Investigations into seed microbiomes have unveiled intricate networks of microbial interactions that promote nutrient mobilization and uptake, significantly contributing to seedling vigor and resilience in diverse environmental conditions. Emerging evidence suggests that seed-borne microbes not only protect against pathogens but also influence plant developmental pathways, providing novel strategies for enhancing crop productivity via microbial inoculation. Moreover, these microbes might interfere with the endobiome of other plants, potentially suppressing competitor species, enhancing seedling growth, and increasing mortality. However, this effect is likely to be species-dependent, influenced by host plant species, microbial community composition, and environmental conditions. This implies that, in natural ecosystems, endobiome interference can be an important factor in plant-plant interactions. If validated through rigorous laboratory experiments and subsequent field trials, leveraging endobiome interference could offer a viable strategy to manage invasive or weedy plant species. This approach would be supported by integrated omics techniques, particularly genomics and metabolomics, to elucidate the genetic and metabolic contributions of seed microbiomes. Such insights could pave the way for precision microbiome engineering, ultimately optimizing agricultural yields while minimizing environmental impacts. This review article underscores the diverse and beneficial roles of seed microbiomes in plant biology, illustrating how ongoing research continues to deepen our understanding of their profound impacts on both plant health and the sustainability of agriculture.
Endobiome, Microbiome, Omics, Engineering, Interactions
One way to characterize the process of seed to seedling transition is as a major limitation on plant biological fitness and microbiome assembly.1 This transformation has significant ramifications in agriculture. Systematic approaches and strategies for preserving plant variety are required in naturally occurring ecological systems.2 Microbial communities living on the surface and within seeds collectively known as the seed microbiota can influence both the quality of seeds meant for consumption and the resilience of seeds grown in agricultural settings.3 Our understanding of seed microbiota lags significantly behind that of other plant-associated microbial communities, such as those in the rhizosphere (root-associated) and phyllosphere (leaf-associated).4 A structured approach has been developed to enhance our understanding of the dynamics and assembly mechanisms of plant-associated microbiota. The seed microbiome, serving as the initial microbial source, significantly influences the plant’s long-term microbiome, potentially affecting its health and resilience over time.5 Bacteria associated with seeds can be acquired horizontally from diverse environmental sources, including air, water, insects, or during seed processing. Alternatively, they can be acquired vertically, passed down from the mother plant across multiple generations.6 Although considerable research has examined pathogen presence on seeds, the roles of other commensal and mutualistic bacteria still require further exploration.7
The predominant seed microbiome consists mostly of unidentified species. Pioneering research established that seed microbiome may impact plant fitness by regulating seed germination, phenotypic traits of seedlings or by facilitating root symbiosis.8 Despite some research efforts, there has been limited investigation into the core seed microbiota that is specific to certain species of plants.9 The concept of core microbiota is similar to the term ‘core’ as it pertains to the portion of the microbiota that colonizes a substantial number of hosts.10 The characterization of the fundamental and adaptable microbiota of plant environment analysis may facilitate the identification of microbial species and their roles may have a significant impact on the fitness of the host.11 Such identification may be accomplished via extensive ongoing data synthesis initiatives, such as meta-analysis, that are yet unfinished for the seed microbiome.12 Further investigation of seed microbial diversity on a worldwide scale is even more evident.13 In the present moment, seeds emerge as a crucial means of advancing sustainable practices.14
In agriculture, seeds can serve a dual role: as a primary vehicle for advancing innovations that leverage seed-borne microbes as essential components, and as valuable biotechnological resources.15 Seeds can act as carriers for microbial bio-stimulants or biocontrol agents, enhancing the precision and efficiency of treatments applied to fields. This approach optimizes both the coverage and quantity of treatments, effectively reducing application costs and minimizing potential environmental impacts.16 Microbiota would expedite innovations and support future methodologies. Facilitating the proliferation of crucial seed microbes for phyllosphere and agricultural output.17 The seed microbiome displays remarkable diversity, encompassing a wide array of microbial taxa that can colonize seed samples. These taxa range from just a few to hundreds of amplicon sequence variants (ASVs), with a median of around one hundred ASVs (including both fungi and prokaryotes).18 Various studies have highlighted this diversity in the assembly of seed microbiota, even at the single-seed level; for instance, individual oak seeds may contain between 2 and 28 bacterial ASVs, while bean seeds may host 1 to 45 bacterial ASVs.4 Similar to other plant compartments, the microbial taxa within seeds predominantly belong to four bacterial phyla and three fungal groups.19 Results indicate that seeds generally show similar levels of microbial (bacterial and fungal) diversity, unlike other plant compartments, which typically display a marked dominance of bacterial diversity.20 This unique balance in seeds may stem from specific environmental conditions, such as extreme desiccation, limited nutrient availability, and high ecological pressures.
Research on various well-characterized plants reveals that the seed microbiota diversity are strongly shaped by the plants taxonomy. This influence is particularly evident in fungal communities, where significant differences appear in the prevalence of certain fungal orders between families; for instance, Dothideomycetes dominate in Brassicaceae, while Fabaceae exhibit a distinct fungal profile.9 Within bacterial communities, Poaceae seeds are notably rich in Actinobacteria. Future research should investigate the underlying mechanisms behind such differences in seed microbiota across plant species, including factors like the seed’s chemical composition, the potential for microbial transfer from mother plants, and the effects of pollination methods and reproductive structures.21 A striking finding of this meta-analysis is the identification of microbial (bacterial and fungal) taxa that are widely shared across various plant species and geographic locations. Among these, some bacterial and fungal genera were highly prevalent, including specific taxa with antibiotic resistance genes.22 Several of these seed-borne microbes positively impact plant health; for example, seed endophytes like Cladosporium may promote root symbiosis, while Sphingomonas can provide resistance to bacterial diseases. The study also showed that fungi (arbuscular mycorrhizal) are rarely passed to seedlings via seeds, as only a small fraction of glomeromycetes ASVs were detected in seeds, suggesting that seeds are an uncommon vector for these potentially nitrogen-fixing microbes.23 This research showed that fungi are seldom transmitted to seedlings via seeds. Only a small number of glomeromycetes ASVs were found.22 To advance research on the potential effects of these core taxa-such as their role in seed development, disease resistance, and germination-it is essential to determine whether these microbes are transmitted vertically (from parent plants) or acquired from the environment. This study identified core microbial taxa from samples containing approximately a thousand seeds, but such large sample sizes may overestimate microbial frequency. The core taxa may not be present in every individual seed, and distinct patterns of co-occurrence could indicate specific adaptations to environmental conditions or selective associations with certain plant hosts.
