.5,16,23 The taxa distribution in all the springs concerning variation in the temperature is depicted in Figure 2A. From the Figure, it is clear that there is a drift in each taxon’s distribution with the deviation in temperature. Proteobacteria was found to be dominant in the spring S1 and shows the decrease in the population with the increase in temperature (S1> S2 > S3, i.e. 43°C > 55°C > 65°C with the phyla abundance of 0.27, 0.19 and 0.15 respectively). Besides the differential temperature, carbon metabolism may also be the second key factor governing their abundance.10,16 Thus the specific distribution of these taxa indicates the influence of the geochemical variables in the different springs of the Deulajhari cluster. Based on the CCA analysis (Figure 2C), the significant taxa present across the samples and their association with the fifteen different physiochemical parameters are studied. The CCA analysis (Figure 2C) depicts the abundance of microflora Aquificae, OS-K and Clostridia highly correlated with potassium and ammonia. Similarly, the microbial diversity representing the Brachyspirae, Gammaproteobacteria, Chthonomonadetes and Chloroflexi seems to be strongly impacted by the abundance of floride, TDS, phosphorus and pH (Figure 2C). While the microbial diversity representing Chlorobia, Ignavibacteria, Spirochaetes, Deinococci, OPB56 and OM190 seems to be influenced by the abundance of the chloride, COD, DO, ORP and temperature. The aqueous concentration of TOC, nitrate, potassium and ammonia seem to affect the population of Planctomycetia (Figure 2C).
Alpha Diversity Study of the Deulajhari Hot spring
Various diversity indices viz, Dominance (D), Chao I, Shannon and Simpson are calculated for the three springs S1, S2 and S3 (Table 2). From the statistical approach, it predicts that the spring S2 has the most dominant diversity as compared to S1 (second most diverse) and S3 (most minor various); the above data approved by the abundance of the observed species, which appears to be highest in the spring S2 (858) followed by S1 (739) and S3 (346) respectively (Table 2). Out of The Chao I, Shannon and Simpson’s indices calculated (Table 2) the Shannon and Simpson indices were observed to be positively correlated with each other (the alpha diversity of both indexes appears to be S3 > S1 > S2) in the prediction of the alpha diversity of the Deulajhari hot spring cluster.
Predicted Functional Analysis of the S1, S2 and S3 Deulajhari Clusters
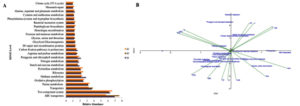
PICRUSt and Tax4Fun software were used to predict the functional analysis of the three springs of the Deulajhari hot spring cluster. Depending upon the 16S rRNA gene frequency, the functional profiling of the three springs showed that cellular processes were abundant in spring S3 (4.9), followed by S1 (4.19) and S2 (4.15). The membrane transport was the most dominant among the various cellular processes with the abundance level of 15% in S2, 14.2% in S1 and 13.09% in S3. Among the ABC transport were highly abundant in S2 (7.61%), followed by S1 (6.94%) and S3 (5.70%) and two-component systems were highly represented in S3 (6.6%), followed by S1 (6.32%), and S3 (6.14%). Other membrane transports like bacterial secretion system, phosphotransferase system (PTS) and secretion system also reported to a shallow extent. Signal transduction among the cellular process was observed to be 2.45% (S1), 2.40% (S3) and 2.14% (S2). Among the signal transduction, various signaling pathways were reported in which the most abundant one was two-component systems (2.29% in S1, 2.24% in S3 and 1.99% in S2). The other pathways, like the calcium signaling pathway, mTOR signaling pathway, were reportedly absent in all three springs (Figure 3A).
