ISSN: 0973-7510
E-ISSN: 2581-690X
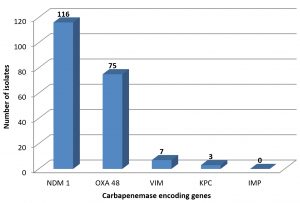
Members of Enterobacteriaceae family are responsible for both community and hospital acquired infections. Because of development of antimicrobial resistance carbapenem has remained as last resort of drug for treatment of infections caused by these bacteria.Mechanism for development of this resistance in carbapenem resistant Enterobacteriaceae (CRE) may due to production of carbapenemases, efflux mechanism or loss of outer membrane porins.The most common carbapenemase enzymes are Class A – KPC, Class B – NDM, VIM and IMP and Class D oxacillinase(OXA-48 like enzymes).In India, most prevalent carbapenemase encoding gene is NDM-1but there is rising threat of OXA-48 prevalence. Unlike the phenotypic methods, the genotypic methods are useful to discriminate the type of carbapenemase enzyme, specifically for OXA-48 like enzymes. Total 170 CRE isolates were subjected for multiplex PCR study for their molecular characterization. Of the 170 CRE isolates,68.2 % (n=116) were positive for NDM-1 gene while 44.1 % (n= 75) of the isolates showed presence of OXA-48 gene. VIM (2.3%), KPC (1.7 %) were responsible for carbapenemase production while none of the isolates showed presence of IMP gene. NDM-1 and OXA-48 coexisted in 21.2 % (n=36) of the total isolates. OXA-48 causes weak hydrolysis of carbapenem because of which it is under reported with routine diagnostic methods. Early detection of OXA-48 and other carbapenemase encoding genes, helps for contact precautions and effective therapy which prevents further escalation and horizontal spread of CRE.
OXA-48, NDM-1, Carbapenem Resistant Enterobacteriaceae, Carbapenemase
Most of microbiome of human intestine consists of Enterobacteriaceae family. The members from this family are responsible for both community and hospital acquired infections which includes wound infections, pneumonia, septicemia, meningitis, peritonitis, pylonephritis, cystitis and various catheter related infections1,2.Because of development of antimicrobial resistance carbapenem has remained as last resort of drug for treatment of infections caused by these bacteria3. But recently a major threat has emerged out as a result of carbapenem resistance leading to therapeutic failure. This has lead to serious healthcare concern especially in association with nosocomial outbreaks. Carbapenem resistance may occur due to production of carbapenemases, efflux mechanism or loss of outer membrane porins4,5.
Carbapenemase enzymes confers resistance to most β-lactam antimicrobials including carbapenem6. Also these isolates are coded with other antimicrobial resistant genes resulting in therapeutic failure. The most common carbapenemase enzymes belongs to three classes : Class A – KPC, Class B – NDM, VIM and IMP and Class D oxacillinase ( OXA -48 like enzymes)6,7.
In India, most prevalent carbapenemase encoding gene is NDM-1 which is been proved in many studies8. But of late the OXA-48 is seen to be closely following the NDM-1 prevalence, though it is more prevalent in Turkey, Morocco, Tunisia, Russia, Germany and other countries compared to India5,9.
Various phenotypic methods are used by many clinical microbiology laboratories for detection of carbapenem resistance10. Major disadvantage of these methods is failure to discriminate the exact type of carbapenemase produced which is very well identified and confirmed by genotypic methods. With development of number of newer resistance mechanisms by bacteria, identifying the most encountered resistance is important in a clinical setting. This will guide for infection control, contact precautions and understanding the prevalent carbapenem resistance mechanism in the geographical area11. In the present study we aimed to identify the common blaNDM1, blaVIM, blaIMP, blaKPC, blaOXA genes which are responsible for worldwide carbapenem resistance in clinical Enterobacteriaceae isolates by multiplex PCR study. Being one of the largest 1125 bedded hospital in this region, the study will provide prevalence of carbapenem resistance pattern from this place.
Type of study
Observational cross-sectional study.
Study population
A total 170 carbapenem resistant Enterobacteriaceae isolates obtained from clinical specimens received for culture and sensitivity in the department of microbiology from June 2016 to May 2018 were included in the study.
Study setting
Department of Microbiology KIMS Karad, Department of Molecular Biology and Genetics, KIMSDU, Karad.