Despite this core microbiome, the majority of seed microbiota falls within a more flexible category, influenced by biotic and abiotic factors in the seed environment. These flexible species, while suited to the seed’s ecological niche, often have lower abundance. This flexible fraction holds importance for passing on microbial diversity suited to particular geographic regions and environmental conditions to the next plant generation.9
In addition to shedding light on seed microbiome composition and diversity, this meta-analysis highlights significant gaps in current knowledge. More research is needed to understand how factors like plant genetics and regional climates influence seed microbiomes. Additionally, seeds differ in physiology, size, structure, and chemistry, yet the effects of these seed-specific characteristics on microbiome composition remain unclear. The data from this meta-analysis are insufficient to fully answer these questions; therefore, further studies exploring these relationships are necessary.
A notable gap in seed microbiota research involves understanding microbial groups beyond bacteria and fungi, such as viruses, oomycetes, and other protists. Although some of these groups have been identified in seeds through cultivation and plant pathology studies, commonly used primers in amplicon sequencing often miss or only partially capture them (such as Archaea). To expand knowledge in this field, future studies should adopt multi-marker approaches on multiple seed samples, submitting their data to public repositories with comprehensive metadata to support the growth of a Seed Microbiota Database.9 Such research efforts could greatly deepen our understanding of seed microbiota and accelerate the use of seeds as a platform for agricultural innovation, especially in developing microbiome-based solutions for crop enhancement.
Molecular mechanisms for microbial inheritance and transmission
Members of the plant microbiota may spread vertically, directly from the parent plant, or horizontally, from the surrounding environment.8 Both horizontal and vertical transmission methods are predicted to play a part in defining the ultimate composition of the seed microbiome, even if their relative importance in plants is yet unknown. Any microbiological entity’s ability to spread, whether horizontally or vertically, is somewhat dependent on the seed transmission pathway that the microorganism uses. As of right now, three main routes of transmission have been identified: (i) the internal route, which passes through the mother plant’s xylem or nonvascular tissue; (ii) the floral route; and (iii) the external route, which passes through the plant’s threshing residue or fruit after the seed comes into contact with a microbial inoculum.24 Depending on the form of the seed, different transmission channels have different relative impact in the composition of the seed microbiota.
The term “floral route” describes how microorganisms are obtained from their surroundings. Nevertheless, the selection enforced by the target plant determines the transmission mechanism-horizontal or vertical. The microorganisms are then established in certain seed regions. It is possible to think of these places as unique microhabitats. Fungal symbionts that are transported vertically have been proven to directly benefit their host plant. For instance, the fungus endophyte Epichloe festucae is thought to contribute to increased resistance to drought in Festuca rubra.22 On the other hand, the variety of bacterial isolates that are vertically transmitted seems to vary greatly across seed samples and could not exist at all in some genotypes of Arabidopsis. However, there is enough data to suggest that bacterial entities are vertically transmitted in a variety of plant species, including populations of Arabidopsis, rice, wheat, maize, and switchgrass. A particularly reliable method for detecting vertical transmission is to follow research where GFP-labeled bacteria penetrate the seed from the parent plant.25 However, for non-model, uncultivable microorganisms, this approach requires genetic alteration of strains and the use of procedures that are not currently accessible.
Functional dynamics of the seed microbiome
Due to their mutualistic connections’ influence on a variety of physiological and biochemical processes, microbes and plants are essential for enhancing plant fitness. It has been shown that seed-associated microbes help host plants establish seedlings and foster their growth and development in settings with nutrient-deficient soils and difficult circumstances. They achieve this by enhancing nutrient acquisition and assimilation through various biological processes. Additionally, they support plant growth by sustaining competitive advantages over other species and providing antagonistic effects, such as biocontrol against plant pathogens. They also contribute to resistance against abiotic stressors like salinity, dehydration, and heavy metal toxicity. Furthermore, they regulate plant metabolism by modulating levels of phytohormones such as auxins, cytokinins, gibberellins, and reactive oxygen species (ROS). Lastly, they improve the interaction between plants, water, and soil, thereby promoting overall plant health and productivity. As a consequence, competitive interactions within a community arise from the seed microbiome’s effect on the host’s overall fitness and ability to outcompete other plants, which in turn defines patterns of community development. An analysis of the extant literature suggests that the primary emphasis is on the utilisation of bacteria associated with seeds to augment agricultural plant development, rather than the many ecological functions that these microbes may fulfill. To ascertain these functions’ contributions to plant species survival, colonization, and fitness under diverse environmental situations, further study is required.31
How important is the seed microbiome for maintaining plants in environment that change over time?
The adaptability of plant hosts to shifting environmental conditions may be facilitated by the diversity of seed microbiological diversity and its impact on the metabolism, behavior, physiology, and growth of the hosts. The advantages that the seed microbiome bestows upon a host plant in the initial phases of its life cycle are especially vital to the plant’s survival and establishment in adverse environments. For example, the quality of seeds is greatly influenced by the seed microbiome. Higher-quality seeds typically exhibit better vigor, a higher germination percentage, and are free of diseases. For example, when harvested before reaching physiological maturity, tall fescue seeds were found to have decreased rates of endophytic fungal infection. It was later discovered that these low infected seeds had poor germination and diminished seedling vigor.32 Furthermore, through controlling the hormone levels in the soil, advantageous microorganisms that live in seeds play a significant role in moderating seed dormancy and setting up a favorable environment for germination. Furthermore, it shields seeds from disease and predators, especially when they are not dormant and have no chemical or physical defenses.33 Numerous research investigations have documented the role of the seed microbiome in promoting seed germination, especially at higher elevations where germination conditions are frequently unfavorable.33
The fundamental processes of germination improvement are associated with the reduction of stressful environments through the regulation of gibberellic acid and cytokinin levels in seed hormones. The seed microbiome is responsible for root growth, drought tolerance during periods of extreme situation, and seed germination.34,35 The seed microbiome must be investigated, and functionally defined for a variety of conservation and restoration goals.
