Legumes are special group of nitrogen-fixing plants that are an essential component of cropping system and important source of food/feed for human/animal consumption. Therefore it is timely to review the current evidence of the benefits of legumes for human health. However Like other crops, the productivity of legumes is threatened by abiotic stresses caused due to global climate change. Abiotic stress tolerance is complex trait involving a suite of genes, the expression of which is controlled by transcription factors including gene/polypeptide sequences. The discovery of microRNAs (miRNAs) as gene regulators has led to a paradigm shift in the understanding of post-transcriptional gene regulation in plants and animals. In addition to protein coding genes, microRNAs (miRNAs) have emerged as important players in plant stress responses. Initial clues suggesting that miRNAs are involved in plant stress responses stem from studies showing stress regulation of miRNAs and target predictions for some miRNAs. Subsequent studies have demonstrated an important functional role for these miRNAs in abiotic stress responses. This review summarizes the current knowledge on the role of different miRNAs in response to main abiotic stresses in legumes.
Abiotic stress, drought, cold, salinity, legume, miRNA.
Legumes belong to the family Fabaceae, previously Leguminosae, which includes some of the world’s most important food and feed crops such as; Cicer arietinum (chickpea), dry cowpea (Vigna ungiculanta), pigeonpea (Cajanus cajun) and lentil (Lens culinaris), black gram (vigna mungo) Glycine max (Soybean), Phaseolus vulgaris (common bean), Pisum sativum (pea), Medicago sativa (alfalfa) and Arachis hypogea (peanut). Together, they account for one third of global primary crop production and are vital to meet the growing population demands. Legumes are rich in protein, oil, fibre and micronutrients, and are highly valued in the cropping cycle due to their ability to fix atmospheric nitrogen and act as a disease break between cereal or oilseed crops. Under conducive environmental conditions, legumes establish symbiotic relationships with arbuscular mycorrhizal (AM) fungi, leading to the formation of arbuscules, sites of phosphorous nutrient exchange 1.
The some of the most studied legume genomes, due to economic significance genome of Medicago truncaluta, a self-fertile plant with a small diploid (~500MB) with a short generation time, Lotus japonicus, which has a diploid genome (about 470MB) and a short life cycle; and Glycine max, with an amphidiploid genome (~1.1Gb).2 These species genomes have been completely sequenced and a multitude of genomics tools are available for each in the public domain (http://www.plantgdb.org/MtGDB/ http://www.plantgdb.org/LjGDB/ and http://www.plantgdb.org/GmGDB/). Recently, the chickpea genome (wild and cultivated species) was sequenced by two different groups.3, 4 Genomic tools developed for chickpea include BAC libraries, cDNA/EST databases, microarrays, high density linkage maps and mutant libraries.
In the last few years, small RNAs were determined to be important regulators of gene expression and plant growth.5, 6, 7 There are two major classes of endogenous small RNAs in plants; microRNAs (miRNAs) and small-interfering RNAs (siRNAs). The miRNAs are ~20-24-nt non-coding single stranded RNAs, processed from imperfectly folded hairpin-like precursors by the Dicer-Like1complex.6, 8 Both miRNAs and siRNAs play important roles in plant growth and development.6, 9 MicroRNAs have been discovered using three basic approaches: direct cloning, forward genetics, and bioinformatic prediction followed by experimental validation. The most direct method of miRNA discovery is to isolate and clone small RNAs from biological samples, and several groups have used this approach to identify small plant RNAs. The miRNAs regulate gene expression in plants by targeting mRNAs for cleavage or through translational repression.10 They affect diverse processes such as leaf morphogenesis, floral organ identity, and root development.7, 11 They also function in the feedback regulation of small RNA pathways and in the biogenesis of trans-acting siRNAs.12 They have been implicated in a wide array of stress responses,13,14,15 enabling plants to survive under adverse conditions such as drought, salinity, and high/low temperature. This involves triggering sophisticated mechanisms governed by complex gene networks. Although there have been significant in depth gene studies of the tolerance mechanisms, relatively little is known about the functional roles of miRNAs within them, particularly in non-model plants.16 Such studies are required to fully understand the mechanisms by which crop plants survive under adverse environmental conditions. Next Generation Sequencing (NGS) technology has greatly accelerated the discovery and characterisation of miRNAs in a range of diverse plant species.17-23 In this review, we focus on the current understanding of miRNA involvement in combating abiotic stresses in legumes. However, up until recently, legumes have been orphaned from the developments in functional genomics.24 Therefore, whilst discussing the role of miRNAs in abiotic stress tolerance in legumes, we also point out important research performed in model crops such Arabidopsis and rice. These miRNAs can be validated in legumes in future studies.
miRNA biogenesis in plants and their mode of action
MicroRNAs (miRNAs) are a class of non-coding RNAs, approximately 21-24 nucleotides long, which serve as post-transcriptional negative gene regulators by guiding target mRNA cleavage or translational inhibition in plants and animals.10, 25, 6 Most of the miRNAs are coded from inter-genic regions.