Bacteriology Workup
Standard methodology was used for processing of samples12. All the clinical specimens received during the period were cultured on blood agar,chocholate agar, and MacConkey agar12. The growth in the culture media was identified by the Vitek. ®2 system (BioMerieux, France). Antimicrobial susceptibility testing was done by Vitek®2 system (BioMerieux, France)13. 170 isolates of Enterobacteriaceae were confirmed resistant to any one or all carbapenems tested in the gram negative antimicrobial panel and defined as carbapenem resistant Enterobacteriaceae isolates14.
Molecular Workup
Plasmid Extraction
Plasmid extraction was used for detection of carbapenemase encoding genes15. All 170 carbapenem resistant Enterobactericeae isolates were processed for plasmid extraction. (QIAGEN® Plasmid Mini Kits). The plasmid DNA were stored at -20°C for further use.
Polymerase Chain Reaction
Detection of carbapenemase encoded genes blaNDM-1, blaKPC, blaOXA-48, blaVIM, and blaIMPwas done with subset of primers. Primers used in the study were as shown in Table 1.
Table (1):
Primers used for carbapenemase encoded genes in the study.
| Gene | Forward/ Reverse |
Primer Sequences | Amplicon size (bp) |
Ref. No. |
|---|---|---|---|---|
| NDM-1 | F | 5’-GGGCAGTCGCTTCCAACGGT-3’ | 475 | 16 |
| R | 5’-GTAGTGCTCAGTGTCGGCAT -3’ | |||
| OXA-48 | F | 5’-GCGTGGTTAAGGATGAACAC-3’ | 307 | 17 |
| R | 5’-CGCTCCGATACGTGTAACTT-3’ | |||
| KPC | F | 5’-GCT CAG GCG CAA CTG TAA G-3’ | 150 | 18 |
| R | 5’-AGC ACA GCG GCA GCA AGA AAG-3’ | |||
| VIM | F | 5’ATTGCCGATGGTGTTTGG-3’ | 523 | 19 |
| R | 5’TGGGCCATTCAGCCAGA-3’ | |||
| IMP | F | 5’- GAAGGCGTTTATGTTCATAC -3’ | 587 | 20 |
| R | 5’- GTAAGTTTCAAGAGTGATGC-3’ |
PCR reactions were processed in a Master Cycler Gradient (Eppendorf, India) in 20 µl mixtures containing 2 U Taq polymerase(GeNei, India), 1 X assay buffer consisting of 10 mMTris-HCL(pH 9.0 at 2.5°C),1.5 mM MgCl2, 50 mM KCL and 0.01 % gelatin, each dNTP at a concentration of 200 µM and 10 pmoles of each oligonucleotides primers per reaction.
Initially PCR was carried out to standardize the protocol for individual genes. For this PCR mixture used was as deionized distilled water -15µl ,buffer-2 µl ,dNTP-0.5 µl, forward primer -0.5 µl, reverse primer 0.5 µl, Taq DNA polymerase – 0.5 µl, Template DNA – 1 making total volume 20 µl. The PCR programs used for amplification were as shown in Table 2.
Table (2):
PCR programs used for standardization of PCR.
| Initial Denaturation | Denaturation | Annealing | Extension | Final Extension | |
|---|---|---|---|---|---|
| Class A | |||||
| 1. KPC (150 bp) | |||||
| 950C | 950C | 550C | 720C | 720C | 40C |
| 10 minutes | 30 seconds | 30 seconds | 30 seconds | 10 minutes | ∞ |
| 30 cycles | |||||
| Class B | |||||
| 2.NDM-1 (475bp) | |||||
| 950C | 950C | 550C | 720C | 720C | 40C |
| 10 minutes | 30 seconds | 30 seconds | 30 seconds | 10 minutes | ∞ |
| 30 cycles | |||||
| 3.VIM (523 bp) | |||||
| 950C | 950C | 450C | 720C | 720C | 40C |
| 10 minutes | 30 seconds | 30 seconds | 45 seconds | 10 minutes | ∞ |
| 35 cycles | |||||
| 4. IMP (587 bp) | |||||
| 950C | 950C | 550C | 720C | 720C | 40C |
| 5minutes | 30 seconds | 1 minute | 45 seconds | 10 minutes | ∞ |
| 35 Cycles | |||||
| Class D | |||||
| 5. OXA-48 (307 bp) | |||||
| 950C | 950C | 550C | 720C | 720C | 40C |
| 10 minutes | 30 seconds | 30 seconds | 30 seconds | 10 minutes | ∞ |
| 30 Cycles | |||||
bp – base pair
Klebsiella pneumoniae ATCC BAA- 2146 was used as control for NDM -1 gene and Klebsiella pneumoniae ATCC BAA-1705 was used for KPC21. For VIM and OXA-48, in house control strains confirmed by gene sequencing were used as control during PCR study. For detection of blaIMP conventional PCR method was used. After standardization of PCR, all DNA plasmids were subjected for multiplex PCR study22. (Qiagen Multiplex PCR Kit). This was used for detection of blaNDM-1,blaKPC , blaOXA-48, and blaVIM. For this reaction mixture was prepared as mastermix (Qiagen) -10 µl, primers NDM-1 , KPC ,OXA- 48, VIM – forward and reverse- 0.5 µl each, RNAse free water- 3 µl, Q buffer – 2µl , template- 1 µl making 20 µl total reaction volume. PCR program used to carry multiplex PCR reaction is as shown in Table 3.