Genetic information processing at level 2 was reported to be 18.61% (S1), 18.64% (S2), and 19.62% (S3). Among these at level 2, the folding, sorting and degradation level was 2.76% (S1), 2.74% (S2), and 2.93% (S3). At level 3, the chaperones and folding catalysts were found to be 1.05% (S1), 1.0% (S2) and 1.09% (S3). At level 2, also the replication and repair were reported to be 7.99% (S1), 7.96% (S2), and 8.35% (S3), while transcription level was observed to be 2.46% (S1), 2.39% (S2), 2.44% (S3), and the translation level was found to be 5.39% (S1), 5.55% (S2) and 5.89% (S3). Similarly, human diseases at level 1 were reported to be 0.98% (S1), 0.82% (S2) and 0.91% (S3). Metabolism was the highest reported functional factor in the Deulajhari hot spring. At level 1, the frequency was reported to be 58.44% (S1), 58.22% (S2), and 58.09% (S3). Under the metabolism factor at level 2, amino acid metabolism was abundant with 11.76% (S1), 11.94% (S2), and 11.61% (S3). Apart from this, other factors at level 2 like biosynthesis of other secondary metabolites, carbohydrate metabolism, glycan biosynthesis and metabolism, lipid metabolism were also reported. Some of the enzymes families (level 2) were reported to be 2.25% in S1, 2.23% in S2 and 2.25% in S3. Cytochrome P450, peptidases and protein kinases are also detected at level 3. Cytochrome P45 was found to be negligible in all three springs. Peptidases are almost equally distributed in the three springs with the abundance of 1.82% (S1), 1.86% (S2) and 1.83% (S3). Protein kinases were found to be 0.43% in S1, 0.37% in S2 and 0.42% in S3 in all the three springs of the Deulajhari hot spring cluster (Figure 3A).
Thus from the functional analysis, it was observed that various vital pathways and genes involved in overall metabolism in the three springs of Deulajhari hot spring were found to be similar, but their profusion expressing the functionality with respect to individual subsystem were found to be different probably because of the variation in the eco-physiochemical characteristics existing among these springs, signifying the occurrence of unique metabolism in between the ecological niche of the three other springs of the Deulajhari hot spring cluster thus encouraging discrepancy in the phylotypic abundance. It helps us in predicting the functional analysis of the metagenomics data of the Deulajhari hot spring cluster. The most common subsystem in all the levels seemed to support resolving the complexity in the prevailing metabolic system in their ecological niche. The PICRUSt mediated interpretation showed the key pathways responsible for the overall functional metabolism. Although the difference in abundance at all three levels 1, 2, 3 may be the outcome of differential ecophysiology of the three springs S1, S2, and S3 of the Deulajhari. This differential parameter supports the distinct metabolism in the ecophysiological niche of all the three springs, S1, S2, and S3, supporting the diverse microbial community composition and structure (Figure 3A).
Figure 3. A. Functional analysis of the three springs (S1, S2 and S3) metagenome of the Deulajhari hot spring cluster and B. CCA (Canonical Correspondences Analysis) triplot of relative abundance of PICRUSt metabolic pathways and fifteen physiochemical parameters. The physiochemical parameters are represented by the green lines; the PICRUSt pathways are shown as filled circles.
Phenotypic Enrichment Study from the Springs (S1, S2 And S3) of the Deulajhari Hot Spring Cluster
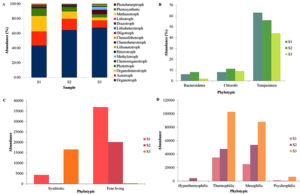
The METAGENassist web server provided a detailed study concerning phenotypic enrichment24 of the three springs S1, S2 and S3. Phylotypic symbiotic and free-living abundance data deduced from the three springs led to the prediction of the highly abundant free-living microbial diversity compared to the symbiotic abundance (Figure 4C). Among the three springs (S1, S2 and S3), the free-living is abundant in the spring S3 followed by S2 and S1; similarly, the symbiotic abundance is high in the order S3 > S2 > S1 (Figure 4C). Differential energy sources of the microbial community in all the three springs, S1, S2 and S3, predict that autotrophs are equally present in S3 and S2 with a slight decrease in S1 (Figure 4A). Whereas organotrophs seem to be dominant in S1, followed by S2 and S3 respectively. Phototrophs shows the almost similar abundance with the autotroph’s population (i.e., S3 > S2 > S1). Chemoorgantotrophs and methylotrophs are highly present in the spring S3, followed by S1 and S2, respectively. Heterotrophs seem to be highest in S2 followed by S3 and S1, respectively (Figure 4A). Lithoautotrophs were highest in S3 and absent in S2 and very few were present in S1. Chemoheterotrophs are highly abundant in the spring S3, while chemolithotrophs are highly abundant in S1 followed by S2 and absent in S3. Oligotrophs, lithoheterotrophs, diazotrophs, lithotrophs, methanotrophs, photosynthetic and photoheterotrophs present with the variation in the abundance (Figure 4A). At the different temperature levels viz. S3 (65°C), S2 (55°C) and S1 (43°C) diverse microbial communities like mesophilic, thermophilic, psychrophilic and hyperthermophilic are present. Spring S3 showed the abundance of hyperthermophilic, thermophilic and mesophilic microflora. Pscryophilic diversity of the microbiota appears to be absent from the spring S3. Spring S2 showed the second most richness of the mesophilic and thermophilic microbiota of the spring S3. Psychrophilic microflora reported to be highest in spring S2 (Figure 4D). Spring S1 has the lowest abundance of mesophilic and thermophilic microbial diversity. Some psychrophilic microflorae were also reported in the spring S1 (Figure 4D).
