ISSN: 0973-7510
E-ISSN: 2581-690X
Oily sludge generated by petroleum industries results in serious hazards to the environment due to its toxic effects. Microorganisms can survive in the harsh environment utilizing the pollutants as their sole source of carbon and energy. The present study was carried out to isolate and characterize the bacterial population present in the refinery wastes collected from Bongaigaon Refinery, Assam, India.The diversity of bacteria was significantly correlated (p< 0.05) with the physicochemical characteristics and level of hydrocarbon content of the studied samples. A total of 36 bacterial strains were isolated from the oily sludge sample. Two potent hydrocarbonoclastic bacteria were screened out based on their ability of degradation of petroleum hydrocarbon. The potent bacterial isolates also produced biosurfactants. The potent strains were identified as Pseudomonas sp. and Bacillus sp. Gravimetric estimation of hydrocarbon degradation and FTIR analysis revealed that these potent bacterial strains could remove different constituents of petroleum hydrocarbon. The indigenous bacterial isolates either alone or in consortium may be applied for degrading different fractions of hydrocarbon contaminants of the oily sludge to remediate the sludge polluted soil environment.
Oily refinery sludge, bacterial diversity, biosurfactant, hydrocarbon degradation, FTIR
Rapid industrialization and the enhanced dependency on petroleum products have led to an unavoidable increase in the daily generation of oily petroleum sludge. Petroleum and oil refineries in India produce 28,220 tons of hazardous sludge per annum1. Oil spillage occurring during the process of production, refining, storage, and transportation releases a huge amount of harmful chemicals and the remaining of the crude oil pollutes the exposed areas2. In most of the refineries in India, the oily sludge generated from the wastewater treatment plant is stored in lagoons and mixed with oily sludge from the crude oil tank and is finally disposed of in low lying areas1. The bio sludge and chemical sludge are dried in sludge drying beds in the presence of sand and gravels, and are often applied as manure in agriculture land. These dried sludge and the lagoons used for storage are inadequate for proper disposal and consequently possess significant risk and adverse impacts on the environment. The amount of sludge generated by a refinery depends on the nature of crude oil and the process of effluent and sludge treatment. Bongaigaon refinery (BRPL), Assam, has 2.350 MMTPA of refining capacity and produces 241.8 MTA of hazardous wastes3. The oily sludge is composed of total petroleum hydrocarbon (TPH) which is a complex mixture of aromatic, aliphatics, asphaltenes, Nitrogen, Sulfur and Oxygen-containing compounds besides harmful pollutants and traces of heavy metals which are treated as hazardous wastes according to the environmental guidelines of the Ministry of Environment & Forests, Government of India4. The crude oil tank bottom sludge mainly consists of about 45% of recoverable oil which may pose serious problems if remains exposed to the environment. These oily contaminants are hydrophobic and persistent in nature and are recalcitrant to degradation due to limited water solubility5. Inefficient handling and disposal of these harmful pollutants have caused an enormous problem to the soil and water bodies around the refineries and oil exploration sites6. Oil contamination has a hazardous effect on flora and fauna and alters the sustainability of the ecosystem7. The negative impact of crude oil affects soil health and consequently decreases soil fertility and seed germination rate8. Crude oil is also responsible for damages to human health such as cancers, nervous disorders, liver diseases, respiratory problems etc9. These necessitates proper disposal of the crude oil wastes for proper maintenance of the environment. Various conventional methods were used for the removal of the contaminants from the environment; however, none of these methods could accomplish this task efficiently and eco-friendly way besides being costly. Most of these techniques produce secondary contaminants that persist in the environment for a longertime.
Several approaches have been implemented for effective management of petroleum sludges, such as incineration, solvent extraction, chemical treatments, land farming, ultrasonic treatment, photocatalysis, and bioremediation10,11. Amongst all these techniques, bioremediation has been established as an efficient method to manage the harmful hydrocarbon pollutants due to their low cost and eco-friendly nature12. Bioremediation is a cost-effective and environmentally friendly technique with no chances of secondary environmental contamination13. The application of bioremediation for the treatment of oily sludge is, however, limited. Earlier studies have established that almost all hydrocarbon components are susceptible to degradation by microorganisms14. However,biodegradation studies of crude oil revealed that intermediate length of saturated hydrocarbons are more easily biodegradable as compared to the high molecular weight long and branched-chain alkanes,resins, asphaltenes, and cyclic aliphatic hydrocarbons15. Relatively little is known about the environmental determinants of microbial population in oily refinery sludge. Soil bacterial communities are mainly affected by the type of soil, physicochemical and biological properties of soil and presence of contaminants. Petroleum hydrocarbons do have strong toxic impacts on the microbial population which are often overlooked16. Earlier studies revealed that petroleum hydrocarbons have inhibited the microbial biomass, decreased phylogenetic diversity, and caused a decline in species richness and evenness which have resulted in the dominance of few species that can survive in the harsh environment17. Indigenous bacteria have adapted to metabolize or degrade the hydrocarbons to meet their requirement of carbon and energy for their growth and development10,18 . The abundance of hydrocarbon-degrading bacteria and their degrading abilities are dependent on the types of contaminants and the surrounding environmental factors19,20. Recent studies have identified several bacteria from a variety of oil-contaminated soils; some of these bacteria such as Achromobacter, Acinetobacter, Staphylococcus, Pseudomonas, Rhodococcus, Bacillus and Dietzia are potent hydrocarbon degraders21,15. Many of these microorganisms secrete extracellular enzymes and produce biosurfactants which improve mineralization of the hydrophobic recalcitrant hydrocarbon pollutants22. Biosurfactant is the surface-active extracellular compounds that are produced by the microorganisms which possess hydrophobic and hydrophilic groups that can accumulate between oil-water phase reducing surface and interfacial tension23. They are considered as a promising alternative to the chemical surfactants due to their eco-friendly and nontoxic nature, effectiveness at extreme environmental conditions, and low production cost24. These compounds have a wide range of applications in different industries like in remediation of petroleum hydrocarbon contaminants and heavy metals from different ecosystems and also serve as a potential agent in microbial enhanced oil recovery22,24. Application of biosurfactant producing bacteria along with potential hydrocarbon-degrading abilities can effectively accelerate the rate of bioremediation of hydrocarbon polluted environments due to their ability to promote bioavailability of nonpolar substrates and enhancing the affinity between the bacterial cell surface and oil-water interfaces24,25. The present study on isolation of bacteria from oily refinery sludge, therefore, will help in understanding the distribution of bacterial communities in harsh environments besides knowing the potent indigenous oil degraders adapted to the harsh local environments.