Seed microbiome: The hidden driver of plant invasion success
According to reports, the alien plants show mutualistic relationships with the microbes that helped the invasion process succeed in the particular region (Figure 1 and Table 1).31 The distribution of the seed microbiome, influence plant capability.36 This is due to the fact that seeds are a biological means of survival and eventual colonization of new environments. Because of their susceptibility to pathogen predation, many plants have substantial seed-stage mortality. It is possible to stop seed pathogenic microorganisms from destroying the seeds by controlling the seed microbiome.33
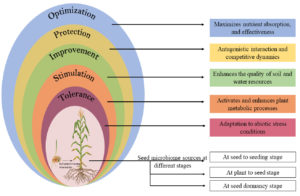
Figure 1. A conceptual depicting the relationships between seeds, microbiomes, and plants have various effects on plant growth, including increased nutrient availability, defense against various stresses, enhanced plant-soil-water relations, defense system stimulation, and tolerance to a variety of abiotic stresses
Table (1):
Table illustrating the differences between horizontal and vertical transmission
| Feature | Horizontal Transmission | Vertical Transmission | Ref. |
|---|---|---|---|
| Source of Microbiota | Surrounding environment (e.g., soil, fruits, threshing residues) | Parent plant (directly from mother plant) | 8 |
| Transmission Channels | External Route: Seed contamination from contact with environmental inoculum Floral Route: Microorganisms enter through the stigma of the flower |
Internal Route: Through xylem or nonvascular tissue | 6 |
| Fungal Vertical Inheritance: Symbionts transmitted vertically, benefiting host plant | 13 | ||
| Microbial Selection | Depends on environmental conditions and microbial availability | Strongly influenced by the parent plant’s microbial selection process | 26 |
| Examples | Acquisition of microorganisms from soil or threshing residues | Epichloe festucae in Festuca rubra, enhancing drought tolerance | 27 |
| Impact on Seed Microbiome | Plays a role in establishing the microbiome through environmental exposure | Contributes to the inheritance and continuity of the seed microbiome across generations | 8 |
| Variability in Microbial Diversity | High variability; depends on environmental factors and seed exposure | Varies significantly; some species exhibit high vertical transmission rates, while others may have low or no transmission | 28 |
| Establishment of Microorganisms in Seeds | Microorganisms settle in specific seed locations, including seed coat and storage tissues (endosperm, perisperm) | Microorganisms can establish in locations such as the embryonic axis, cotyledons, and other internal seed structures | 29 |
| Technical Challenges in Study | Overlapping microbiota from the environment makes tracking difficult | Difficult to quantify due to overlapping microbiota and the need for specialized tracking techniques | 30 |
| Research Examples | Rhizosphere soils’ microbes, oxygen, etc. | Vertical transmission has been studied in crops like rice, wheat, maize, switchgrass, and Arabidopsis | 6 |
Seed-associated microbial assemblages in Abutilon theophrasti, for instance, generate a variety of diffusible phenolic chemicals that may be crucial in maintaining the longevity of seed banks by shielding seeds from microbial antagonists.37 It is thought that Lolium rigidium invades Mediterranean-type environments mostly because of this dormancy, which is brought on by levels of cytokinin and abscisic acid-mediated by endogenous seed endophytes.36 Additionally, it has been discovered that Phragmites australis seed endophytes improve germination and reduce seedling mortality, which improves host establishment, growth, and invasiveness.31 Furthermore, microbes associated with seeds have the ability to either directly produce secondary metabolites or, by developing elicitors, promote the production of secondary metabolites. These secondary metabolites, including ethylene, jasmonate, and salicylic acid, subsequently strengthen host defense against insects or illnesses through the induced systemic resistance (ISR) and systemic acquired resistance (SAR) pathways.38 Additionally, several invasive plants show enhanced competitive capacity in introduced regions.39 These substances aid in the invading alien species ability to outcompete the diversity of native plants while shielding them from vertebrate and invertebrate herbivores. The allelochemical production is influenced by the corresponding seed microbiota of these plants. The ability of Phragmite seed bacterial colonies to produce secondary metabolites may be the mechanism underlying this. Consequently, a plant with robust symbiotic partners is more likely to outcompete rivals and take over the ecosystem.39
In invasion biology, the roles of mutualism and symbiosis are well established; however, the contribution of the seed microbiome to the invasiveness of alien species has received less attention.40 Since the native microbiome of alien species is disseminated by their seeds and is probably crucial to their establishment, colonization, and spread. However, to date, this knowledge gap remains unaddressed and requires further investigation.
Microbiome engineering and modulation
Microbiome engineering and modulation refer to the deliberate manipulation and optimization of the microbial communities associated with seeds to enhance plant health, growth, and stress resilience. Seeds naturally harbor a diverse array of microorganisms, including bacteria, fungi, and other microbes, collectively known as the seed microbiome (Table 2). These microbes can influence seed germination, nutrient uptake, and the plant’s ability to withstand biotic and abiotic stresses. Through microbiome engineering, beneficial microbial strains can be selectively introduced or promoted within the seed’s environment to improve agricultural outcomes. By harnessing the power of the seed microbiome, plants can be better equipped to thrive in challenging environments, ultimately leading to more sustainable and resilient agricultural systems. Prior research has focused mostly on seed-borne diseases, but increasing attention has lately been drawn to the impact of the entire seed microbiome on plant health.9 Due to their role in the generational transfer of both potentially advantageous and harmful microbes, seeds are particularly interesting as microbial carriers.41
Table (2):
Different crops and their corresponding microbiota (bacterial and fungal) isolates
| Source | Microbiota | Ref. |
|---|---|---|
| Brassica napus seed | Alphaproteobacteria (32.6%), Betaproteobacteria (21.5%), Gammaproteobacteria (11.9%), Bacilli (7.9%), Actinobacteria. Erysiphe, Cladosporium, Cryptococcus, Colletotrichum and Rhizoctonia | 54 |
| Cucurbita pepo seed | Proteobacteria (83%), Firmicutes (11%, 8% and 6%) and Actinobacteria (2%, 17% and 15%) | 43 |
| Zea mays ssp. mays L. rhizosphere and seed transmission |
Bacteroidetes and Proteobacteria (Burkholderia, Pantoea/Enterobacter, Stenotrophomonas/Pseudomonas, Massilia/Telluria, Sphingobium/Sphingomonas, Agrobacterium/Rhizobium, Pseudorhodoferax, Bradyrhizobium and Ochrobacterium) | 41 |
| Oryza sativa L. Hybrid seeds, Salvia miltiorrhiza seed | Capnodiales and Pleosporales for fungi, Pseudomonadales and Enterobacteriales for bacteria, Thaumarchaeota and Euryarchaeida for archaea | 55 |
| Bacterial genera: Enterobacter, Curtobacterium, Erwinia, Xanthomonas, Rhodanobacter, Roseomonas and Leuconostoc. Fungal genera: Davidiellaceae, Auerobasidium, Protomyces, Septoria and Microdochium. | 56 | |
| Zea mays seed | Luteibacter, Klebsiella, Tatumella, Acinetobacter, Serratia, Shigella and Stenotrophomonas, Enterobacter, Methylobacterium. | 56 |
| Phaseolus vulgaris seed | Actinobacteria, Bacteroidetes, Firmicutes, Alphaproteobacteria, Betaproteobacteria, Cyanobacteria. Fungal genera: Basidiomycota and Ascomycota | 56 |
| Oryza sativa seed, Triticum and Brassica seeds | Gammaproteobacteria and Deltaproteobacteria | 56 |
| Erwinia herbicola, Pseudomonas syringae, Telluria mixta, Xanthomonas axonopodis, Pantoea agglomerans, Pantoea stewartii, Pyrenophora tritici-repentis, Xanthomonas fuscans, Porphyrobacter sanguineus, Sphingobium japonicum, Sphingomonas wittichii, Massilia timonae, Pseudomonas fluorescens | 57 | |
| Urochloa ramosa L. seed Hordeum vulgare | Curtobacterium sp., B. amyloliquefaciens, and others | 58 |
| Enterobacteriaceae (Unclassified members) Curtobacterium, Paenibacillus, Pantoea, Sanguibacter and Saccharibacillus | 59 |