Table (3):
PCR program used for Multiplex PCR.
| Initial Denaturation | Denaturation | Annealing | Extension | Final Extension | |
|---|---|---|---|---|---|
| VIM (523 bp), NDM -1 (475 bp), OXA-48 (307bp),KPC (150 bp) | |||||
| 950C | 950C | 450C | 720C | 720C | 40C |
| 10 minutes | 30 seconds | 1 minute | 1 minute | 10 minutes | ∞ |
| 30 cycles | |||||
The amplification product of multiplex PCR were analyzed by electrophoresis in 2.0% agarose gel at 100 V for 1 h in 1X Tris acetate EDTA (TAE) buffer stained with 0.01 mg/ml ethidium bromide. For molecular weight marker 1 Kb DNA ladder was loaded in a gel along with the samples to confirm specific size of the corresponding gene. For staining of the gel ethidium bromide (10mg/ml) was used and UV transilluminator was used for visualization and results were photographed in gel documentation system (Bio- Rad Laboratories).
Table (4):
Genotype of CRE isolates.
| Patterns of Genotype |
NDM -1 | OXA -48 | KPC | VIM | IMP | Number of CRE isolates |
|---|---|---|---|---|---|---|
| 1 | + | – | – | – | – | 74 |
| 2 | + | + | – | – | – | 36 |
| 3 | + | + | + | – | – | 1 |
| 4 | + | – | – | + | – | 5 |
| 5 | – | + | – | – | – | 38 |
| 6 | – | – | + | – | – | 2 |
| 7 | – | – | – | + | – | 2 |
| 8 | – | – | – | – | – | 12 |
| Total | 170 | |||||
DNA Sequencing
DNA sequencing was done for OXA-48 and VIM gene as no control strains were available for the same. Two clinical isolates from the study encoded with likely OXA-48 and VIM gene with PCR amplification of 307 bp fragments and 523 bp fragments respectively were used for sequencing purpose. PCR products were first purified and were sent for DNA sequencing by direct cycle sequencing23. DNA sequencing was carried out using ABI PRISM 310 Analyzer from commercial source by dideoxy nucleotide chain termination method. DNA sequencing of the core region was performed using the sense and antisense primers separately.
The analysis of derived nucleotide sequences for identification of sequence similarity was performed using the NCBI/BLAST network service of National Centre for Biotechnology Information System (NCBI) website (www.ncbi.nlm.nih.gov/BLAST). The obtained DNA sequences from isolates were aligned with representative of the sequences for identification of sequence similarity from gene bank database (NCBI blast search) with the help of nucleotide alignment program. The DNA sequence of both OXA-48 and VIM genes were sent for publication in National Centre for Biotechnology Information (NCBI) database and were assigned with GenBank accession number MK183750.2 and MK183751.2 respectively24, 25.
Statistical Analysis
Data were filled in the MS Excel Software. Analyzed results were expressed as percentage and p values by Chi square test using GraphPad Instat Software. If the probability is less than 0.05, the association or difference is said to be significant.
Of the total 170 CRE isolates,158 (93 %) of the isolates confirmed carbapenemase activity by presence of at least one or more carbapenemase encoded plasmid gene in the multiplex PCR study. Remaining 12 (7%) of the isolates were negative for presence of any one of the gene tested in the multiplex study.