Taxonomic Correlation between the Bacteriodetes and Chlorobi among the three Springs S1, S2 and S3
Various studies suggest that the phylogenetically cultured representative of the phyla Chlorobi shows the common root with the Bacteriodetes.25-27 In our study, the differential population of Chlorobi was also positively correlated with the abundance of Bacteriodetes in all three springs, which supports the above studies. The phyla Bacteriodetes and Chlorobi are also observed to be linked with the temperature (Figure 4B). The population of Chlorobi is highest in spring S2 (55°C) with an abundance of 10.82% followed by 9.52% in S3 (65°C) and observed to be lowest in the S1 (43°C) with the population frequency of 8.32%. Bacteriodetes, on the other hand, follow the same trend of abundance shown by Chlorobi, i.e., S2> S3> S1, with the plenty of 4.65%, 3.10% and 1.51%, respectively. The two phyla Chlorobi and Bacteriodetes (Figure 4B) also relate to each other in conserved proteins and indels, as evident from the previous research.23, 26 Thus, the presence of various conserved indels in numerous proteins like FtsK, UvrB and ATP synthase alpha subunit also justifies the typical ancestral relationship between Chlorobi and Bacteriodetes. From our study, since the Bacteriodetes feeds on the waste produced by the Chlorobi and are phylogenetically related23,26 both the population are abundant at the temperature of 55°C, it concluded that there exists the taxonomic correlation between the two taxa (Figure 4B).
Figure 4. A. Phylotypic abundance of the community at the energy source of the three springs S1, S2, and S3 of the Deulajhari hot spring cluster, B. Abundance of Bacteriodetes and Chlorobi in relation with temperature, C. Phylotypic symbiotic and free-living abundance of the three springs and D. Phylotypic abundance of the community at the temperature level of the Deulajhari hot spring cluster.
Analysis of the Functional Characteristics of Microbial Structure Concerning the Physiochemical Parameters
The existing physiochemical environmental conditions have an extensive impact on the functional composition of the microbial constitution.15,23,28 The triplot generated for canonical correspondence analysis (CCA) represents the direct link between the physiochemical parameters and the relative abundance of the metabolic pathways. The starch and sucrose metabolism were found positively correlated with the pH, fluoride, TDS and potassium. In contrast, fructose and mannose metabolism, transporter, ABC transporters, glycine, serine and threonine, arginine and proline metabolism are positively associated with physiological factors like ammonia, COD and phosphorus (Figure 3B). The Peptidoglycan biosynthesis, pyrimidine metabolism, purine metabolism two-component system oxidative phosphorylation linked with temperature, ORP, DO and chlorine, while the nitrogen metabolism, carbon fixation pathway and chlorophyll metabolism and methane metabolism correlated with TOC, nitrate, and ammonia (Figure 3B). Thus, the physiochemical parameters (ammonia, potassium, TOC, nitrate, nitrogen, COD, chloride, pH, TDS, phosphorus, DO, fluoride and ORP) govern the microbial structures. They also play a leading role in the community’s diffusion of organic function and adaptive retort in the Deulajhari hot spring niche.