Sample collection
Refinery sludge samples were collected from the sludge waste pit of Bongaigaon oil refinery owned by Bongaigaon Refinery and Petrochemicals Ltd. (BRPL), Assam, India (26°31’00.81’’N; 90° 31’ 53.85’’ E). The sludge samples collected included Oily Sludge (SAB1), Biological sludge mixed with treatment water (SAB2), and treated sludge samples from sludge drying bed which contain very little quantity of crude oil (SAB3). Each sludge sample was collected in sterilized polybags with the help of a sterile spatula and was immediately transported to the laboratory and stored at 4°C, until further analysis. The collected samples were crushed and separated from large debris. A non-contaminated soil sample (SAB4) was collected as a control sample to study the differences in the bacterial diversity.
Physicochemical analysis
The temperature of the samples was measured on-site with the help of a soil thermometer. The moisture content was measured gravimetrically by the oven drying method26. The pH of the sample was measured in soil and water suspension (1:5 ratio) using a digital pH meter27. The total organic carbon (TOC), available phosphorous (AP), total nitrogen (TN), and available potassium (AK) contents were determined by following standard methods28,29. Estimation of total petroleum hydrocarbon (TPH) content in the sludge sample was determined gravimetrically 29.
Isolation and enumeration of bacteria
Different culture media such as Nutrient Agar (NA), Reasoner’s 2A (R2A) agar, (HiMedia Laboratories, India), and Soil Extract Agar (SEA) were used for the isolation of heterotrophic bacteria. About 1 g of sample was dissolved in normal saline solution and serial dilutions were prepared (10-6). 0.1 ml aliquot was placed on the agar plates containing different media using the spread plate method. Bacterial colonies in agar plates were determined by spread plate method30. Bacterial population was determined as colony forming units after 48 h of incubation at 30°C. The bacterial colonies on R2A agar were determined after 5 days of incubation. Enrichment culture method using the Bushnell Hass medium BHM (HiMedia Laboratories, India) supplemented with 2% crude oil as the sole carbon source was used to isolate the cultivable hydrocarbonoclastic bacteria. Sterilization of crude oil was done separately by autoclaving the crude oil at 121°C for 15 minutes. Enrichment was done by incubating the BHM flasks on a rotary shaker at 150 rpm for 7 days at 30°C. After 7 days of incubation, the first stage of inoculums were transferred to another set of flask containing fresh media with crude oil as the sole carbon source and incubated for three days. After three subsequent transfers, liquid culture was serially diluted and each dilution was inoculated in Bushnell Haas Agar plates. All plates were incubated at 30±1° C for 24-48 h and colonies were counted. Triplicate plates were maintained for each plate assay. The identification of the isolates was carried out based on their colony and cellular morphology and biochemical characteristics based on Bergey’s Manual of determinative bacteriology31.
Screening of the bacterial isolates for hydrocarbon degradation
The isolated bacteria were screened for their ability to degrade petroleum hydrocarbon by assessment of bacterial growth on Petroleum Agar plates using Bushnell Hass agar media. 2, 6- Dichlorophenol Indophenol (2,6-DCPIP) oxidation test was used for hydrocarbon degradation32. The change in colour from blue to colorless (reduction) of DCPIP, an electron acceptor dye, indicated the degradation of crude oil by bacterial isolates. The ability of the isolates to grow in Bushnell Hass Broth supplemented with 2% crude oil was observed and their Optical Density was determined.
Screening of the isolates for biosurfactant production
The isolates were screened for their ability to produce biosurfactant by Drop collapse test 33, Emulsification assay34, and CTAB methylene blue agar test35. The emulsification activity of the bacterial strains was obtained in terms of their stability in hydrophobic sources. The stability of the emulsion in oil-water was calculated by adding 2ml of culture supernatant in 2 ml of kerosene and allowing it to stand for 24h to determine the height of the emulsion layer. Emulsification assay for the isolates were calculated as emulsification index using the following formula-
Emulsification index=[ Height of emulsified layer / Total height of the mixture ]×100
Selection of Efficient Hydrocarbon Degrading Bacterial Isolates
Efficient crude oil-degrading bacteria were selected depending on their growth in crude oil-enriched condition with slight modification of methods described earlier36. Bacterial isolates were cultured in nutrient broth and their optical density (OD) was measured. For the screening 5 ml of bacterial culture with OD600=1, of each isolate that showed positive results for DCPIP test and growth on petroleum agar plates (Table 3) were inoculated into 250 ml Erlenmeyer flask containing 200 ml of Bushnell Haas medium(BHM) enriched with 2% crude oil and incubated at 30°C in a shaking incubator at 200 rpm shaking conditions for 7 days. Another set of flask with the same composition without the inoculation of bacteria were maintained as control. Bacterial growth was determined in the medium of each flask at 0th day and 7th days by taking the optical density at 600nm with the help of UV- Vis-spectrophotometer. Sets of flasks were inoculated to study the degradation rate of crude oil after an interval of 15 days up to 60 days of incubation. Degradation studies of the bacterial strains were determined as the percentage of degradation by gravimetric estimation of residual hydrocarbon after an interval of 15 days29. After incubation for 60 days, the residual crude oil was extracted by following the extraction procedure using DCM as the solvent, and then the extract was centrifuged for 5 min at 10,000g/4°C. The supernatant was rota-evaporated and the gravimetric estimation of the residual oil after a definite incubation period was calculated. Degradation of TPH was estimated using the following equation:
Percentage Reduction of TPH(%)= [ (Intial weight of TPH-final weight of TPH)/(initial weight of TPH) ] ×100
Analysis of crude oil degradation by FTIR analysis
Residual oil, after degradation assay, was subjected to FTIR analysis to study the breakdown of complex hydrocarbons. Liquid hydrocarbon samples were milled with potassium bromide (KBr) to form a very fine powder. This powder was then compressed into a thin pellet which was analysed using Fourier transform infrared (FTIR) in the region of mid IR 400–4000 cm-1 with 16-scan speed37. FTIR was performed to determine the functional group present in hydrocarbon before and after degradation in the sample.