Multiplex PCR study showed presence or absence of different carbapenemase encoded genes. Of the 170 CRE isolates, 116 (68.2 %) had presence of NDM -1 gene.OXA-48 genes were seen in 75 (44.1 %) of the isolates. Only 7 (2.9 %) were positive for VIM gene (Fig. 1). All 170 isolates were negative for presence of IMP gene.
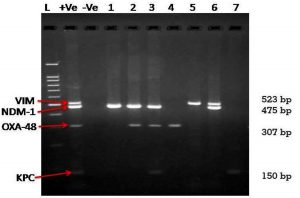
Total 74 CRE isolates showed presence of only NDM-1genes, while 36 isolates were having presence of NDM-1 and OXA-48 like genes (Table 4). Total 48 % of OXA- positive isolates (36/75) were having presence of NDM-1 gene which is extremely significant. (χ2= 22.09 and p< 0.0001). NDM-1 and VIM gene combination was present in 5 of the total 170 CRE isolates. One isolate had presence of NDM, OXA 48 and KPC genes. Twelve isolate were negative for any of the carbapenemase gene studied in the project. Fig. 2 is representative agarose gel image of amplification of VIM- 523bp, NDM-1-475 bp, OXA-48-307 bp and KPC gene-150 bp.
Fig. 2. Multiplex gene specific PCR – Agarose gel image
Lane L: 100 bp DNA ladder, Lane +Ve- Positive control, Lane -Ve-Negative control and Lanes 1 to 8:Clinical isolates for Carbapenem Resistant Enterobacteriaceae family showing presence or absence of carbapenemase encoding genes.
Lane 1- NDM-1 Positive, Lane 2-NDM-1 & OXA-48 Positive, Lane 3- NDM-1,OXA-48 & KPC Positive, Lane 4 –OXA -48 Positive, Lane 5- VIM positive, Lane 6 – VIM & NDM-1 positive, Lane 7 – KPC positive
Members of Enterobacteriaceae got main focus of attention especially around year 2000 because of sudden rise of Extended Spectrum β- Lactamase (ESBL) producing isolates. This resistance scenario got worsened with rapid spread of CTX-M enzyme producing Enterobacteriaceae isolates. But in last decade or so the focus has shifted to the rise of carbapenem resistance caused by carbapenemase producing Enterobacteriaceae isolates which inactivates almost all types of β-lactam antimicrobials26.
Molecular methods are gold standard method for detection of carbapenemase production3. Also it discriminates different types of carbapenemase production unlike phenotypic methods which fails to discriminate the types of carbapenemase3,27. Of the 170 CRE isolates 158 isolates were encoded by at least one carbapenemase producing gene. According to Centre for Disease Control (CDC) definition,remaining 12 isolates were CRE14 but negative for blaNDM-1, blaOXA -48, blaKPC, blaVIM, blaIMP14. These 12 isolates might be having other mechanism for carbapenem resistance like target site mutation, OMP alteration and efflux pumps28. Chromosomal mediated genes like blaIMI, blaSME, blaNMC are also having carbapenem resistant activity29. In the present project, plasmid mediated genes were focused and because of low prevalence of other mechanisms, they were not studied. Multiplex PCR study is one of the reference method for simultaneous detection of genes encoded for carbapenemases of different classes21.NDM-1 has been predominant carbapenemase in Indian CRE isolates8. In a SENTRY Antimicrobial Surveillance Program, 2006-2007 Castanheira M. et al. retrospectively found a prevalence rate of 38.5 % of NDM-1 among Enterobacteriaceae isolates collected from multiple Indian cities30. In one of the pioneer study from India, Kumarasami K. found it as 31.2 % and 55.3 % in isolates from Chennai and Haryana respectively31. A higher prevalence of 91.6 % was found by Deshpande P. et al. in clinical isolates from a tertiary care hospital in Mumbai but the study included only 24 isolates32. In a study from Mumbai Kazi M. et al. have found prevalence of 75.2% in 2012.33In blood stream Enterobacteriaceae isolates Mohanti et al. found 65.6 % of the CRE isolates conferred with NDM-1 gene34. Almost similar but slightly higher that is 68.2 % of the CRE isolates in present study were encoded with blaNDM-1 gene.
Next to NDM-1 gene, multiplex PCR study showed OXA-48 (44.1 %), VIM (2.3%), KPC (1.7 %) responsible for carbapenemase production among 170 CRE isolates in the present study. Mohanti et al. found OXA-48 in 24.7%, OXA-181 in 23.6%; VIM in 6.4%; and KPC in 2.1% of the isolates in their study34. In a study conducted by Khajuria A. et al. found 42 % (19/45) of E.coli isolates showed presence of OXA-48 gene35.