An emerging subject called “beneficial plant-microbe interactions”42 is dedicated to breeding plants for advantageous plant-microbe interactions. The influence of the seed microbiome, however, is yet entirely unknown. A study conducted an in-depth analysis of the microbiome using 16S rRNA gene sequencing to investigate bacterial and partial archaeal diversity across 14 Cucurbita pepo genotypes. This research marks an important step toward better understanding genotype-dependent plant-microbe interactions in Styrian oil pumpkins, with the ultimate goal of incorporating beneficial plant-microbe relationships into breeding programs. Although the relative abundance of phyla and overall diversity varied, the results showed that various bacterial communities were present in all microhabitats that were studied. The least diverse microbiomes were found in seeds, which were primarily composed of Enterobacteriaceae, which includes possible pathogens such as Pectobacterium and Erwinia. Significant numbers of potentially helpful bacteria, including Lactococcus, Paenibacillus, and Lysobacter, were also found. Notably, rhizospheres did not exhibit unique genotype-specific microbiomes, but seeds did. According to the study, the C. pepo genotype has a major impact on the composition of the seed microbiome, which should be taken into account when breeding new cultivars to better use advantageous native microbial communities.43
Many genetic technologies have been developed and used in recent decades to increase output and lessen the damage caused by pests and diseases.44 Because selected microbial species can be genetically engineered directly to introduce individual heterologous features into well-characterized microorganisms, this technique has the potential to be both quick and reasonably effective for agricultural applications. The breakthrough in RNA interference (RNAi) is one of the most recent biotechnological advancements in genetic tools, enabling researchers to change genes at the expression level. Double-stranded RNA (dsRNA) initiates RNAi by activating the ribonuclease protein Dicer, which produces small interfering RNA (siRNA). The complex that is created when these siRNA attach to particular proteins is added to the RNA-induced silencing complex (RISC). The catalytic component of RISC, a cleavage reaction, is set off when one strand of integrated siRNA binds to the complementary messenger RNA (mRNA) sequence.31 This process inhibits translation or gene expression. The usefulness of RNA interference for anti-pathogen applications in agriculture has been established. The ease of editing genes and genomes has increased with the introduction of the CRISPR and CRISPR/Cas9 technologies.45
Sequence-specific single guide RNA (sgRNA), engineered to interact with Cas9, functions as an RNA-guided DNA endonuclease within the cell. This enables precise genome editing by inserting or removing genetic material at a targeted site. Numerous studies have demonstrated the effectiveness of this technique, showing its potential applications in agriculture, such as modifying crop genes and increasing resistance to pathogens.46 With these molecular tools, we can extract molecular insights at both the genetic and transcriptional levels, understanding gene functions and expressions, and also manipulate genes to achieve desired traits, such as enhanced nutrient mobilization and improved defense against pathogens. These gene-editing technologies also open the door to the use of genetically modified microorganisms in agriculture, offering a potential solution to the rapid decline of introduced microbial populations and ultimately benefiting crop health (Figure 2). Transgenic or genetically modified bacteria can increase predictability and dependability, although their use in agricultural systems is still debatable. The limited field survival of individual genotypes (clones) of microbes and the potential for gene transfer between strains are drawbacks. These factors raise serious questions about the effectiveness, survivability, and environmental risks of any newly introduced genetically modified organism.47 Furthermore, constant observation of the fate and behaviors of genetically modified organisms is necessary as a crucial part of their introduction. This is a significant drawback because the monitoring techniques are costly, necessitate highly skilled workers, and are subject to biosafety regulations. The widespread release of genetically modified organisms into the environment is restricted by this and legal bans in many nations; to get beyond these obstacles, policy decisions based on sound science are required.
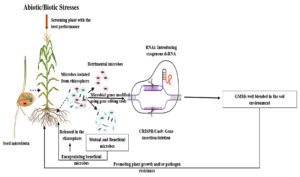
Figure 2. Strategies to enhance microbial inoculants: from isolating beneficial rhizosphere microbes and screening through plant phenotyping, to genetic modification (e.g., CRISPR, RNAi) and encapsulated delivery for improved plant growth and stress resistance
Utilizing metabolic engineering, which involves the role of chemicals naturally produced by plants and microorganisms to allure and sustain essential microbiota or other beneficial microbes, is a biochemical strategy for manipulating indigenous microbiomes. For instance, it is well established that root exudates can modify microbiome composition in the plant rhizosphere by attracting beneficial microbes.48 The production of exudates is also known to be enhanced by environmental stressors. According to a recent study, volatile organic compounds released from plants can draw pathogen-antagonistic soil bacteria from a wide area. This finding raises the possibility of using specific volatile organic substances to engineer the soil microbiome in order to improve plant defense.49 To enhance a plant’s ability to attract and sustain beneficial microbes and their functions within the rhizosphere, researchers can identify and either isolate or synthesize specific root metabolites that are linked to the proliferation of certain microbial components. Using quorum sensing-like microbial systems is another way to encourage advantageous microbes in the plant microbiome. It is a population-density-dependent regulation of microbial gene expression, which facilitates effective communication through the release of signaling molecules. This process can shape collective microbial behaviors, such as enhancing community survival in response to population stress, improving defense mechanisms against competitors, and aiding in nutrient acquisition or niche adaptation.50 Additionally, during stress, microbial signals like heat shock proteins and reactive oxygen species emerging from the rhizosphere can be detected and positively utilized by plants. These signaling molecules may be applied to improve microbial nutrient cycling (e.g., fixation, mineralization, mobilization), selectively enhance beneficial bacterial populations, or trigger microbe-mediated responses against pathogens in the context of in situ microbiome manipulation. Hence, these signal molecules offer promising tools for managing plant-microbe interactions to optimize resource use and plant defense. However, our understanding of the identities, roles, and molecular mechanisms of these microbial or plant signals remains limited, making it challenging to fully harness their potential. Improved sensitivity in spectroscopy-based detection technologies, combined with the integration of metabolomics, is necessary to better characterize the diversity and specificity of these interactions.51
In plants, the host’s genetic makeup plays a crucial role in shaping the composition, abundance, and function of the microbiome. Given the strong connection between a plant’s genome and its associated microbiome, it may be feasible to develop plants with enhanced microbiomes by leveraging the interactions between the host genome and its microbiome. Tools such as quantitative trait loci (QTL) mapping allow for the identification of genes or genetic regions responsible for key biological traits (phenotypes) in organisms of interest. Identifying genetic loci that impact particular microbial taxa or pathways, as well as connecting environmental factors to changes in microbial composition, have been accomplished through the widespread use of this strategy in mouse and human microbiome research.52 This shows that there is a lot of promise for this technology to produce better plant types through conventional plant breeding methods or genetic engineering. Upon identification of QTLs, or the genetic characteristics of crops that mediate the interactions between the crop and beneficial microbiomes, new and better crop varieties that have the ability to draw in and utilize beneficial indigenous microbiota can be produced. Since the development of CRISPR/Cas9 technology, these methods have the potential to significantly enhance or stimulate plants capacity to attract beneficial bacteria in a preferred manner53 (Figure 2). A comparable method might be employed to control the hub of microbiota early colonization, which could ultimately mold the core and entire plant microbiomes in predictable ways. Furthermore, in order to induce a positive response from plant microbiomes, genes related to biochemical processes or microbe-microbe signaling may be inserted into crops.