Unlike most of the animal miRNA genes which are mainly localized within introns or exons, majority of the plant miRNA genes are intergenic. They form fold-back imperfect hairpin structure which is also more variable than animal miRNA genes. Plant miRNA genes rarely occur in tandem in contrast to animal miRNA genes which are usually clustered and sometimes co-transcribed. 26
miRNA genes are transcribed by RNA Pol II and produces pri-miRNAs with 5’cap and polyadenylation in both plants and animals. In plants, pri-to-pre-miRNA conversion also requires interaction of an RNA-binding protein HYPONASTIC LEAVES1 (HYL1) and a C2H2-zinc finger protein SERRATE in nucleus.27,28 After the dual processing of miRNA by DCL-1, mature miRNA duplexes forms which are stabilized by a methyl transferase, Hua Enhancer 1 (HEN1) which is dependent on the S-adenosyl methionine for methylation. This enzyme methylates all plant-silencing small RNAs. Methyl groups are added to the 3’ terminal nucleotides of each strand. This prevents 3’ uridylation and degradation. 29, 30 An exportin-5homolog in plant, HASTY is required for miRNA biogenesis and function of miRNA. HASTY helps in the exporting of methylated miRNA/miRNA* to the cytoplasm but the exact exported form is not clear.31
miRNAs are finally loaded to the RISC for target cleavage or translational inhibition of target mRNAs. RISC is a RNA-induced silencing complex, a multiprotein complex. Single stranded 21 nt miRNAs serves as the template for RISC to recognise complementary mRNA. RISC is associated with a class of proteins called ARGONAUTE which activates and cleaves the mRNA. Arabidopsis contains 10 Argonaute protein paralogs.32 Among them, AGO1 is involved in miRNA mediated gene regulation either by cutting miRNA target mRNAs 33 or repressing their translation. 34 AGO1 contain 2 domains, the PAZ domain containing a cleft which binds mRNA and a PIWI domain with ribonuclease activity.
Stress responsive miRNAs
Several studies have convincingly demonstrated the role for miRNAs in stress responses. miRNAs have emerged as important links between control of plant growth and development during stress responses. The expression of several miRNAs is altered during stress responses (both biotic and abiotic) and included drought, salinity, cold, heat ABA, oxidative and hypoxia 35 (figure 2). One particular feature that has emerged from these studies is that the attenuation of plant growth and development during stress might result from conserved miRNAs that target transcription factors, particularly the miRNAs that regulate auxin perception and signalling. The studies on stress-responsive miRNAs and their target genes will provide better understanding of miRNA target networks operational in a plant cell. The better understanding is likely to provide techniques/methodologies to enhance stress tolerance in plants.
Overview of the role of miRNAs in plant abiotic stress responses
Plants are sessile organisms that must endure stressful environments. A large proportion of plant genes are regulated by stresses such as drought, soil salinity and extreme temperatures, 36, 37, 38 Of the many gene regulatory mechanisms such as transcriptional, post-transcriptional and post-translational regulation, transcriptional regulation is the most widely studied mechanism. The action of specific transcription factors that bind to conserved cis-acting promoter elements is well documented as a cause of changes in gene expression, particularly those induced by abiotic Stress. 37 Furthermore, post-transcriptional gene regulation under stress conditions has been documented before, although the underlying mechanism was not known.39 Considering the important roles of small RNAs in guiding post-transcriptional gene silencing, their involvement in stress-regulated gene expression seemed likely 40,41. The discovery that stress can regulate miRNA levels, coupled with the identification of stress-associated genes as miRNA targets provided clues about the role of miRNAs in stress responses. Functional analyses have demonstrated that several plant miRNAs play vital roles in plant resistance to abiotic as well as biotic stresses.13,42-44 Understanding small RNA-guided stress regulatory networks should provide new tools for the genetic improvement of plant stress tolerance (Figure 1). Indeed, it has been shown recently that manipulation of miRNA-guided gene regulation can help to engineer plants that will be more stress.44
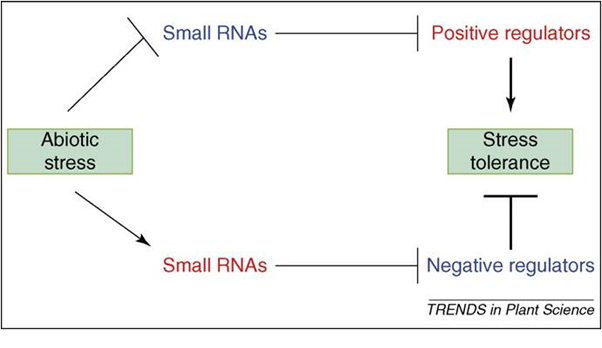 Fig. 1. Two possible modes of small RNA-guided target gene regulations under abiotic stress and their impact on plant stress tolerance
Fig. 1. Two possible modes of small RNA-guided target gene regulations under abiotic stress and their impact on plant stress tolerance 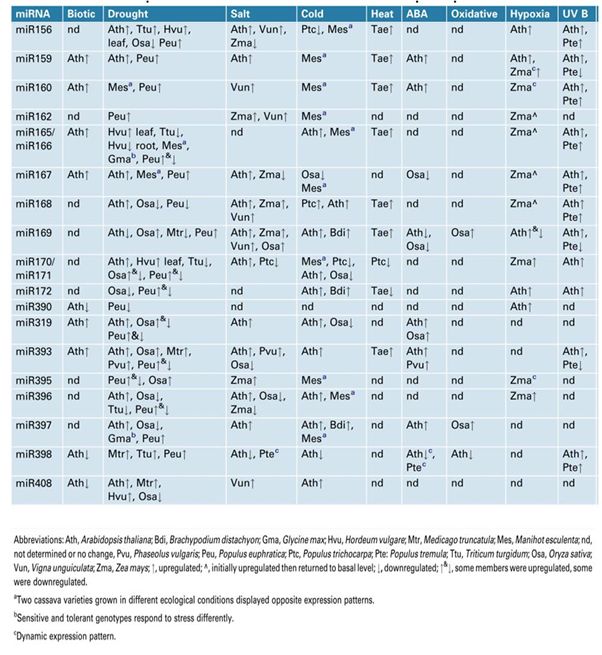 Fig. 2. MicroRNAs responsive to biotic and abiotic stresses in diverse plant species
Fig. 2. MicroRNAs responsive to biotic and abiotic stresses in diverse plant speciesmiRNA expression in response to drought, cold and salinity