Molecular identification of potent hydrocarbon-degrading bacterial isolate
Bacterial genomic DNA was extracted from the bacterial isolates grown on nutrient broth using standard protocol38. PCR amplification of the genomic DNA was carried out using the universal primers 27F and 1492R having reaction conditions set at 94°C for 3 min, followed by 35 cycles of 94° for 30 sec, 55°C for 30 sec and 72°C for 90 sec and then final extension at 72°C for 5 min. Amplification products were analysed on 1.5% (w/v) agarose gels in TAE buffer. 16S rDNA amplicon from each strain was sequenced, and the data were searched using NCBI-BLAST search tool for identification of the strain type. The sequence was submitted to the NCBI GeneBank .
Statistical analysis
Bacterial population number was transformed into log10cfu before performing the statistical analysis. All tests were performed in triplicates and results are expressed as mean ± Standard Deviation (n=3). One way analysis of Variance was calculated to analyse the differences among the mean of different assays. Statistically significant difference between means was determined by calculating the least significant differences (LSD).
Physicochemical analysis
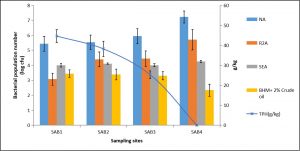
Physicochemical properties and microbiological characterization of the oily sludge sample are summarized in Table 1. Oil content in the sludge sample is considered as one of the most hazardous content in the oily sludge. The temperature of the samples was higher (36.03±0.49°C) than the average temperature of the region (33.16±0.28°C).Oil content i.e. the extractable Total Petroleum Hydrocarbon (TPH) was present in a higher amount in the sample SAB1, followed by SAB2, SAB3, and absent in the control sample SAB4 (Table 1). Moisture content was highest in the SAB2, followed by SAB1, SAB4, and SAB3. The pH of the studied sample was found to be basic in nature and varied significantly (p<0.05) among the samples. Electric conductivity (EC) varied significantly among the study sites and it was highest for the control soil and decreased with the rise in the level of contamination. Total organic carbon was higher in the oily sludge due to higher content of petroleum hydrocarbons and significant variations were found among the samples. Total nitrogen, available phosphorous and potassium content varied significantly amongst the studied samples. The ratio between total carbon and nitrogen determines the fertility of the soil and a reduction of the value was observed with higher contamination level in the samples.
Table (1):
Physicochemical and biological properties of samples ( Values are mean of three replicates ±SD) (* BP= Bacterial population number are represented as the average of the log cfu obtained in different media ) ; Means with different letters in the superscript within a column are significantly different (p < 0.05).
SAMPLE |
TEMP (°C) |
pH |
MC(%) |
TPH(g/kg) |
EC |
TOC(%) |
TN(%) |
AP(%) |
K(%) |
C:N ratio |
BP (log cfu) |
|---|---|---|---|---|---|---|---|---|---|---|---|
SAB1 |
35.3±0.7a |
8.2±0.03a |
23.28±3.2a |
443.66±2.4a |
0.21±0.005a |
3.21±0.01a |
0.119±0.003a |
0.41±0.01a |
0.212 ±0.02a |
26.97±1.3 a |
3.985±0.14a |
SAB2 |
34.5±0.5a |
8.0±0.2b |
44.21±3.2b |
381.44±0.8b |
0.52±0.005b |
2.8±0.02b |
0.122±0.01a |
0.484±0.005b |
0.178±0.01b |
22.95±0.19 b |
4.35±0.22b |
SAB3 |
36.03±0.4ab |
7.86±0.3c |
18.27±1.7c |
252.9±0.3c |
0.86±0.02c |
2±0.1c |
0.173±0.01b |
0.546±0.02c |
0.109±0.01c |
11.76±0.88 c |
4.42±±0.44b |
SAB4 |
33.16±0.2c |
6.9±0.1d |
22.4±0.3c |
0 |
1.29±0.01d |
1.58±0.2d |
0.346±0.03c |
0.641±0.04d |
0.113±0.01d |
4.56±0.8 d |
4.452±0.83b |
LSD |
0.96 |
0.375 |
4.84 |
2.465 |
0.028 |
0.29 |
0.038 |
0.05 |
0.028 |
1.731 |
0.215 |
Isolation and enumeration of bacteria
A total of 36 bacterial strains were isolated from the collected samples using NA, R2A, SEA, and BHM culture media. The total number (CFU counts per g of soil) of heterotrophic bacteria was highest in the control soil (SAB4)when the NA medium was used and it varied from 2.7× 105 to 16.9 × 106 in the samples. The bacterial population number in the same varied from 1.5 × 104 to 5.2 × 105and 10 × 102 to 1.8 × 103 respectively when R2A and SEA medium were used for isolation (Fig.1). However, the highest population number of hydrocarbon-degrading bacteria in enrichment culture in BHM medium amended with 2% crude oil was 2.8 × 103, 2.4× 103, 2.0 × 103 and 0.22 × 103 respectively in SAB1, SAB2, SAB3 and SAB4.The refinery sludge with 40% crude oil (SAB1) harboured the lowest bacterial population number. NA and R2A were the most suitable media for the isolation of heterotrophic bacteria from contaminated as well as non-contaminated soil. The use of SEA media enabled-isolation of most of the hydrocarbon-degrading bacteria from the oily sludge. Bacterial population number decreased with the increasing level of hydrocarbon content in different samples (Fig. 1). Studies on the physicochemical characteristics of the collected samples indicated a significant negative correlation between TPH and bacterial population number (p<0.05). A significant positive correlation was obtained between bacterial population number and temperature, moisture content, EC, TN, and AP. However, the correlation between bacterial population number and pH, TOC, and AK was negative though significant (p <0.05).