Harnessing endobiome interference: A new strategy for managing invasive weeds and phytopathogens
The microorganisms found in the endobiome of a certain plant are acclimated to its internal environment, and the conditions within plant endospheres might vary between different plant species. The extraction of endobiome microorganisms from their natural hosts and their transference to unadapted seedling hosts may lead to: 1) internal colonisation; 2) disruption of activities of other endobiome microbes; 3) interruption in plant development; or 4) an enhancement in seedling mortality. “Endobiome interference” may lead to impaired seedling development due to colonisation by non-adapted microorganisms.60 Such relationships are unique.61 Certain endophytic bacteria can suppress seedling growth and even kill them when they are introduced into plants that are not their natural hosts.60 This is an alternative method of managing weeds and invasive plants without using chemical herbicides or uprooting plants manually. “Endobiome interference” refers to the suppression of plant growth and perturbation of endophyte-host symbiotic interactions caused by the introduction of non-adapted microbial endophytes into plant cells and tissues.60,61 Additionally, Micrococcus luteus (bacterial endophyte) after being isolated from tomato seeds and seedlings, was introduced into seedlings of various species. There, it invaded the root cells of several plant species, such as Poa annua, Phragmites australis, Fallopia japonica, Taraxacum officinale, and Rumex crispus, thereby decreasing the population of native endophytic bacteria and slowing down the growth of the seedlings.
In natural plant communities, endobiome interference may be a frequent occurrence and a means by which plants inhibit the growth of rival plants. Similarly, endobiome interference may be able to decrease the invasive nature of weedy and invasive plant species when applied as a management strategy.62 In order to suppress the non-native Phragmites australis (common reed), an agenda was recently designed for evaluating the use of microbial community in this. This technique could also be used to target other invasive and weedy plant species. The symbiotic associations between P. australis and its endophytic microbes provide it with a competitive advantage over native species. However, the specific locations and functions of individual microbes within the plant remain uncertain. Plants can become infected with a variety of microorganisms, including bacteria, fungus, viruses, and nematodes, which can cause biotic stress. The first line of defence in plants is made up of physical barriers such trichomes, wax, and cuticle.63 The primary way by which endophytic bacteria regulate plant diseases are by increasing nutrient absorption and availability, stress tolerance, and disease resistance.64 Bacillus, Enterobacter, Serratia, Pseudomonas, Actinobacteria and Paenibacillus are the most often reported bacterial genera that exhibit biocontrol activity.65 These activities can be broadly grouped into two groups viz., direct and indirect biocontrol mechanisms. Direct biocontrol entails the synthesis of substances that hinder growth, including quorum-sensing inhibitors, siderophores, hydrogen cyanide, and enzymes that break down cell walls.
A number of studies have focused on identifying endophytic bacteria that can produce siderophores, promote plant growth, and help control phytopathogens.66-69 The production of hydrogen cyanide by these bacteria plays a crucial role in inhibiting phytopathogen growth through a mechanism known as biogenic cyanogenesis. Cyanide acts as a metabolic inhibitor, helping plants combat soil-borne diseases by disrupting the metalloenzymes of pathogens, including cytochrome C oxidase.70-74 A strain of Bacillus subtilis that produces hydrogen cyanide and other compounds, effectively suppressing the phytopathogen Fusarium oxysporum.75 Additionally, hydrogen cyanide-producing endophytes isolated from Glycine max demonstrated antagonistic effects against several phytopathogens, such as Rhizoctonia solani, Sclerotium rolfsii, Alternaria alternata, Colletotrichum truncatum, Macrophomina phaseolina, and F. oxysporum.67-71 Quorum sensing regulates various processes, including pathogenicity and biofilm formation. Endophytic bacteria can disrupt quorum sensing that limit biofilm development and bacterial infections.76 The several endophytic bacteria produce lactonases and acylases that inactivate or degrade N-acyl homoserine lactones, the key signaling molecules involved in quorum sensing, thereby promoting quorum quenching.77
Moreover, endophytic bacteria can trigger ethylene and jasmonic acid-mediated induced systemic resistance, leading to an immune response that helps defend plants against future phytopathogen attacks.78 This results in the production of phytoalexins, pathogenesis-related proteins, and defense enzymes such as phenylalanine ammonia lyase and polyphenol oxidase (PPO), as well as the formation of physical barriers like cuticles and modifications to the cell wall.79 The endophytic bacterial strain REB01 can lower malondialdehyde (MDA) content, enhance PPO and peroxidase (POD) enzyme activity, and confer disease resistance against rice sheath blight caused by R. solani.80 Some endophytic bacteria engage in rhizophagy, a biphasic cycle that alternates between an intracellular phase in roots (where nutrients are extracted by plants) and a free-living soil phase (where bacteria acquire nutrients).81 The term “endobiome interference” describes a situation in which certain endophytes impede rhizophagy, competing for nutrients from native microorganisms after colonization. Through endobiome interference, stress levels in the host plant can be heightened, inhibiting the proliferation of its endobiome and serving as an environmentally friendly biocontrol strategy.82,83
In commercial crop plants, microbial endophytes and soil microorganisms could be used to directly to improve plant health and productivity. Endophytic microbes have become a growing trend as biocontrol agents in recent years because of their superior acclimatization potential and high colonization efficiency. Endophytes prevent insect damage, diseases, and weedy plant competition. In natural plant communities, endobiome interference is thought to play a role in plant-plant interactions. One novel way to maximise the benefits of endophytes in plant disease management is to screen for suitable bacteria that may colonise inside host tissues and pass on to the next generation of plants through seeds. Analysing the relationship between screened endophytes and the plant microbiota in detail will also help us understand the benefits and drawbacks of applying endophytes to plant microbiomes. Endophytes may make it possible to grow crops without the need for pesticides, fungicides, fertilisers, or herbicides. Reducing the growth of competing weeds and increasing microbial diversity in soils and plants by adding microbe supplements that lower pathogen virulence can lead to reduced environmental contamination and more ecologically friendly agriculture practices. Endophytes have many advantages in the control of plant diseases, despite this, their applications in the field are restricted. Initiatives should also be made to teach end users how to use this technology for effective and environmentally friendly management of weeds and plant diseases. Additional study in this area may lead to the development of endobiome interference, a method for maximising microbial activities to improve crop protection and productivity.