Plants suffer from variety of abiotic stresses, however drought, heat, cold and salt stress are more frequently encountered. Drought is one of the most ubiquitous environmental stresses affecting plant growth and development as majority of crops are grown under rainfed conditions. Recent studies in various plants species suggest that miRNAs play important role in drought tolerance. These include conserved miRNAs such as miR164, miR169, miR171, miR396, miR398, miR399, miR408 and miR2118.45 Their expression patterns vary across species. For example, miR169 was down-regulated in Arabidopsis and M. truncatula 46, 47 but up-regulated in common bean (in response to abscisic acid treatment) and rice (Arenas-Huertero et al., 2009; Zhao et al., 2009)48, 49. In M. truncatula, miR398a,b and miR408 were strongly up-regulated in shoots and roots under drought stress.47 The miR398 and miR408 repress the COX5b, CSD1 and plantacyanin genes. 47
Recently, 50 identified 22 members of 4 miRNA families were upregulated and 10 members of 6 miRNA families were down-regulated in response to drought stress in M. truncatula. Among the 29 new miRNAs/new members of known miRNA families, 8 miRNAs were responsive to drought stress with 4 miRNAs each being up- and down-regulated. The known and predicted targets of the drought-responsive miRNAs were found to be involved in diverse cellular processes including development, transcription, protein degradation, detoxification, nutrient status and cross adaptation. Further, a number of novel legume miRNA were also identified in Phaseolus vulgaris. For instance, pvu-miRS1, pvu-miR1514a, miR159.2, pvu-miR2118 and pvu-miR2119 accumulated upon drought and ABA treatments. Novel miRNAs may target regulatory elements for cellular processes that may be unique to legumes.48 Using a different approach, small RNAs were sequenced from two cowpea genotypes (CB46, drought-sensitive, and IT93K503-1, drought-tolerant) that grew under well-watered and drought stress conditions. About 157 miRNA genes that belong to 89 families were identified by mapping small RNA reads to cowpea genomic sequences. Among the 44 drought-associated miRNAs, 30 were up-regulated in drought condition and 14 were down-regulated. Although miRNA expression was in general consistent in two genotypes, 9 miRNAs were predominantly or exclusively expressed in one of the two genotypes and 11 miRNAs were drought-regulated in only one genotype, but not the other.51
In a similar study in soybean, drought tolerant and sensitive genotypes were subjected to drought stress and miRNAs that were differentially expressed characterised. By sequencing drought tolerant and sensitive genotypes as well as rust tolerant and sensitive seedlings, they identified a total of 24 families of novel miRNAs that had not been reported before, six families of conserved miRNAs that exist in other plants species, and 22 families previously reported in soybean.52 They observed the presence of several isomiRNAs during the analyses. A striking feature however was that majority of the miRNAs (miR166-5p, miR169f-3p, miR1513c, miR397ab and miR-Seq13), were up-regulated during water deficit stress in the sensitive plants whilst, for the tolerant genotypes, these miRNAs were down-regulated.52 The miRNAs that were differentially expressed in the tolerant/sensitive genotypes under drought stress may potentially be regulating genes associated with drought tolerance/sensitivity and should be further investigated. Salt stress is also responsible for decline in crop productivity and approximately 6% of the global arable land is affected by excess salt. 53 Several studies have demonstrated that plants express a variety of miRNAs in response to salt stress. 40, 54, 48 Soybean miRNAs searches have also identified some potential candidates. 55, 56 In one study, soybean miRNAs associated with abiotic stresses (drought, salinity, and alkalinity) were identified and analyzed with deep sequencing. One hundred and thirty three conserved miRNAs representing 95 miRNA families were expressed in soybeans under these treatments. 23 Out of these, 71, 50, and 45 miRNAs are either uniquely or differently expressed under drought, salinity, and alkalinity, respectively, suggesting that many miRNAs are inducible and are differentially expressed in response to certain stress. In addition, other genome-wide studiesin Arabidopsis, rice, soybean, maize and Populus have identified salt responsive miRNAs such as miR393, miR394, miR396 and miR156. 45, 57, 58, 23 Recently, the expression profiles of nine different miRNAs were analysed in Phaseolus vulgaris seedlings in response to 0.4 M NaCl and drought stress. The miR395 was most sensitive to both stresses and was up-regulated by 616 and 2810-folds by 1.00% PEG and 0.4 M NaCl, respectively.59 Further, miR396 and miR172 were up-regulated after exposure to both the stresses. The miR396 has been shown to function in leaf development,60 and expression of miR396 has been shown to be induced under high salt, cold, and drought stresses.45 Interestingly, over-expression of miR396 leads to an increased tolerance to drought stress. 61 The authors found that individual miRNA expression profiles varied between the two different stresses, indicating that salt and drought stresses induce differential miRNA expression through different mechanisms, such as oxidative stress or inhibition of plant growth. They also reported that salt and drought conditions induced the expression of APX and ADH, two stress-related plant genes, in Phaseolus vulgaris.