In the present investigation, we analyzed the unexplored diversity with their predicted function of the three springs (S1, S2 and S3) of the Deulajhari hot spring cluster through the 16S rRNA amplicon profiling. The physiochemical parameters analysis of collected sediment samples indicated the changes in BOD, COD, pH, phosphorus properties etc. Community structure among the bacterial population indicated dominance of Chloroflexi and Proteobacteria (at phylum level) and Dehalococcoidetes, Alphaproteobacteria, Deltaproteobacteria and Chloroflexi (at class level). The OTUs with no BLAST hits with known 16S rRNA in the database could be novel microbes in these habitats which needs to be further established. ABC transporter, two-component system and other transporter were expected to be the most prevalent microbial processes in sediment, while organotroph, autotroph and free-living organism were expected to occur highest in all the hot spring sediments sample. Furthermore, the co-occurrence of the Bacteriodetes and Chlorobi, along with the variant temperature, was also successfully established. Our results provide valuable insights into the putative microbial diversity and the function with their physiochemical correlation by sediment microbial communities in the hot springs sample. Further studies on in vitro culture followed by physiological analysis of these important microbes would be required to determine their precise functional roles within these communities.
Additional file: Additional Table S1-S3 and Figure S1.
ACKNOWLEDGMENTS
The authors would like to thank Siksha ‘O’ Anushandhan (Deemed to be University) for providing the basic infrastructure and facilities. Authors are also thankful to their colleagues for delivering their constant support to this work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
AS, ES, RKS, MG, SD Conceptualized the study. AS, MG, RKS applied methodology. AS, RKS performed Investigation. AS, MG performed formal analysis.AS, SD performed visualization. AS, ES wrote original draft. AS, ES, RKS, MG, SD reviewed and edited the manuscript. ES performed supervision, resources & project administration. All authors read and approved the final manuscript for publication.
FUNDING
This work was supported by the Siksha ‘O’ Anushandhan (Deemed to be University), India.
DATA AVAILABILITY
All the sequencing data and the corresponding metadata of all the three springs are deposited in NCBI’s SRA database and available under the accession SRX1459732 (S1), SRX1459733 (S2), SRX1459734 (S3).
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed by any of the authors.
- Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A. 1998;95(12):6578-6583.
Crossref - Prosser JI, Bohannan BJM, Curtis TP, et al. The role of ecological theory in microbial ecology. Nat Rev Microbiol. 2007;5(5):384-392.
Crossref - Xiong J, Liu Y, Lin X, et al. Geographic distance and pH drive bacterial distribution in alkaline lake sediments across Tibetan Plateau. Environ Microbiol. 2012;14(9):2457-2466.
Crossref - Chan CS, Chan KG, Tay YL, Chua YH, Goh KM. Diversity of thermophiles in a Malaysian hot spring determined using 16S rRNA and shotgun metagenome sequencing. Front Microbiol. 2015;6:177.
Crossref - Inskeep WP, Rusch DB, Jay ZJ, et al. Metagenomes from high-temperature chemotrophic sys-tems reveal geochemical controls on microbial community structure and function. PLoS One. 2010;5(3):e9773.
Crossref - Horner-Devine MC, Lage M, Hughes JB, Bohannan BJM. A taxa-area relationship for bacteria. Nature. 2004;432(7018):750-753.
Crossref - Ramette A, Tiedje JM. Multiscale responses of microbial life to spatial distance and environmental heterogeneity in a patchy ecosystem. Proc Natl Acad Sci U S A. 2007;104(8):2761-2766.
Crossref - Hollister EB, Engledow AS, Hammett AJM, Provin TL, Wilkinson HH, Gentry TJ. Shifts in microbi-al community structure along an ecological gradient of hypersaline soils and sediments. ISME J. 2010;4(6):829-838.
Crossref - Swingley WD, Meyer-Dombard DR, Shock EL, et al. Coordinating environmental genomics and geochemistry reveals metabolic transitions in a hot spring ecosystem. PLoS One. 2012;7(6):e38108.