ACKNOWLEDGMENTS
All authors are thankful to their parent institutions.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
Not applicable.
ETHICS STATEMENT
Not applicable.
- Shade A, Jacques MA, Barret M. Ecological patterns of seed microbiome diversity, transmission, and assembly. Curr Opin Microbiol. 2017;37:15-22.
Crossref - Paiola A, Assandri G, Brambilla M, Zottini M, Pedrini P, Nascimbene J. Exploring the potential of vineyards for biodiversity conservation and delivery of biodiversity-mediated ecosystem services: A global-scale systematic review. Sci Total Environ. 2020;706:135839.
Crossref - Sudhakar N, Karthikeyan G, RajhaViknesh M, Saranya AS, Shurya R. Technological Advances in Agronomic Practices of Seed Processing, Storage, and Pest Management: An Update. In: Tiwari AK, ed. Advances in Seed Production and Management. Springer Singapore. 2020:359-398.
Crossref - Abdelfattah A, Wisniewski M, Schena L, Tack AJM. Experimental evidence of microbial inheritance in plants and transmission routes from seed to phyllosphere and root. Environ Microbiol. 2021;23(4):2199-2214.
Crossref - Berg G, Kusstatscher P, Abdelfattah A, Cernava T, Smalla K. Microbiome Modulation-Toward a Better Understanding of Plant Microbiome Response to Microbial Inoculants. Front Microbiol. 2021;12:650610.
Crossref - L’Hoir M, Nasslahsen B, Ferhout H, et al. Endophytic Seed Bacteria: A Relevant Pool of Microorganisms with the Ability to Promote Plant Growth. In: Arora NK, Bouizgarne B, eds. Microbial BioTechnology for Sustainable Agriculture Volume 1. Vol 33. Microorganisms for Sustainability. Springer Nature Singapore. 2022:105-141.
Crossref - Mannaa M, Seo YS. Plants under the Attack of Allies: Moving towards the Plant Pathobiome Paradigm. Plants. 2021;10(1):125.
Crossref - Abdelfattah A, Tack AJM, Lobato C, Wassermann B, Berg G. From seed to seed: the role of microbial inheritance in the assembly of the plant microbiome. Trends Microbiol. 2023;31(4):346-355.
Crossref - Simonin M, Briand M, Chesneau G, et al. Seed microbiota revealed by a large scale meta analysis including 50 plant species. New Phytol. 2022;234(4):1448-1463.
Crossref - Neu AT, Allen EE, Roy K. Defining and quantifying the core microbiome: Challenges and prospects. Proc Natl Acad Sci. 2021;118(51):e2104429118.
Crossref - Trivedi P, Batista BD, Bazany KE, Singh BK. Plant-microbiome interactions under a changing world: responses, consequences and perspectives. New Phytol. 2022;234(6):1951-1959.
Crossref - Bauermeister A, Mannochio-Russo H, Costa-Lotufo LV, Jarmusch AK, Dorrestein PC. Mass spectrometry-based metabolomics in microbiome investigations. Nat Rev Microbiol. 2022;20(3):143-160.
Crossref - Samreen T, Naveed M, Nazir MZ, et al. Seed associated bacterial and fungal endophytes: Diversity, life cycle, transmission, and application potential. Appl Soil Ecol. 2021;168:104191.
Crossref - Peschard K, Randeria S. ‘Keeping seeds in our hands’: the rise of seed activism. J Peasant Stud. 2020;47(4):613-647.
Crossref - Fu Y, Ma L, Li J, et al. Factors Influencing Seed Dormancy and Germination and Advances in Seed Priming Technology. Plants. 2024;13(10):1319.
Crossref - Jindo K, Goron TL, Pizarro-Tobias P, et al. Application of biostimulant products and biological control agents in sustainable viticulture: A review. Front Plant Sci. 2022;13:932311.
Crossref - Parasuraman P, Pattnaik S, Busi S. Phyllosphere Microbiome: Functional Importance in Sustainable Agriculture. New and Future Developments in Microbial Biotechnology and Bioengineering. 2019:135-148.
Crossref - Van Dingenen J, Garcia Mendez S, Beirinckx S, et al. Flemish soils contain rhizobia partners for Northwestern Europe adapted soybean cultivars. Environ Microbiol. 2022;24(8):3334-3354.
Crossref - Bajpai A, Rawat S, Johri BN. Fungal Diversity: Global Perspective and Ecosystem Dynamics. In: Satyanarayana T, Johri BN, Das SK, eds. Microbial Diversity in Ecosystem Sustainability and Biotechnological Applications. Springer Singapore. 2019:83-113.
Crossref - Hacquard S, Garrido-Oter R, Gonzalez A, et al. Microbiota and Host Nutrition across Plant and Animal Kingdoms. Cell Host Microbe. 2015;17(5):603-616.
Crossref - Patel JK, Archana G. Diverse culturable diazotrophic endophytic bacteria from Poaceae plants show cross-colonization and plant growth promotion in wheat. Plant Soil. 2017;417(1-2):99-116.
Crossref - Wang D, Huang Z, Billen J, Zhang G, He H, Wei C. Structural diversity of symbionts and related cellular mechanisms underlying vertical symbiont transmission in cicadas. Environ Microbiol. 2021;23(11):6603-6621.
Crossref - Luo DL, Huang SY, Ma CY, et al. Seed-borne bacterial synthetic community resists seed pathogenic fungi and promotes plant growth. J Appl Microbiol. 2024;135(4):lxae073.
Crossref - Jonkers W, Gundel PE, Verma SK, White JF, eds. Seed Microbiome Research. Frontiers Media SA. 2022.
Crossref - Jha DK. Problems and prospects of utilization of bacterial endophytes for the management of plant diseases. Indian Phytopathol. 2023;76(1):3-20.