Interestingly, four miRNAs associated with cold tolerance in Arabidopsis (miR319, miR393, miR397, miR402) were analysed for similar role in sweet pea (Pisum sativum). Primers to these miRNAs were designed and their role in pea was investigated using RT-PCR. They showed that miR319, miR393, miR397, and miR402 probably exist in pea, and the level of their expressions increased after the cold treatment. 62 Lu et al 63 found that miR169a and miR169c were substantially down-regulated by drought, leading to the enhanced resistance to drought in Arabidopsis because one of the miR169’s targets, NFYA5 (Nuclear Factor YA5), is a crucial transcription factor regulating the expression of a number of drought stress-responsive genes.
Wang et al. 64 identified 283 and 293 known miRNAs from the control and drought stress libraries, respectively. Under drought stress, they found that there was up-regulation of miR2089 and miR2118, whose targets may be proteins associated with disease resistance. It is envisaged that these miRNAs may enhance the ability of drought tolerance through unknown mechanisms associated with cross adaptation in plants. They identified 32 known members of 10 miRNA families and 8 new miRNAs/new members of known miRNA families that were responsive to drought stress by high-throughput sequencing of small RNAs from M. truncatula.
miRNAs involved in ABA-mediated stress responses
The phytohormone ABA is involved in plant responses to environmental stresses. The first indication that miRNAs may be involved in ABA-mediated responses came from observations of ABA hypersensitivity in an Arabidopsis mutant containing a “pleiotropic recessive Arabidopsis transposon insertion mutation,” hyl1. 65 Recently, two research groups independently found that either ABA or gibberellin (GA) treatment regulated miR159 expression 40, 66 and controlled floral organ development. 40 In germinating Arabidopsis seeds, miR159 was upregulated in ABAtreated seedlings. 67 Sunkar and Zhu reported that the expression of miR393, miR397b and miR402 was upregulated by ABA treatment. In contrast, miR389a appears to be downregulated by ABA.40 Other studies in Arabidopsis have also reported upregulation of miR160 68 and miR417 69and downregulation of miR169 70 in response to ABA. In rice, miR319 was upregulated, whereas miR167 and miR169 were downregulated in ABA-mediated responses.60 In Phaseolus vulgaris, miR159.2, miR393, and miR2118 were induced under ABA treatments whereas miRS1, miR1514, and miR2119 were moderately upregulated in response to ABA.48
miRNAs in biotic stress
In plants, Arabidopsis miR393 was the first miRNA reported to play a role in plant antibacterial PTI (pattern triggered immunity) by regulating the auxin signaling pathway. miR393 was induced by a bacterial PAMP (pattern associated molecular pattern) peptide flg22 post-treatment.71 Using small RNA-expression profiling on Arabidopsis leaves collected at 1 and 3 h postinoculation (hpi) with Pst DC3000 hrcC,19 identified three miRNAs miR160, miR167, andmiR393) that were highly induced and one that was downregulated (miR825) after infection. The role of miRNAs in plant basal defense was further supported by the finding that Arabidopsis miRNA-deficient mutants dcl1 and hen1 showed enhanced growth of the bacterium Pst DC3000 hrcC and several bacteria strains that are non-pathogenic to Arabidopsis, including Pseudomonas syringae pv. phaseolicola, P. fluorescens, and E. coli.72
In a recent study in chickpea plants Based on the significant upregulation of miR530 in response to Fusarium infection and its unique target genes in the chickpea, it appears to be involved in the response to pathogen attack. The three legume-specific miRNAs (miR2111, miR2118 and miR5213) play critical roles during pathogen attack. In the chickpea, miR2111 targets a Kelch repeat-containing F-box protein. F-box proteins are responsible for the controlled ubiquitin-dependent degradation of cellular regulatory proteins and are involved in defense responses, auxin responses and floral organ development.73, 74 Targets of F-box proteins are central regulators of key cellular events and include G1 cyclins and inhibitors of cyclin-dependent kinases.75 It appears that miR2111 and F-box proteins act together to regulate the defense response in chickpea following biotic stress. Other than F-box proteins, miR2111 also targets TIR domain-containing NBS-LRR disease resistance proteins. miR2118 and miR5213 also target the same class of R genes. Interestingly, the chickpea miR2118 was upregulated in response to wilt infection and down regulated following salt stress.76 miR2118 has also been shown to be suppressed after Verticillium fungal attack in cotton.77 Fusarium wilt leads to symptoms that are similar to those of Verticillium wilt, whose common host plant is cotton. miR2118 functions through three novel target transcripts encoding TIRNBS- LRR disease resistance proteins, but its functional regulation remains unclear. In the soybean, miR2118 targets the protein family that is associated with disease resistance in addition to zinc finger proteins 78 and replication termination factor 2 in response to biotic (Asian soybean rust) and abiotic (water deficiency) stresses.52