Fig. 1. Variation in bacterial population number (log cfu) when different culture media were used for isolation along with the level of TPH content in the studied samples
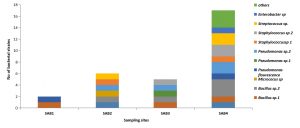
All the contaminated sites were mainly dominated by Pseudomonas sp and Bacillus sp
(Fig. 2). The relative abundance of bacterial isolates was lowest in the oily sludge samples SAB1, followed by SAB3 and SAB2 compared to the control sample SAB4 (Fig. 2). It was observed that the diversity of heterotrophic bacteria decreased with the increasing concentration of crude oil in the oily sludge.
Screening of the bacterial isolates for hydrocarbon degradation
All the 36 bacterial strains were screened for the ability to degrade hydrocarbon. Ten bacterial strains showed positive growth response on petroleum agar plates and were also positive for other screening tests carried out for hydrocarbon degradation (Table 2). These potent isolates (10) were further tested for their ability to degrade crude oil in culture broth with 2% crude oil as the sole carbon source. The strain SAB09 exhibited 80% degradation in 60 days and maximum degradation being observed after 15 days of incubation while the degradation rate of strain SAB06 varied from 29.26 to 62.13% in 60 days (Table 3).
Table (2):
Screening of bacterial isolates for hydrocarbon degradation abilities and biosurfactant production (‘+++’ maximum growth; ‘++’ average growth; ‘+’ slow growth; ‘-’ no growth ).
| Bacterial strains | Growth on Petroleum Agar plates | DCPIP activity | Growth on Bushnell Haas Agar media + 2% crude oil (OD at 600nm) | Biosurfactantproduction | ||
|---|---|---|---|---|---|---|
| Drop collapse test | CTAB Methylene blue agar test | E24% | ||||
| SAB02 | +++ | + | 0.36 | +ve | -ve | 30 |
| SAB06 | +++ | + | 1.32 | +ve | +ve | 78 |
| SAB07 | ++ | – | 0.65 | +ve | -ve | 45 |
| SAB09 | +++ | + | 1.20 | +ve | +ve | 70 |
| SAB10 | + | + | 0.24 | -ve | -ve | 20 |
| SAB13 | + | + | 0.43 | -ve | -ve | – |
| SAB15 | ++ | – | 0.05 | +ve | -ve | – |
| SAB24 | ++ | – | 0.56 | +ve | -ve | – |
| SAB29 | ++ | + | 0.62 | +ve | -ve | 35 |
| SAB36 | ++ | – | 0.08 | +ve | -ve | – |
While screening the isolates for biosurfactant production, it was observed that all the biosurfactant producing isolates didn’t possess oil degrading abilities (Table 2). Hence, based on oil degradation and biosurfactant production abilities isolates SAB06 and SAB09 were selected as the potent hydrocarbon degraders (Table 2, Table 3).
Degradation of crude oil by the two potent isolates
Out of the ten isolates that grew on petroleum agar plates, 08 isolates showed positive results for drop collapse test and two isolates showed positive results on CTAB agar plates indicating the production of rhamnolipids. Only 6 isolates showed emulsification index (%) ranging from 20% – 78%, while no emulsification was observed in the case of 4 isolates (Table 2). Depending upon the results of the screening tests, isolates SAB06 and SAB09 were selected as the potent biosurfactant producing isolates.
These two isolates not only produced biosurfactant but also showed the maximum emulsification index (78% and 70% respectively) (Table 2) as well as a high rate of petroleum hydrocarbon degradation. The isolate SAB09 exhibited a high rate of degradation of hydrocarbon (80.27 %) followed by SAB06 showing 62.13% degradation during 60 days of incubation (Table3). The two isolates were identified as Pseudomonas sp.( MT250346) and Bacillus sp.(MT250345).
Table (3):
Percentage degradation of crude oil by four bacterial strains at different interval of time after incubation (Data mean of 3 replicates ± SE).
Strains (culture ID) |
Days of incubation |
Initial concentration(I) (g) |
Final concentration(F)(g) |
% TPH degradation (I-F)/I×100 |
|---|---|---|---|---|
Pseudomonas strain I |
0 |
2.246±0.06 |
2.246±0.56 |
– |
(SAB09) |
15 |
2.246±0.06 |
1.453±0.24 |
35.29 |
30 |
2.246±0.06 |
1.084±0.27 |
51.72 |
|
45 |
2.246±0.06 |
0.893±0.09 |
60.21 |
|
60 |
2.246±0.06 |
0.443±0.22 |
80.27 |
|
Pseudomonas strain II |
0 |
2.525 ± 0.82 |
– |
– |
(SAB02) |
15 |
2.525 ± 0.82 |
2.031±0.20 |
19.56 |
30 |
2.525 ± 0.82 |
1.887±0.56 |
25.24 |
|
45 |
2.525 ± 0.82 |
1.767±0.66 |
30.01 |
|
60 |
2.525 ± 0.82 |
1.381±0.78 |
45.28 |
|
Bacillus sp |
0 |
2.321 ± 0.26 |
– |
– |
(SAB06) |
15 |
2.321 ± 0.26 |
1.641±0.75 |
29.26 |
30 |
2.321 ± 0.26 |
1.270±0.45 |
45.24 |
|
45 |
2.321 ± 0.26 |
1.103±0.74 |
52.46 |
|
60 |
2.321 ± 0.26 |
0.879±0.23 |
62.13 |
|
Micrococcus sp(SAB26) |
0 |
2.841± 0.78 |
– |
– |
15 |
2.841± 0.78 |
2.268±0.75 |
20.16 |
|
30 |
2.841± 0.78 |
2.088 ±0.23 |
26.48 |
|
45 |
2.841± 0.78 |
1.461 ±0.74 |
48.56 |
|
60 |
2.841± 0.78 |
1.384 ±0.45 |
51.26 |
Analysis of crude oil degradation by FTIR analysis
Degradation of crude oil by the two potent bacterial isolates was confirmed by FTIR analysis. The results of the un-inoculated control sample were compared with that of the inoculated (SAB06 and SAB09) samples. The Infrared (IR) spectral data were obtained after 60 days of incubation of the inoculated samples using Spectrum 2 Perkin-Elmer FTIR spectrometer. The spectra were scanned in the 400-4000 cm-1range.The spectra were plotted as intensity versus wavenumber(cm-1). The peaks and the representing compounds in the FTIR graph are studied. Functional groups before and after degradation study were also identified. Most of the compounds are assigned as the aliphatic groups. Although all the samples possessed an absorption bands in similar regions; however, certain differences were observed in the FTIR spectra of the treated and non-treated samples. The control crude oil sample showed a significant strong absorption peak at 3134.33 cm-1 indicating the presence of O-H stretching of the Carboxylic group. Peak at 2980.01 was due to C-H stretching of alkane and a weak peak at 2381.39 corresponded to the O=C=O stretching (Supplementary Fig. 1). A strong absorption band at 1394.05 indicated C-H bending in the aliphatic compounds. The medium absorption peak at 1600.92 was assigned to the C=C stretching of conjugated alkene and a medium band at 723.31 cm-1 indicated the presence of the C=C alkene group.