Crossref - Nerva L, Sandrini M, Moffa L, Velasco R, Balestrini R, Chitarra W. Breeding toward improved ecological plant-microbiome interactions. Trends Plant Sci. 2022;27(11):1134-1143.
Crossref - Decunta FA, Perez LI, Malinowski DP, Molina-Montenegro MA, Gundel PE. A Systematic Review on the Effects of Epichloe Fungal Endophytes on Drought Tolerance in Cool-Season Grasses. Front Plant Sci. 2021;12:644731.
Crossref - Walsh CM, Becker-Uncapher I, Carlson M, Fierer N. Variable influences of soil and seed-associated bacterial communities on the assembly of seedling microbiomes. ISME J. 2021;15(9):2748-2762.
Crossref - Mokrani K, Tarchoun N. Development, dormancy and germination of seeds metabolism, hormonal control and genetic control. Int J Agric Environ Biores. 2022;07(01):27-75.
Crossref - Shi H, Grodner B, De Vlaminck I. Recent advances in tools to map the microbiome. Curr Opin Biomed Eng. 2021;19:100289.
Crossref - Shearin ZRC, Filipek M, Desai R, Bickford WA, Kowalski KP, Clay K. Fungal endophytes from seeds of invasive, non-native Phragmites australis and their potential role in germination and seedling growth. Plant Soil. 2018;422(1-2):183-194.
Crossref - Hill NS, Bouton JH, Hiatt EE, Kittle B. Seed Maturity, Germination, and Endophyte Relationships in Tall Fescue. Crop Sci. 2005;45(3):859-863.
Crossref - Dalling JW, Davis AS, Schutte BJ, Elizabeth Arnold A. Seed survival in soil: interacting effects of predation, dormancy and the soil microbial community. J Ecol. 2011;99(1):89-95.
Crossref - Hanna AL, Hamouda HM, Goda HA, et al. Biosynthesis and Characterization of Silver Nanoparticles Produced by Phormidium ambiguum and Desertifilum tharense Cyanobacteria. Milea D, ed. Bioinorg Chem Appl. 2022;2022(1):1-14.
Crossref - Reuver M, Maher J, Wilson AM. Ocean Restoration and the Strategic Plan of the Marine Microbiome. In: Stal LJ, Cretoiu MS, eds. The Marine Microbiome. Vol 3. The Microbiomes of Humans, Animals, Plants, and the Environment. Springer International Publishing. 2022:731-766.
Crossref - Goggin DE, Emery RJN, Kurepin LV, Powles SB. A potential role for endogenous microflora in dormancy release, cytokinin metabolism and the response to fluridone in Lolium rigidum seeds. Ann Bot. 2015;115(2):293-301.
Crossref - Chee-Sanford JC, Williams MM, Davis AS, Sims GK. Do microorganisms influence seed-bank dynamics? Weed Sci. 2006;54(3):575-587.
Crossref - War AF, Bashir I, Reshi ZA, Kardol P, Rashid I. Insights into the seed microbiome and its ecological significance in plant life. Microbiol Res. 2023;269:127318.
Crossref - White JF, Kingsley KI, Kowalski KP, Elmore MT, Verma SK. Disease protection and allelopathic interactions of seed-transmitted endophytic Pseudomonads of invasive reed grass (Phragmites australis). Plant Soil. 2018;422(1-2):195-208.
Crossref - Morin L, Paini DR, Randall RP. Can Global Weed Assemblages Be Used to Predict Future Weeds? Moora M, ed. PLoS ONE. 2013;8(2):e55547.
Crossref - Johnston-Monje D, Lundberg DS, Lazarovits G, Reis VM, Raizada MN. Bacterial populations in juvenile maize rhizospheres originate from both seed and soil. Plant Soil. 2016;405(1-2):337-355.
Crossref - Bakker MG, Manter DK, Sheflin AM, Weir TL, Vivanco JM. Harnessing the rhizosphere microbiome through plant breeding and agricultural management. Plant Soil. 2012;360(1-2):1-13.
Crossref - Adam E, Bernhart M, Muller H, Winkler J, Berg G. The Cucurbita pepo seed microbiome: genotype-specific composition and implications for breeding. Plant Soil. 2018;422(1-2):35-49.
Crossref - Godfray HCJ, Beddington JR, Crute IR, et al. Food Security: The Challenge of Feeding 9 Billion People. Science. 2010;327(5967):812-818.
Crossref - Cong L, Ran FA, Cox D, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339(6121):819-823.
Crossref - Ali Z, Abul-faraj A, Li L, et al. Efficient Virus-Mediated Genome Editing in Plants Using the CRISPR/Cas9 System. Mol Plant. 2015;8(8):1288-1291.
Crossref - Wang S, O’Brien TR, Pava-Ripoll M, St. Leger RJ. Local adaptation of an introduced transgenic insect fungal pathogen due to new beneficial mutations. Proc Natl Acad Sci. 2011;108(51):20449-20454.
Crossref - Stringlis IA, Yu K, Feussner K, et al. MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc Natl Acad Sci. 2018;115(22):e5213-e5222.
Crossref - Schulz-Bohm K, Gerards S, Hundscheid M, Melenhorst J, De Boer W, Garbeva P. Calling from distance: attraction of soil bacteria by plant root volatiles. ISME J. 2018;12(5):1252-1262.
Crossref - Papenfort K, Bassler BL. Quorum sensing signal-response systems in Gram-negative bacteria. Nat Rev Microbiol. 2016;14(9):576-588.
Crossref - Qiu Z, Egidi E, Liu H, Kaur S, Singh BK. New frontiers in agriculture productivity: Optimised microbial inoculants and in situ microbiome engineering. Biotechnol Adv. 2019;37(6):107371.
Crossref - Kurilshikov A, Wijmenga C, Fu J, Zhernakova A. Host Genetics and Gut Microbiome: Challenges and Perspectives. Trends Immunol. 2017;38(9):633-647.
Crossref - Schaeffer SM, Nakata PA. CRISPR/Cas9-mediated genome editing and gene replacement in plants: Transitioning from lab to field. Plant Sci. 2015;240:130-142.
Crossref - Rybakova D, Mancinelli R, Wikstrom M, et al. The structure of the Brassica napus seed microbiome is cultivar-dependent and affects the interactions of symbionts and pathogens. Microbiome. 2017;5(1):104.
Crossref - Liu Y, Xu P, Yang F, et al. Composition and diversity of endophytic bacterial community in seeds of super hybrid rice ‘Shenliangyou 5814’ (Oryza sativa L.) and its parental lines. Plant Growth Regul. 2019;87(2):257-266.
Crossref - Chen H, Wu H, Yan B, et al. Core Microbiome of Medicinal Plant Salvia miltiorrhiza Seed: A Rich Reservoir of Beneficial Microbes for Secondary Metabolism? Int J Mol Sci. 2018;19(3):672.