miRNAs in bacterial pathogenesis
On contact with pathogens, host plants can recognize the nature of the pathogen based on pathogen-associated molecular patterns, and mounta defense response that involves rapid changes in gene, hormone and metabolite levels. miRNAs are also part of such defense mechanisms. Several miRNAs, such as miR393, miR319, miR158, miR160, miR167, miR165/166 and miR159, are upregulated, whereas miR390, miR408 and miR398 are downregulated in Arabidopsis leaves challenged with a virulent form of the bacterium, P. syringae pv. tomato DC3000.79 miR393, miR160 and miR167 were upregulated in leaves challenged with the PstDC3000 hrcC pathogen.19 Similarly, treatment with flagellin-22 induced miR393 expression in Arabidopsis.71 These studies illustrate that miRNAs that target genes involved in auxin perception and signaling are upregulated during the pathogenesis of bacterial infection. The resulting repression of auxin signaling promotes host plant resistance against such infection,80 suggesting that miRNAs play a vital role in protecting plants against pathogenic bacteria.
miRNAs in biological Nitrogen fixation
miRNAs also play critical and diverse roles in symbiotic N fixation. To identify potential regulators early in nodule development, Subramanian et al., 81 inoculated soybean (Glycine max) roots with Bradyrhizobium japonicum and identified differentially expressed miRNAs. For example, miR168 and miR172 were upregulated at 1 or 3 hpi but returned to basal levels by 12 hpi; miR159 and miR393 were upregulated by 3 hpi and continued to maintain these levels to 12 hpi; miR160 and miR169 were downregulated in response to rhizobia. Also, rhizobial infection changed the levels of miR160, miR393, miR164, and miR168, which target ARFs (ARF10, ARF16, and ARF17), TIR1, NAC1, and AGO1, respectively. Li et al, 82 examined the gene expression levels of six families of novel miRNAs and investigated their functions in nodule development in soybean. The results suggested that miR482, miR1512, and miR1515 might have specific and important functions during soybean nodulation. Cloning and sequencing of miRNAs from functional nodules of soybean revealed conserved miRNAs (miR167, miR172, miR396, and miR399), whereas four other families had sequences homologous to Gm-miR1507, GmmiR1508, Gm-miR1509, and Gm-miR1510, which play a role in N fixation.83 In Medicago truncatula, the functions during nodulation have been studied for two conserved miRNAs; miR169 regulated MtHAP2-1, which altered nodule development,84 while overexpression of miR166 reduced the number of symbiotic nodules and lateral roots.85
A complete understanding of the actions of miRNAs depends on the identification of the target genes. Identification of entire sets of miRNAs and their targets will lay the foundation that is needed to unravel the complex miRNA-mediated regulatory networks controlling development and other physiological processes. Given that miRNAs are crucial components of in gene regulatory networks, we believe that a complete understanding of the functions of miRNAs will greatly increase our understanding of plant tolerance to biotic and abiotic stresses. Also, the artificial microRNA (amiRNA)-mediated approach should have broad applicability for engineering multiple stress-responsive genes in crop plants.
In summary, an understanding of post-transcriptional gene regulation by miRNAs under biotic and abiotic stress is crucial for understanding and improving stress tolerance in crop plants. In silico identification of miRNAs in diverse plant species suggests that these miRNAs are conserved across species boundaries. Conservation of these miRNAs implies that they have conserved biological functions. Appropriate manipulation of miRNA target genes should help overcome post-transcriptional gene silencing and, thus, might lead to better expression of engineered traits in transgenic plants.
- Parniske, M. Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol., 2008; 6:763-75.
- Sato, S., Isobe, S., Tabata, S. Structural analyses of the genomes in legumes. Curr Opin Plant Biol., 2010; 13:146-52.
- Varshney, R. K., Song, C., Saxena, R. K., Azam, S., Yu. S., Sharpe, A. G., Cannon, S. et al., Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nature Biotechnology, 2013; 31:240–246.