FTIR spectra of the crude oil sample inoculated with SAB06 (Supplementary
Fig. 2) showed a strong intensity band at 1394.05 indicating the presence of an aliphatic group. The peaks in the region 2840-3000 which corresponds to the C-H stretching in the aliphatic group was absent in both the treated sample. The assigned peaks at 3166.59 and 3129 of the treated samples by bacteria corresponds to the O-H stretching of primary alcohol. Absorption band of both the treated samples at 728.57 and 720.79 represents CH2 bending of aliphatic compounds. New peaks were observed in both treated samples at 1109.51 indicating the formation of C=O stretching of secondary alcohol which might be due to degradation of n alkane. Band at 1394.05 which was observed in the control sample as a strong peak was also found in the sample inoculated with SAB06, however, the peak disappeared after the treatment of SAB09 (Supplementary Fig.3) and a new strong peak was formed at 1401.81 indicating O-H bending of alcohol. A new peak was observed at 3166.59 which corresponds to =C-H stretching in aromatic compounds. It was observed in the study that there is a significant change in transmittance in the treated sample which represents the degradation of complex hydrocarbon compounds to simpler compounds with the help of bacteria.
Soil is a vital resource for all kinds of life forms. Addition of hydrocarbon to soil increases the bulk density of soil, decreases pore space, reduces water infiltration into soil consequently inhibiting plant growth39. Polyaromatic hydrocarbon (PAH), which is the main constituent of oily refinery sludge possess a great threat to the environment due to its toxic effect, mutagenicity, and carcinogenicity29. Owing to the increased awareness of the environmental protection, an effective eco-friendly method for the treatment of these hazardous compounds is the need of the hour. The oily sludge in the present study is characterized by basic nature, high content of hydrocarbon (TPH), low nitrogen, higher organic carbon, lower level of potassium, and phosphorus40. A high amount of TPH content in the oily sludge marked the level of contamination in the studied samples. The physicochemical characteristics varied significantly amongst the study sites (p< 0.05). Soil pH and TOC were positively correlated with hydrocarbon content which is similar to the results obtained in other studies41. Higher total organic carbon content in the oily sludge may be attributed to the addition of carbon content to the soil in the form of oily sludge compared to the control soil. Total nitrogen content (TN) and available phosphorous (AP) are important macronutrients present in soil which is important for the plants as well as microorganisms. Our results indicated a decline in the content of both nitrogen and phosphorous with an increase in contamination which was also reported in other studies42. Study sites SAB1 and SAB 2 didn’t differ significantly as regards to total nitrogen content. The variation, however, was significant among other sites. The ratio of total organic carbon and nitrogen increased with the increase in the content of crude oil in the soil. The higher C:N ratio indicated degradation of soil quality in the studied sites. Available phosphorous varied significantly among the study sites. Decrease in total nitrogen and available phosphorus in the contaminated sites may be due to an increase in carbon content in the soil which might have altered the nutritional balance in the soil indicating low soil quality. It was noted that the oily sludge sample shared commonality in characteristics concerning the different physicochemical characteristics compared to the control soil sample.
Soil microorganisms are omnipresent and they play an important role in maintaining the quality of the soil. Bacterial diversity in oily sludge was studied to gain knowledge of the bacterial community inhabiting the polluted environment. Isolation and characterization of the bacterial isolates are required to implement efficient bioremediation strategies.Bacterial population number and diversity varied significantly with the level of hydrocarbon content in the studied samples. Hydrocarbon content harmed the heterotrophic bacterial population number, however, the population number of hydrocarbonoclastic bacteria varied positively with the level of contamination. It was observed that the bacterial diversity was reduced with an increase in the oil content in the samples compared to the control sample (Fig. 2). However, with the increase in hydrocarbon content, the abundance of particular bacterial genera increased limiting the growth of other bacterial genera43.Studies revealed that the organisation of the bacterial communities in the contaminated sites is dependent on the level of contamination and their responses to the contaminants44. Our study on bacterial diversity is inconsistent with the previous studies which indicated that the presence of hydrocarbons in soil has resulted in the occurrence of certain groups of bacteria that have adapted to the harsh environment45. Although hydrocarbon contaminants are found to affect microbial diversity, some of these bacteria can utilize the harsh hydrocarbon pollutants as the source of carbon and energy which results in the development of hydrocarbon resistant bacteria communities46. The genus Pseudomonas have been reported as potent hydrocarbon degraders in many oil degradation studies45,47,48. Earlier studies reported that strains of Pseudomonas aeruginosa can degrade about 90-95% of diesel and kerosene in 21 days and 72 % TPH was degraded over a period of 4 weeks49. Degradation by Bacillus sp was also reported by earlier studies50. The isolation of culturable bacteria from refinery sludge enabled us to understand the hydrocarbon-degrading ability of the indigenous bacteria 51. Our studies revealed that the enrichment culture studies enhanced the isolation of hydrocarbonoclastic bacteria form the hydrocarbon contaminated soil and it is positively correlated with the level of hydrocarbon content.