Crossref - Links MG, Demeke T, Grafenhan T, Hill JE, Hemmingsen SM, Dumonceaux TJ. Simultaneous profiling of seed associated bacteria and fungi reveals antagonistic interactions between microorganisms within a shared epiphytic microbiome on Triticum and Brassica seeds. New Phytol. 2014;202(2):542-553.
Crossref - Verma SK, White JF. Indigenous endophytic seed bacteria promote seedling development and defend against fungal disease in browntop millet (Urochloa ramosa L.). J Appl Microbiol. 2018;124(3):764-778.
Crossref - Bziuk N, Maccario L, Straube B, et al. The treasure inside barley seeds: microbial diversity and plant beneficial bacteria. Environ Microbiome. 2021;16(1):20.
Crossref - White JF, Kingsley KL, Zhang Q, et al. Review: Endophytic microbes and their potential applications in crop management. Pest Manag Sci. 2019;75(10):2558-2565.
Crossref - Bell TH, Hockett KL, Alcala-Briseno RI, et al. Manipulating Wild and Tamed Phytobiomes: Challenges and Opportunities. Phytobiomes J. 2019;3(1):3-21.
Crossref - Kowalski KP, Bacon C, Bickford W, et al. Advancing the science of microbial symbiosis to support invasive species management: a case study on Phragmites in the Great Lakes. Front Microbiol. 2015;6:956.
Crossref - Iqbal Z, Iqbal MS, Hashem A, Abd Allah EF, Ansari MI. Plant Defense Responses to Biotic Stress and Its Interplay With Fluctuating Dark/Light Conditions. Front Plant Sci. 2021;12:631810.
Crossref - Hamilton CE, Gundel PE, Helander M, Saikkonen K. Endophytic mediation of reactive oxygen species and antioxidant activity in plants: a review. Fungal Divers. 2012;54(1):1-10.
Crossref - Ek-Ramos MJ, Gomez-Flores R, Orozco-Flores AA, Rodriguez-Padilla C, Gonzalez-Ochoa G, Tamez-Guerra P. Bioactive Products From Plant-Endophytic Gram-Positive Bacteria. Front Microbiol. 2019;10:463.
Crossref - Lacava PT, Silva-Stenico ME, Araujo WL, et al. Detection of siderophores in endophytic bacteria Methylobacterium spp. associated with Xylella fastidiosa subsp. pauca. Pesqui Agropecuaria Bras. 2008;43(4):521-528.
Crossref - Pandey PK, Samanta R, Yadav RNS. Plant Beneficial Endophytic Bacteria from the Ethnomedicinal Mussaenda roxburghii (Akshap) of Eastern Himalayan Province, India. Adv Biol. 2015;2015(1):1-8.
Crossref - Passari AK, Mishra VK, Leo VV, Gupta VK, Singh BP. Phytohormone production endowed with antagonistic potential and plant growth promoting abilities of culturable endophytic bacteria isolated from Clerodendrum colebrookianum Walp. Microbiol Res. 2016;193:57-73.
Crossref - Walitang DI, Kim K, Madhaiyan M, Kim YK, Kang Y, Sa T. Characterizing endophytic competence and plant growth promotion of bacterial endophytes inhabiting the seed endosphere of Rice. BMC Microbiol. 2017;17(1):209.
Crossref - Voisard C, Keel C, Haas D, Defago G. Cyanide production by Pseudomonas fluorescens helps suppress black root rot of tobacco under gnotobiotic conditions. EMBO J. 1989;8(2):351-358.
Crossref - Blumer C, Haas D. Mechanism, regulation, and ecological role of bacterial cyanide biosynthesis. Arch Microbiol. 2000;173(3):170-177.
Crossref - Dheeman S, Maheshwari DK, Dubey RC, Kumar S, Baliyan N, Dhiman S. Harnessing Beneficial Bacillus in Productivity Improvement of Food Security Crops of Himalayan Agro-Climatic Zones. In: Maheshwari DK, Dheeman S, eds. Field Crops: Sustainable Management by PGPR. Vol 23. Sustainable Development and Biodiversity. Springer International Publishing. 2019:105-143.
Crossref - Maheshwari R, Bhutani N, Bhardwaj A, Suneja P. Functional diversity of cultivable endophytes from Cicer arietinum and Pisum sativum: Bioprospecting their plant growth potential. Biocatal Agric Biotechnol. 2019;20:101229.
Crossref - Swarnalakshmi K, Rajkhowa S, Senthilkumar M, Dhar DW. Influence of Endophytic Bacteria on Growth Promotion and Protection against Diseases in Associated Plants. Singh DP, Prabha R, eds. Microbial Interventions in Agriculture and Environment. Springer Singapore. 2019:263-287.
Crossref - Dubey RC, Khare S, Kumar P, Maheshwari DK. Combined effect of chemical fertilisers and rhizosphere-competent Bacillus subtilis BSK17 on yield of Cicer arietinum. Arch Phytopathol Plant Prot. 2014;47(19):2305-2318.
Crossref - Zhou L, Zhang Y, Ge Y, Zhu X, Pan J. Regulatory Mechanisms and Promising Applications of Quorum Sensing-Inhibiting Agents in Control of Bacterial Biofilm Formation. Front Microbiol. 2020;11:589640.
Crossref - Rashid S, Charles TC, Glick BR. Isolation and characterization of new plant growth-promoting bacterial endophytes. Appl Soil Ecol. 2012;61:217-224.
Crossref - Miliute I, Buzaite O, Baniulis D, Stanys V. Bacterial endophytes in agricultural crops and their role in stress tolerance: a review. Zemdirb-Agric. 2015;102(4):465-478.
Crossref - Wiesel L, Newton AC, Elliott I, et al. Molecular effects of resistance elicitors from biological origin and their potential for crop protection. Front Plant Sci. 2014;5:655.
Crossref - Mao J, Gong M, Guan Q. Induced disease resistance of endophytic bacteria REB01 to bacterial blight of rice. 2019:020017.
Crossref - Dudeja SS, Suneja Madan P, Paul M, Maheswari R, Kothe E. Bacterial endophytes: Molecular interactions with their hosts. J Basic Microbiol. 2021;61(6):475-505.
Crossref - Verma H, Kumar D, Kumar V, et al. The Potential Application of Endophytes in Management of Stress from Drought and Salinity in Crop Plants. Microorganisms. 2021;9(8):1729.
Crossref - White JF, Kingsley KL, Butterworth S, et al. Seed-Vectored Microbes: Their Roles in Improving Seedling Fitness and Competitor Plant Suppression. In: Verma, S., White, Jr, J. (eds) Seed Endophytes. Springer, Cham. 2019:3-20.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.