- Jain, M., Misra, G., Patel, R.K., Priya, P., Jhanwar, S., Khan, A.W., Shah, N., Singh, V.K., Garg, R, et al., A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.). Plant J., 2013; doi: 10.1111/tpj.12173.
- Chen, X. MicroRNA biogenesis and function in plants. FEBS Lett., 2005; 579:5923-31.
- Jones-Rhoades, M. W., Bartel, D. P., Bartel, B. MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol., 2006; 57:19-53.
- Mallory, A. C., Vaucheret, H. Functions of microRNAs and related small RNAs in plants. Nat Genet., 2006; 38 Suppl, S31-6.
- Ramachandran, V., Chen, X. Small RNA metabolism in Arabidopsis. Trends Plant Sci., 2008; 13:368-74.
- Khraiwesh, B., Zhu, J. K., Zhu, J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta., 2012; 1819,:137-48.
- Bartel, D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116: 281-97.
- Sunkar, R., Chinnusamy, V., Zhu, J., Zhu, J. K. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci., 2007; 12: 301-9.
- Allen, E., Xie, Z. Gustafson, A. M., Carrington, J. C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005; 121:207-21.
- Fujii, H., Chiou, T. J., Lin, S. I., Aung, K., Zhu, J. K., A miRNA involved in phosphate starvation response in Arabidopsis. Curr Biol., 2005; 15: 2038-43.
- Zhang, B., Pan, X., Cannon, C. H., Cobb, G. P., Anderson, T. A. Conservation and divergence of plant microRNA genes. Plant J., 2006a; 46,: 243-59.
- Yang, Q., Zhang, R., Horikawa, I., Fujita, K., Afshar, Y., Kokko, A., Laiho, P., Aaltonen, L. A., Harris, C. C. Functional diversity of human protection of telomeres 1 isoforms in telomere protection and cellular senescence. Cancer Res., 2007; 67: 11677-86.
- Zhang, B., Pan, X., Cobb, G. P., Anderson, T. A. Plant microRNA: a small regulator molecule with big impact. Dev Biol., 2006b; 289: 3-16.
- Lu, C., Tej, S. S., Luo, S.,, Haudenschild, C. D. Meyers, B. C., Green, P. J. Elucidation of the small RNA component of the transcriptome. Science. 2005; 309: 1567-9.
- Rajagopalan, R., Vaucheret, H., Trejo, J., Bartel, D. P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006; 20: 3407-25.
- Fahlgren, N., Howell, M. D., Kasschau, K. D., Chapman E. J., Sullivan, C. M., Cumbie, J. S., Givan, S. A., Law, T. F., Grant, S. R., Dangl J. L., Carrington, J. C. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS One. 2007; 2: e219.
- Sunkar, R., Jagadeeswaran G. In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol. 2008; 8: 37.
- Zhao, C. Z., Xia, H., Frazier, T. P., Yao, Y., Bi, Y., Li, Y. P., Li, A. Q., Li, M. J., Zhang B. H., Wang X. J. Deep sequencing identifies novel and conserved microRNAs in peanuts (Arachis hypogaea L.). BMC Plant Biol., 2010; 10: 3.
- Chen, L., Zhang, Y., Ren, Y., Xu, J., Zhang Z., Wang Y. Genome-wide identification of cold-responsive and new microRNAs in Populus tomentosa by high-throughput sequencing. Biochem Biophys Res Commun. 2011; 417: 892-6.
- Li, H., Dong, Y., Yin, H., Wang, N., Yang, J., Liu, X., Wang, Y., Wu J., Li X. Characterization of the stress associated microRNAs in Glycine max by deep sequencing. BMC Plant Biol., 2011; 11:170.
- Varshney, R.K., Ribaut, J.M., Buckler, E.S., Tuberosa, R., Rafalski, J.A., Langridge, P. Can genomics boost productivity of orphan crops? Nat Biotechnol., 2012; 30(12): 1172-1176.
- Carrington, J.C., Ambros, V. Role of microRNAs in plant and animal development. Science. 2003; 301: 336–338.
- Kim, V.N. MicroRNA biogenesis: coordinated cropping and dicing. Nature Reviews:Molecular Cell Biology. 2005; 6: 376–385.
- Fang, Y., Spector, D.L. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Current Biology. 2007; 17: 818–823.
- Kurihara, Y., Takashi, Y., Watanabe, Y. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA. 2006; 12: 206–212.
- Li, J., Yang, Z., Yu, B., Liu, J. Chen, X. Methylation protects miRNAs and siRNAs from a 3’-end uridylation activity in Arabidopsis. Current Biology. 2005; 15: 1501–1507.
- Yang, Z., Ebright, Y. W., Yu, B., Chen, X. HEN1 recognizes 21–24 nt small RNA duplexes and deposits a methyl group onto the 2’ OH of the 3’ terminal nucleotide. Nucleic Acids Research. 2006; 34: 667–675.
- Park, M. Y., Wu, G., Gonzalez-Sulser, A., Vaucheret, H., Poethig, R.S. Nuclear processing and export of microRNAs in Arabidopsis. Proceedings of Natural Academy of Science USA. 2005; 102: 3691–3696.