One of the limitations in the degradation of petroleum hydrocarbon is the bioavailability of these contaminants to bacteria due to hydrophobic nature and low water solubilities. In such cases, the application of potent biosurfactant producing bacteria can emulsify or solubilize the hydrocarbons making the substrates available to the microorganisms and decreases the surface tension of oil-water interface, thereby, enhancing the rate of degradation52. Some of the hydrocarbonoclastic bacterial strains isolated in this study are potent producers of biosurfactants. Earlier studies have reported the increased rate of degradation of n –hexadecane by Bacillus sp in the presence of rhamnolipid biosurfacatant53. The application of rhamnolipid produced by Pseudomonas strain also enhanced the rate of degradation of hydrocarbon54. Pseudomonas and Bacillus strain have biosurfactant producing abilities which are found to be effective for efficient removal of crude oil from contaminated sites which is consistent with other studies55. Promising results were achieved in many studies for enhancement of hydrocarbon degradation by application of biosurfactant producing bacteria which is a solution for decontamination of petroleum polluted soil56,38,57. However it has been studied that all the biosurfactant producing strains don’t have an efficient degradation ability which is similar to the present study58.
The efficiency of biodegradation depends on the biodegrading capabilities of the indigenous bacteria as foreign bacteria that are applied exogenously sometimes fail to adapt to the new environment and perform degradation activities59. The degradation of hydrocarbons was characterized by the difference in the initial and final concentration of hydrocarbon in the culture media52. Biodegradation of crude oil in the present study was visible after 7 days of incubation. Degradation of crude oil components by the potent bacterial strains was further confirmed by FTIR analysis. FTIR analysis has been used by various researchers to classify the dominant petroleum hydrocarbon fractions in the contaminated sites based on the carbon chain length60,61,37. Application of FTIR study was also reported in the study of microbial degradation of hydrocarbon contaminants62,63. The presence of functional groups such as alcohols, phenols, aliphatic hydrocarbons were due to the presence of petroleum hydrocarbons in the studied samples62. It was observed from the FTIR spectra that although some similar characteristics peaks were observed in all the samples but their intensity decreased in the bacteria inoculated samples and many peaks disappeared after the treatment with bacteria which is similar to other studies63. Some new bands were also observed which may correspond to new compounds arising due to bacterial oxidation of the hydrocarbon compounds. In the present study, the alkene compounds were not degraded totally which was observed in the IR peaks of the treated samples. The presence of O-H and C-H bonds indicated the presence of alkane groups of hydrocarbon in the samples. The results of FTIR analysis showed the degradation of most of the compounds in the treated sample. It can be attributed that degradation using Pseudomonas sp and Bacillus sp could be used as efficient remediation techniques as these bacteria could remove up to 80% of crude oil in 60 days of treatment as observed in the present study. Hence, the potential of these hydrocarbonoclastic bacteria could be applied for bioremediation of the hazardous oily wastes.
The results of the present study established that crude oil content of soil negatively influence the indigenous bacterial population number as well as diversity. The crude oil contamination also significantly affects the soil physicochemical properties e.g. moisture content, TOC, available nitrogen, and available phosphorus which finally affects the nutritional properties of soil. Two potent strain Pseudomonas sp. (MT250346) and Bacillus sp. (MT250345) isolated during the present study showed the capability of degrading crude oil by utilizing the same as the source of carbon and energy. The potent hydrocarbon degraders isolated in the present study may be applied for biodegradation purposes because they have already been adapted to the harsh environment. Biosurfactant production abilities of these potent bacterial isolates have aided in their beneficial role in the bioremediation of contaminated sites. The potential of these hydrocarbonoclastic bacterial isolates as degrading agents of petroleum hydrocarbons indicates the importance of biological treatment as the most promising alternative for the removal of hazardous pollutants from the environment.
Additional file: Additional Figure S1 and S2.
ACKNOWLEDGMENTS
Sashi Prava Devi is thankful to the Department of Science and Technology (DST), Government of India, for providing INSPIRE fellowship (Award no : IF140734) . Authors would also like to thank the Head, Department of Botany, Gauhati University for allowing them to use the necessary facilities to carry out the experiments.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors designed the experiments. SPD carried out the experiments, analysed the data and prepared the manuscript. DKJ supervised the work and edited the manuscript. All authors read and approved the manuscript.
FUNDING
The study was supported by Department of Science and Technology (DST), Government of India, in the form of INSPIRE fellowship to SP Devi (Award no : IF 140734).
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All the data are included in the supplementary files and the manuscript includes the accession number of the bacterial isolates.
- Bhattacharyya JK, Shekdar AV. Treatment and disposal of refinery sludges: Indian scenario. Waste Manage Res. 2003;21:249-61.
Crossref - Paisse S, Coulon F, Goni-Urriza M, Peperzak L, McGenity T, Duran R. Structure of bacterial communities along a hydrocarbon contamination gradient in coastal sediment. FEMS Microbiol Ecol. 2008;66:295-305.
Crossref - Ministry of Petroleum and Natural Gas (2019) India’s Hydrocarbon Outlook: 2018-19,Government of India. http://dghindia.gov.in/assets/downloads/ar/2018-19.pdf
- Ministry of Environment & Forests, GOI New Delhi (1985) Environmental Guidelines for Siting Industry.
- Ding A, Sun Y, Dou J, et al. Characterizing Microbial Activity and Diversity of Hydrocarbon-Contaminated Sites; Kutcherov V, Kolesnikov A (eds.), Hydrocarbon, IntechOpen, London, United Kingdom. 2013. 10.5772/50480.
- Atlas RM, Philp J. Bioremediation: applied microbial solutions for real-world environmental cleanup. 2005; pp 78–105. American Society for Microbiology (ASM) Press, Washington, D.C.
Crossref - Militon C, Boucher D, Vachelard C, et al. Bacterial community changes during bioremediation of aliphatic hydrocarbon-contaminated soil. FEMS Microbiol Ecol. 2010;74: 669-681.
Crossref - Saikia N, DekaBarua HP, Das AK, Yein R. Efficacy of bioformulation in germination of seed in crude oil contaminated soil samples. Ind J Environ Prot. 2009;29(12):1072-1078.