In life threatening or invasive infections carbapenems are most used antimicrobials because of their bactericidal effect which is concentration independent36,37. Also because of their broad spectrum activity which includes action against Gram-positive, Gram-negative bacteria including anaerobes, their use is widespread28. The widespread use, selection pressure of antimicrobial surviving resistant strains, horizontal plasmid spread among Enterobacteriaceae isolates might be contributing factors for raised prevalence of NDM-1 and OXA-48 in the present study. Also being tertiary care centre many patients referred have already received antibiotics, β –lactams being preferred one.
There is variation among predominant carbapenemase encoding gene among CRE isolates around the globe. Pollet S. et al. found 78.3 % of isolates were having blaKPC in a study from USA.38 Predominant gene reported were OXA-48 (86%)39, KPC-2 (54.9%)40 NDM (50 %)41 in studies from Turky39,China40 and Oman41 respectively.
In the present study NDM-1 and OXA-48 coexisted in 21.2 % (36/170)of the isolates Simultaneous presence of blaOXA-48 and blaNDM-1 among E. coli in the study by Khajuria A et al. was found to be (25/300) 8.3%35. Certainly there have been increasing reports of outbreaks and case reports seen across the globe42. This rapid spread of OXA-48 like gene is mediated by plasmid mediated transfer of genetic elements which is seen quite common among Enterobacteriaceae family43. OXA-48 like enzymes causes weak hydrolysis of carbapenems. As a result minimum inhibitory concentration of carbapenems remains on lower side44. Also routine phenotypic methods fails to detect presence of OXA-48 like enzymes. Therefore molecular detection of OXA-48 like genes is a must at present time. Early detection of the OXA-48 and other genes helps for active management and prevents further horizontal spread7.
None of the isolates showed presence of bla IMP in the study. Similar findings were observed by Mohanti et al.34 and Solankhi et al.20 among Indian studies. IMP type MBL gene is endemic in Japan and Taiwan though rest of world does show some sporadic cases45. Similarly VIM is more prevalent in Greece but outbreaks and single reports are from all over world46.
blaNDM-1 carrying clinical isolates is characterized by presence of other resistance factors.29 Total 42 (24.2%) of the isolates positive for NDM-1 were carrying at least one other carbapenemase encoded gene. Mohanti et al. found multiple gene in 39.4 % of the isolates34. Such co-occurrence of multiple carbapenemase gene was detected in 5.9%, 10% and 13% from studies in Sultanate of Oman41,China40and Turkey39 respectively. Higher percentage of multiple gene co-occurrence in NDM-1 strains specifically in combination with OXA- 48 in the present study is a cause of concern.
Though NDM-1 is leading carbapenemase encoded gene in carbapenem resistant Enterobacteriaceae isolates, the spread of OXA-48 is also quite noticeable. As there is no well defined phenotypic method available for detection of OXA-48-like producer emphasis should be on its molecular detection. This will help in contact precautions and to prevent further escalation of carbepenem resistance. Further considering a raised prevalence of coexistence of OXA-48 and NDM-1 gene,a study on role of NDM-1in transfer of plasmid mediated transfer of OXA-48 is warranted.
ACKNOWLEDGMENTS
We thank all the staff at the department of Microbiology, KIMS and at Department of Molecular Biology and Genetics, KIMSDU, Karad for their support.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
SKP and STM designed the research project. SKP performed the bacteriology workup. SKP performed molecular workup along with KDD and MNP. SKP analyzed the data along with SVK. SKP wrote the manuscript. STM supervised and reviewed the manuscript. STM, KDD and SKP approved the manuscript.
FUNDING
Krishna Institute of Medical Sciences, “Deemed to be University”, Karad KIMSDU / DR / 81/2016.
ETHICS STATEMENT
Informal consent was taken before the study from each patient. Study approval was issued and maintained by the Institutional Ethical Committee of KIMSDU, Karad before starting the research project.
AVAILABILITY OF DATA
All datasets generated or analyzed in relation to aims and objective of above research project are included in the manuscript.
- Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med. 2012;18(5):263-72.
Crossref - Uskudar-Guclu A, Guney M, Sig AK, Kilic S, Baysallar M. Arising Prevalence of OXA-48 producer Escherichia coli and OXA-48 with NDM co-producer Klebsiella pneumoniae Strains. Revista Romana de Medicina de Laborator. 2019;27(3):319-26.