- Vaucheret, H. Plant ARGONAUTES. Trends Plant Science. 2008; 13: 350–358.
- Baumberger, N., Baulcombe, D.C. Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proceedings of Natural Academy of Science USA. 2005; 102: 11928–11933.
- Brodersen, P., Sakvarelidze-Achard, L., Bruun-Rasmussen, M., Dunoyer, P., Yamamoto, Y.Y., Sieburth, L. Voinnet, O. Wide spread translational inhibition by plant miRNAs and siRNAs. Science. 2008; 320: 1185–1190.
- Sunkar, R., Li, Y.F., Jagadeeswaran, G. Functions of microRNAs in plant stress responses. Trends in Plant Science. 2012; 17:196–203.
- Fowler, S., Thomashow, M. F. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell. 2002; 14: 1675–1690.
- Zhu, J-K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002; 53: 247–273.
- Shinozaki, K., Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot., 2007; 58: 221–227.
- Kawaguchi, R. et al. Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J., 2004; 38: 823–839.
- Sunkar, R., Zhu, J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004; 16: 2001–2019.
- Jones-Rhoades, M. J., Bartel, D. P. Computational identification of plant microRNAs and their targets, including a stress induced miRNA. Mol. Cell. 2004; 14: 787–799.
- Chiou, T.J. et al. Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 2006; 18: 412–421.
- Aung, K. et al. pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol., 2006; 14: 1000–1011.
- Sunkar, R., Kapoor, A., Zhu, J. K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell. 2006; 18: 2051-65.
- Liu, H. H., Tian, X., Li, Y. J., Wu C. A., Zheng, C. C. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA. 2008; 14: 836-43.
- Li, H., Deng, Y., Wu, T., Subramanian, S., Yu, O. Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiol., 2010; 153: 1759-70.
- Trindade, I., Capitao, C., Dalmay, T., Fevereiro, M. P., Santos, D. M. miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta. 2010; 231: 705-16.
- Arenas-Huertero, C., Pérez, B., Rabanal, F., Blanco-Melo, D., De la Rosa, C., Estrada-Navarrete, G., Sanchez, F., Covarrubias,. A, Reyes, J. Conserved and novel miRNAs in the legume Phaseolus vulgaris in response to stress. Plant Mol. Biol., 2009; 70:385–401.
- Zhao, B., Ge, L., Liang, R., Li, W., Ruan, K., Lin, H., Jin Y. Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol Biol., 2009; 10: 29.
- Wang, T., Chen, L., Zhao, M., Tian, Q., Zhang, W. H. Identification of drought responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. BMC Genomics. 2011; 12: 367.
- Barrera-Figueroa, B. E., Gao, L., Diop, N. N., Wu, Z., Ehlers, J. D., Roberts, P. A., , Close, T. J., Zhu, J., Liu, R. Identification and comparative analysis of drought-associated microRNAs in two cowpea genotypes. BMC Plant Biology. 2011; 11:127.
- Kulcheski, F.R., de Oliveira, L.F., Molina, L.G., Almerao, M.P., Rodrigues, F.A. et al. Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genomics. 2011; 12: 307.
- Munns, R., Tester, M. Mechanisms of salinity tolerance. Annu Rev Plant Biol., 2008; 59: 651- 81.
- Lu, S., Sun, Y. H., Chiang, V. L. Stress-responsive microRNAs in Populus. Plant J., 2008; 55: 131-51.
- Zhang, B., Pan, X. Stellwag, E. J. Identification of soybean microRNAs and their targets. Planta. 2008; 229: 161-82.
- Chen, R., Hu, Z., Zhang, H. Identification of microRNAs in wild soybean (Glycine soja). J Integr Plant Biol., 2009; 51:1071-9.
- Ding, D., Zhang, L., Wang, H., Liu, Z., Zhang, Z., Zheng, Y. Differential expression of miRNAs in response to salt stress in maize roots. Ann Bot., 2009; 103: 29-38.
- Gao, P., Bai, X., Yang, L., Lv, D., Pan, X., Li, Y., Cai, H., Ji, W., Chen, Q., Zhu, Y. osa- MIR393: a salinity- and alkaline stress-related microRNA gene. Mol Biol Rep. , 2011; 38: 237-42.
- Nageshbabu, R., Jyothi, M.N. Profile of small interfering RNAs from French Bean Phaseolus vulgaris under abiotic stress conditions. International Journal of Pharma and Bio Sciences. 2013; 4(2): 176 – 185.
- Liu, Q., Zhang, Y. C., Wang, C. Y., Luo, Y.C., Huang, Q. J., Chen, S.Y., Zhou, H., Qu, L.H, Chen, Y.Q. Expression analysis of phytohormone-regulated microRNAs in rice, implying their regulation roles in plant hormone signaling. FEBS Lett.; 2009; 583: 723–728.
- Feng-Xi, Y., Di-Qiu, Y. Overexpression of Arabidopsis Mir396 enhances drought tolerance in transgenic tobacco plants. Acta Botanica Yunnanica, 2009; 31(5): 421-426.