- Chen C S, Hseu Y C, Liang S H, Kuo J Y, Chen S C. Assessment of genotoxicity of methyl-tert-butyl ether, benzene, toluene, ethylbenzene, and xylene to human lymphocytes using comet assay. J Hazard Mater. 2008;153(1-2):351-356.
Crossref - Das N, Chandran P. Microbial degradation of petroleum hydrocarbon contaminants: an overview, SAGE-Hindawi Access to Research Biotechnology Research International. 2011;1-13.
Crossref - Johnston JE, Lim E, Roh H . Impact of upstream oil extraction and environmental public health: A review of the evidence. Science of The Total Environment. 2019;657:187-199.
Crossref - Guerra AB, Oliveira JS, Silva-Portela RC, Araujo W, et al., Metagenome enrichment approach used for selection of oil-degrading bacteria consortia for drill cutting residue bioremediation. Environ Pollut. 2018;235:869-880.
Crossref - Chikere CB, Okpokwasili GC, Chikere BO. Bacterial diversity in a tropical crude oil-polluted soil undergoing bioremediation. Afr J Biotech. 2009;8:2535-2540.
- Head IM. Bioremediation: towards a credible technology. Microbiology. 1998;144(3):599.
Crossref - Varjani SJ. Microbial degradation of petroleum hydrocarbons. Bioresour Technol. 2017;223:277-286.
Crossref - Overholt WA, Marks KP, Romero IC, Hollander DJ, Snell TW, Kostka JE. Hydrocarbon degrading bacteria exhibit a species specific response to dispersed oil while moderating ecotoxicity. Appl Environ Microbiol. 2015;82:518-527.
Crossref - Labud V, Garcia C, Hernandez T. Effect of hydrocarbon pollution on the microbial properties of a sandy and a clay soil. Chemosphere. 2007;66:1863-1871.
Crossref - Baruah R, Mishra SK, Kalita DJ, Silla Y, Chauhan PS, Singh AK, DekaBoruah HP. Assessment of bacterial diversity associated with crude oil-contaminated soil samples from Assam. Int J. Environ Sci Technol. 2017;14:2155-2172.
Crossref - Yang Y, Wang J, Liao J, Xie S, Huang Y. Abundance and diversity of soil petroleum hydrocarbon-degrading microbial communities in oil exploring areas. Appl. Microbiol. Biotechnol. 2015;99:1935-1946.
Crossref - Varjani SJ, Gnansounou E. Microbial dynamics in petroleum oilfields and their relationship with physiological properties of petroleum oil reservoirs. Bioresour. Technol. 2017;245:1258-1265.
Crossref - Margesin R, Labbe D, Schinner F, Greer CW, Whyte LG. Characterization of hydrocarbon-degrading microbial populations in contaminated and pristine Alpine soils. Appl Environ Microbiol. 2003;69:3085-3092.
Crossref - Margesin R, Moertelmaier C, Mair J. Low-temperature biodegradation of petroleum hydrocarbons (n-alkanes, phenol, anthracene, pyrene) by four actinobacterial strains. Int Biodeterior Biodegrad. 2013;84:185-191.
Crossref - Banat IM, Satpute SK, Cameotra SS, Patil R, Nyayanit NV. Cost effective technologies and renewable substrates for biosurfactants’ production. Front Microbiol. 2014;5:697.
Crossref - Banat IM, Franzetti A, Gandolfi I, et al. Microbial biosurfactants production, applications and future potential. Appl. Microbiol. Biotechnol. 2010;87:427-444.
Crossref - Mnif S, Sayadi S, Chamkha M. Biodegradative potential and characterization of a novel aromatic-degrading bacterium isolated from a geothermal oil field under saline and thermophilic conditions. Int Biodeterior Biodegrad. 2014;86:258-264.
Crossref - Wilke BM. Determination of chemical and physical soil properties. In: Margesin R et al. (eds.). Manual for Soil Analysis Monitoring and Assessing Soil Bioremediation (Soil Biology, Vol. 5). Berlin Heidelberg, Germany: SpringerVerlag, 2005;47-95 William and Wilkins. Baltimore. 1994;10:112-135.
- Jackson ML. Soil Chemical Analysis. Prentice Hall of India Ltd, New Delhi,1973;23.
- Walkley A, Black IA. An examination of the Degtjareff method for determining organic carbon in soils: effect of variations in digestion conditions and of inorganic soil constituents. Soil Sci. 1934;63:251-263.
- Mishra S, Jyoti J, Kuhad RC, Lal B. In situ bioremediation potential of an oily sludge degrading bacterial consortium. Curr. Microbiol. 2001;43:328-335.
Crossref - Cappuccino JG, Sherman N. In: Microbiology: A laboratory manual. The Benjamin/Cummings Publishing Company Inc. Melopark, California. 1996;186(4th Ed.).
- Holt JG, Krieg NR, Sneath PH. A Bergey’s manual of determinative bacteriology. J Basic Microbiol. 1994;48:1-8.
- Hanson KG, Desai JD, Desai AJ. A Rapid and Simple Screening technique for potential crude oil Degrading microorganisms. Biotechnol Tech. 1993;7(10):745-748.
Crossref - Tugrul T, Cansunar E. Detecting Surfactant-producing Microorganisms by the Drop-collapse Test. World J Microbiol Biotechnol. 2005;21:851-853.
Crossref - Cooper DG, Goldenberg BG. Surface-active agents from 2 Bacillus species. Appl Environ Microbiol.1987;53:224-229.
Crossref - Satpute SK, Bhawsar BD, Dhakephalkar PK, Chopade BA. Assessment of different screening methods for selecting biosurfactant producing marine bacteria. Ind J Mar Sci. 2008;37:243-250.
- Rahman KSM, Thahira-Rahman J, Lakshmanaperumal-samy P, Banat IM. Occurrence of crude oil degrading bacteria in gasoline and diesel station soils. J Basic Microbiol. 2002;42:286-293.
Crossref - Wang L, Cheng Y, Lamb D, Dharmarajan R, Chadalavada S, Naidu R. Application of infrared spectrum for rapid classification of dominant petroleum hydrocarbon fractions for contaminated site assessment. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy. 2019;207:183-188.
Crossref - Sambrook J, Fritschi EF, Maniatis T. Molecular cloning: a laboratory manual, Cold Spring Harbor Laboratory Press, New York, 1989.