Crossref - Datta P, Gupta V, Garg S, Chander J. Phenotypic method for differentiation of carbapenemases in Enterobacteriaceae: Study from north India. Indian J Pathol Microbiol. 2012; 55:357-360.
Crossref - Falagas ME, Lourida P, Poulikakos P, Rafailidis PI, Tansarli GS. Antibiotic treatment of infections due to carbapenem-resistant Enterobacteriaceae: systematic evaluation of the available evidence. Antimicrobial agents and chemotherapy. 2014;58(2):654-656.
Crossref - Kazi M, Khot R, Shetty A, Rodrigues C. Rapid detection of the commonly encountered carbapenemases (New Delhi metallo-β-lactamase, OXA-48/181) directly from various clinical samples using multiplex real-time polymerase chain reaction assay. Indian J Med Microbiol. 2018;36(3):369.
Crossref - Poirel L, Pitout JD, Nordmann P. Carbapenemases: Molecular diversity and clinical consequences. Future Microbiol 2007;2:501-512.
Crossref - Bakthavatchalam YD, Anandan S, Veeraraghavan B. Laboratory detection and clinical implication of oxacillinase-48 like carbapenemase: the hidden threat. J global infect dis. 2016;8(1):41-50.
Crossref - Chatterjee B, Khanduri N, Kakati B, Kotwal A. Universal Presence of blaNDM-1 Gene in Carbapenem-Resistant Gram-Negative Bacilli in an Indian Hospital in 2015. J Clin Diagn Res. 2017;11(9):DL01-DL02.
Crossref - Nordmann P. Carbapenemase-producing Enterobacteriaceae: overview of a major public health challenge. Medicine et Maladies Infectieuses. 2014;44(2):51-56.
Crossref - Pragasam AK, Veeraraghavan B, Bakthavatchalam YD, Gopi R, Aslam RF. Strengths and limitations of various screening methods for carbapenem-resistant Enterobacteriaceae including new method recommended by clinical and laboratory standards institute, 2017: A tertiary care experience. Indian J Med Microbiol. 2017;35(1):116-119.
Crossref - Grundmann H, Glasner C, Albiger B, et al. Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. The Lancet infect dis. 2017;17(2):153-63.
Crossref - Collee JG, Miles RS, Watt B. Tests for identification of bacteria. In Collee JG, Fraser AG, Marmion BP, Simmons A. Mackie and McCartney’s Practical medical microbiology, 14th ed. Churchill Livingstone. 2006.
- Pawar S, Mohite ST, Datkhile K, Patil MN, Durgawale PP, Patil SR. Closing the Gap Between Phenotypic and Genotypic Detection of Carbapenem Resistant Enterobacteriaceae by New Modified Carbapenem Inactivation Method. J Clin Diag Res. 2018;12(11),DC01-DC04.
Crossref - vanDuin, David. “Carbapenem-resistant Enterobacteriaceae: What we know and what we need to know.” Virulence. 2017;8(4):379-82.
Crossref - Pesesky MW, Hussain T, Wallace M, et al. KPC and NDM-1 genes in related Enterobacteriaceae strains and plasmids from Pakistan and the United States. Emerg Infect Dis. 2015;21(6):1034-1037.
Crossref - Deshpande P, Rodrigues C, Shetty A, Kapadia F, Hedge A, Soman R. New Delhi Metallo-lactamase (NDM-1) in enterobacteriaceae: treatment options with carbapenems compromised. J Assoc Physician India. 2010;58:147-149.
- Damavandi MS, Gholipour A, Pour ML. Prevalence of Class D Carbapenemases among Extended-Spectrum β-Lactamases Producing Escherichia coli Isolates from Educational Hospitals in Shahrekord. J Clin Diagn Res. 2016;10(5):DC01-DC05.
Crossref - Shanmugam P,Meenakshisundaram J, JayaramanP. blaKPC gene Detection in Clinical Isolates of Carbapenem Resistant Enterobacteriaceae in a Tertiary Care Hospital. J Clin Diagn Res. 2013;7(12):2736-2738.
Crossref - Ellington MJ, Kistler J, Livermore DM, Woodford N. Multiplex PCR for rapid detection of genes encoding acquired metallo-β-lactamases. J Antimicrob Chemother. 2007; 59(2):321-322.