- Wang, Y., Long, L. Identification and isolation of the cold-resistance related miRNAs in Pisum sativum Linn. Journal of Liaoning Normal University (Natural Science Edition), 2010; 2.
- Li, W.X., Oono, Y., Zhu, J., He, X.J, Wu, J.M, Iida, K., Lu, X.Y., Cui, X., Jin, H., Zhu, J.K. The Arabidopsis NFYA5 Transcription Factor Is Regulated Transcriptionally and Posttranscriptionally to Promote Drought Resistance. Plant Cell. 2008; 20: 2238–2251.
- Wang, T., Chen, L., Zhao, M., Tian, Q., Zhang, W. H. Identification of drought responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. BMC Genomics. 2011; 12: 367.
- Lu, C., Fedoroff, N. A Mutation in the Arabidopsis HYL1 Gene Encoding a dsRNA Binding Protein Affects Responses to Abscisic Acid, Auxin, and Cytokinin. Plant Cell. 2000; 12: 2351–2366.
- Achard, P., Herr, A., Baulcombe, D.C., Harberd, N.P. Modulation of floral development by a gibberellin-regulated microRNA. Development. 2004; 131: 3357–3365.
- Reyes, J.L., Chua, N.H. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J., 2007; 49: 592–606.
- Liu, P.P., Montgomery, T.A., Fahlgren, N., Kasschau, K.D., Nonogaki, H., Carrington, J.C. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post germination stages. Plant J., 2007; 52: 133–146.
- Jung, H.J., Kang, H. Expression and functional analyses of microRNA417 in Arabidopsis thaliana under stress conditions. Plant Physiol. Biochem., 2007; 45: 805–811.
- Li, W. X., Oono, Y. Zhu, J., He, X. J. J., Wu, M., Iida, K., Lu, X. Y., Cui, X., Jin H., Zhu, J.K. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell. 2008; 20: 2238-51.
- Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., Estelle, M., Voinnet, O., Jones, J.D. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006; 312:436–439.
- Navarro, L., Jay, F., Nomura, K., He, S.Y., Voinnet, O. Suppression of the MicroRNA Pathway by Bacterial Effector Proteins. Science. 2008; 321: 964–967.
- Xie, D.X., Feys, B.F, James, S., Nieto-Rostro, M., Turner, J.G. COI1: an Arabidopsis gene required for jasmonate regulated defense and fertility. Science. 1998; 280: 1091–1094.
- Samach, A., Klenz, J.E., Kohalmi, S.E., Risseeuw, E., Haughn, G. et al. The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J., 1999; 20: 433–445.
- Craig, K.L., Tyers, M. The F-box: a new motif for ubiquitin dependent proteolysis in cell cycle regulation and signal transduction. Prog Biophys Mol Biol., 1999; 72: 299–328.
- Kohli, D., Joshi, G., Deokar, A.A., Bhardwaj, A.R., Agarwal, M. et al. Identification and characterization of wilt and salt stress-responsive microRNAs in chickpea through high-throughput sequencing. PLOS ONE. 2014; 9: e108851.
- Yin, Z., Li, Y., Han, X., Shen, F. Genome-wide profiling of miRNAs and other small non-coding RNAs in the Verticillium dahliae inoculated cotton roots. PLoS ONE, 2012; 7: e35765.
- Wong ,C.E., Zhao, Y.T., Wang, X.J., Croft, L., Wang, Z.H. et al. MicroRNAs in the shoot apical meristem of soybean. J Exp Bot., 2011; 62: 2495–2506.
- Zhang, W. et al. Bacteria-responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol. Biol., 2011; 75: 93–105.
- Ruiz-Ferrer, V. Voinnet, O. Roles of plant small RNAs in biotic stress responses. Annu. Rev. Plant Biol., 2009; 60: 485–510.
- Subramanian,. S, Fu, Y., Sunkar, R., Barbazuk, W.B., Zhu, J.K, Yu, O. Novel and nodulation-regulated microRNAs in soybean roots. BMC Genomics. 2008; 9:160.
- Li, H., Deng, Y., Wu, T., Subramanian, S., Yu, O. Mis-expression of miR482, miR1512, and miR1515 Increases Soybean Nodulation. Plant Physiol., 2010; 153:1759–1770.
- Wang, Y., Li, P., Cao, X., Wang, X., Zhang, A., Li, X. Identification and expression analysis of miRNAs from nitrogen-fixing soybean nodules. Biochem. Biophys. Res. Commun. 2009; 378:799–803.
- Combier, J.P., Frugier, F., de Billy, F., Boualem, A., El-Yahyaoui, F., Moreau, S., Vernié, T., Ott, T., Gamas, P., Crespi, M., Niebel, A. MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev., 2006; 20: 3084– 3088.
- Boualem, A., Laporte, P., Jovanovic, M., Laffont, C., Plet, J., Combier, J.P., Niebel, A., Crespi, M., Frugier, F. MicroRNA166 controls root and nodule development in Medicago truncatula. Plant J., 2008; 54: 876–887.
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