Crossref - Dixit S, Behera DU, Gaur M, et al. Evaluation of Community Structures and their Physicochemi-cal Correlation with Five Hot Springs in India. Geomicrobiol J. 2021;38(8):655-671.
Crossref - Lewin A, Wentzel A, Valla S. Metagenomics of microbial life in extreme temperature environ-ments. Curr Opin Biotechnol. 2013;24(3):516-525.
Crossref - Zeldes BM, Keller MW, Loder AJ, Straub CT, Adams MWW, Kelly RM. Extremely thermophilic microorganisms as metabolic engineering platforms for production of fuels and industrial chemicals. Front Microbiol. 2015;6:1209.
Crossref - Jyoti V, Narayan KD, Das SK. Gulbenkiania indica sp. nov., isolated from a sulfur spring. Int J Syst Evol Microbiol. 2010;60(5):1052-1055.
Crossref - Narayan KD, Pandey SK, Das SK. Characterization of Comamonas thiooxidans sp. nov., and Comparison of Thiosulfate Oxidation with Comamonas testosteroni and Comamonas composti. Curr Microbiol. 2010;61(4):248-253.
Crossref - Najar IN, Sherpa MT, Das S, Das S, Thakur N. Diversity analysis and metagenomic insights into the antibiotic resistance and metal resistances among Himalayan hot spring bacteriobiome-insinuating inherent environmental baseline levels of antibiotic and metal tolerance. J Glob Antimicrob Resist. 2020;21:342-352.
Crossref - Badhai J, Ghosh TS, Das SK. Taxonomic and functional characteristics of microbial communities and their correlation with physicochemical properties of four geothermal springs in Odisha, India. Front Microbiol. 2015;6(OCT).
Crossref - Bartram AK, Lynch MDJ, Stearns JC, Moreno-Hagelsieb G, Neufeld JD. Generation of multimil-lion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end Illumina reads. Appl Environ Microbiol. 2011;77(11):3846-3852.
Crossref - Andrews S, Krueger F, Seconds-Pichon A, Biggins F, Wingett S. FastQC. A quality control tool for high throughput sequence data. Babraham Bioinformatics. Babraham Inst. 2015;1(1):1. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/%0Ahttp://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/
- Podar PT, Yang Z, Bjornsdottir SH, Podar M. Comparative Analysis of Microbial Diversity Across Temperature Gradients in Hot Springs From Yellowstone and Iceland. Front Microbiol. 2020;11:1625.
Crossref - Langille MGI, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31(9):814-821.
Crossref - Abhauer KP, Wemheuer B, Daniel R, Meinicke P. Tax4Fun: Predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics. 2015;31(17):2882-2884.
Crossref - Hammer O, Harper DAT, Ryan PD. Past: Paleontological statistics software package for educa-tion and data analysis. Palaeontol Electron. 2001.
- Choure K, Parsai S, Kotoky R, et al. Comparative Metagenomic Analysis of Two Alkaline Hot Springs of Madhya Pradesh, India and Deciphering the Extremophiles for Industrial Enzymes. Front Genet. 2021;12:643423.
Crossref - Liang Q, Zhang X, Lee KH, et al. METAGENassist: a comprehensive web server for compara-tive metagenomics. World J Microbiol Biotechnol. 2015;31(11):88-95.
Crossref - Gupta RS. The phylogeny and signature sequences characteristics of Fibrobacteres, Chlorobi, and Bacteroidetes. Crit Rev Microbiol. 2004;30(2):123-143.
Crossref - Gupta RS, Lorenzini E. Phylogeny and molecular signatures (conserved proteins and indels) that are specific for the Bacteroidetes and Chlorobi species. BMC Evol Biol. 2007;7.
Crossref - Hiras J, Wu YW, Eichorst SA, Simmons BA, Singer SW. Refining the phylum Chlorobi by resolv-ing the phylogeny and metabolic potential of the representative of a deeply branching, uncultivated lineage. ISME J. 2016;10(4):833-845.
Crossref - Jimenez DJ, Andreote FD, Chaves D, et al. Structural and Functional Insights from the Meta-genome of an Acidic Hot Spring Microbial Planktonic Community in the Colombian Andes. PLoS One. 2012;7(12).
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.