- Abosede EE. Effect of crude oil pollution on some soil physical properties. Journal of Agriculture and Veterinary Science. 2013;6(3):14-17.
Crossref - Das R, Kazy SK. Microbial diversity, community composition and metabolic potential in hydrocarbon contaminated oily sludge: prospects for in situ bioremediation. Environ Sci Pollut Res. 2014;21(12):7369-7389.
Crossref - Alrumman SA, Standing DB, Paton GI. Effect of hydrocarbon contamination on soil microbial community and enzyme activity. J King Saud Univ Sci. 2015;27:31-41.
Crossref - Wang X, Feng J, Wang J. Petroleum hydrocarbon contamination and impact on soil characteristics from oilfield in Momoge wetland. Environmental Science. 2009;30(8):2394-2401.
- Santos dos HF, Cury JC, do Carmo FL, et al., Mangrove bacterial diversity and the impact of oil contamination revealed by pyrosequencing:bacterial proxies for oil pollution. PLoS ONE. 2011;6:e16943.
Crossref - Mittal A, Singh P. Isolation of Hydrocarbon degrading bacteria from soils contaminated with crude oil spills. Indian J Expt Biol. 2009;47:760-765.
- Roy AS, Baruah R, Borah M, et al. Bioremediation potential of native hydrocarbon degrading bacterial strains in crude oil contaminated soil under microcosm study. Int Biodeterior Biodegrad. 2014;94:79-89.
Crossref - Das D, Baruah R, Sarma Roy A, et al. Complete genome sequence analysis of Pseudomonas aeruginosa N002 reveals its genetic adaptation for crude oil degradation. Genomics. 2015;105(3):182-190.
Crossref - Wongsa P, Tanaka M, Ueno A, Hasanuzzaman M, Yumoto I, Okuyama H. Isolation and characterization of novel strains of Pseudomonas aeruginosa and Serratiamarcescens possessing high efficiency to degrade gasoline, kerosene, diesel oil, and lubricating oil. Curr Microbiol. 2004:49:415-422.
Crossref - Yan P, Lu M, Yang Q, Zhang H, Zhang Z, Chen R. Oil recovery from refinery oily sludge using a rhamnolipid biosurfactant-producing Pseudomonas. Bioresour Technol. 2012;116:24-28.
Crossref - Jacquiod S, Cyriaque V, Riber L, et al., Long-term industrial metal contamination unexpectedly shaped diversity and activity response of sediment microbiome. J Hazard Mater. 2018;344:299-307.
Crossref - Ventorino V, Sannino F, Piccolo A, Cafaro V, Carotenuto R, Pepe O. Methylobacterium populiVP2: Plant Growth-Promoting Bacterium Isolated from a Highly Polluted Environment for Polycyclic Aromatic Hydrocarbon (PAH) Biodegradation. The Scientific World Journal. 2014;1-11.
Crossref - Jain PK, Gupta VK, Gaur RK, Lowry M, Jaroli DP, Chauhan UK. Bioremediation of petroleum oil contaminated soil and water. Res J Environ Toxicol. 2011;5(1):1-26.
Crossref - Rahman KSM, Rahman TJ, Kourkoutas Y, Petsas I, Marchant R, Banat IM. Enhanced bioremediation of n-alkane in petroleum sludge using bacterial consortium amended with rhamnolipid and micronutrients. Bioresource Technology. 2003;90:159-168.
Crossref - Chen YJ., Wang HQ, Wang R, Yun Y. Effects of rhamnolipid on the biodegradation of n-hexadecane by microorganism and the cell surface hydrophobicity. Environ Sci. 2007;28:2117-2122.
- Obayori OS, Ilori MO, Adebusoye SA, Oyetibo GO, Omotayo AE, Amund OO. Degradation of hydrocarbons and biosurfactant production by Pseudomonas sp. strain LP1 . World J Microbiol Biotechnol. 2009;25:1615-1623.
Crossref - Das K, Mukherjee AK. Crude petroleum-oil biodegradation efficiency of Bacillus subtilis and Pseudomonas aeruginosa strains isolated from a petroleum-oil contaminated soil from North-East India. Bioresour Technol. 2007;98:1339-1345.
Crossref - Moran AC, Olivera N, Commendatore M, Esteves JL, Sineriz F. Enhancement of hydrocarbon waste biodegradation by addition of a biosurfactant from Bacillus subtilis O9. Biodegradation. 2000;11:65-71.
Crossref - Matvyeyeva OL, Vasylchenko ОА, Aliievа OR. Microbial Biosurfactants Role in Oil Products Biodegradation. Int J Environ Bioremediat Biodegrad. 2014;2(2):69-74.
- Inakollu S, Hung HC, Shreve G. Biosurfactant Enhancement of Microbial Degradation of Various Structural Classes of Hydrocarbon in Mixed Waste Systems. Environ Eng Sci. 2004;21:463-469.
Crossref - Venosa AD, Zhu X. Biodegradation of crude oil contaminating marine shorelines and freshwater wetlands Spill. Science and Technology Bulletin. 2003;8(2):163-178.
Crossref - Abdulkadir I, Uba S, Salihu A, Almustapha MN. A Rapid Method of Crude Oil Analysis Using FT-IR Spectroscopy. Nigerian Journal of Basic and Applied Science. 2016;24(1):47-55.
Crossref - Ahmad I, Sohail SM, Khan H, Khan R, Ahmad W. Characterization of Petroleum Crude Oils by Fourier Transform Infrared (FT-IR) and Gas Chromatography-Mass Spectrometery. Pet Petro Chem Eng J. 2018;2(2):000148.
- Bhat MM, Shankar S, Sikha, Yunus M, Shukla RN. Remediation of Hydrocarbon Contaminated Soil through Microbial Degradation- FTIR based prediction. Adv Appl Sci Res. 2011;2(2):321-326.
- Dominguez-Rosado E, Pichtel J. Chemical characterization of fresh, used and weathered motor oil via GC/MS,NMR, and FTIR techniques. Proceedings of Indiana Academy of Science, 2003;112(2):109-116.
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.