Crossref - Solanki R, Vanjari L, Sreevidya Subramanian AB, Nagapriyanka E, Lakshmi V. Comparative evaluation of multiplex PCR and routine laboratory phenotypic methods for detection of carbapenemases among Gram negative bacilli. J Clin Diagn Res. 2014;8(12): DC23-DC26.
Crossref - Centers for Disease Control and Prevention. Multiplex real-time PCR detection of Klebsiella pneumoniae Carbapenemase (KPC) and New Delhi metallo-β-lactamase (NDM-1) genes. Atlanta. 2011;500:6-7.
- Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119-123.
Crossref - Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U.S.A. 74(12):5463-5467.
Crossref - Pawar SK, Mohite ST, Datkhile KD, Patil SR, Karande GS. Klebsiella pneumoniae subsp. pneumoniae plasmid OXA family beta-lactamase (blaOXA) gene, partial cds National Centre for Biotechnology Information (NCBI) database.GenBank Accession Number : MK183750.2, 2018.
- Pawar SK, Mohite ST, Datkhile KD ,Patil SR, Karande GS. Klebsiella pneumoniae subsp. pneumoniae plasmid VIM family beta-lactamase (blaVIM) gene, partial cdsNational Centre for Biotechnology Information (NCBI) databaseGenBank Accession Number : MK183751.2, 2018.
- Canton R, Akova M, Carmeli Y, et al. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin Microbiol Infect. 2012;18:413-431.
Crossref - Cuzon G, Naas T, Truong H, et al. Worldwide diversity of Klebsiella pneumoniae that produce β-lactamase blaKPC-2 gene. Emerg Infect Dis. 2010;16:1349-1356.
Crossref - Codjoe FS, Donkor ES. Carbapenem resistance: a review. Medical Sciences. 2018;6(1):1.
Crossref - Diene SM, Rolain JM. Carbapenemase genes and genetic platforms in Gram-negative bacilli: Enterobacteriaceae, Pseudomonas and Acinetobacter species. Clin Microbiol Infect. 2014;20:831-838.
Crossref - Castanheira M, Deshpande LM, Mathai D, Bell JM, Jones RN, Mendes RE. Early dissemination of NDM-1-and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006-2007. Antimicrob Agents Chemother. 2011;55(3):1274-1278.
Crossref - Kumarasamy KK, Toleman MA, Walsh TR, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis.2010;10:597-602.
Crossref - Deshpande P, Rodrigues C, Shetty A, Kapadia F, Hedge A, Soman R. New Delhi Metallo-lactamase (NDM-1) in enterobacteriaceae: treatment options with carbapenemscompromised. J Assoc Physician India .2010; 58:147-149.
- Kazi M, Drego L, Nikam C, et al. Molecular characterization of carbapenem-resistant Enterobacteriaceae at a tertiary care laboratory in Mumbai. Eur J ClinMicrobiol Infect Dis. 2015;34(3):467-472.
Crossref - Mohanty S, Gajanand M, Gaind R. Identification of carbapenemase-mediated resistance among Enterobacteriaceae bloodstream isolates: A molecular study from India. Indian J Med Microbiol. 2017;35(3):421-425.
Crossref - Khajuria A, Praharaj AK, Kumar M, Grover N. Emergence of Escherichia coli, co-producing NDM-1 and OXA-48 carbapenemases, in urinary isolates, at a tertiary care centre at central India. Journal of clinical and diagnostic research: JCDR. 2014;8(6):DC01.
Crossref - Abbott I, Cerqueira GM, Bhuiyan S, Peleg AY. Carbapenem resistance in Acinetobacterbaumannii: laboratory challenges, mechanistic insights and therapeutic strategies. Expert Rev Anti-Infect Ther. 2013;11:395-409.
Crossref - Watkins RR, Bonomo RA. Increasing prevalence of carbapenem-resistant Enterobacteriaceae and strategies to avert a looming crisis. Expert Rev Anti-Infect Ther. 2013;11:543-545.
Crossref - Pollett S, Miller S, Hindler J, Uslan D, Carvalho M, Humphries RM. Phenotypic and molecular characteristics of carbapenem resistant Enterobacteriaceae in a Los Angeles health care system, 2011 to 2013. J Clin Microbiol. 2014;52:4003‑4009.
Crossref - Iraz M, Ozad Duzgun A, Sandalli C, et al. Distribution of β‑lactamase genes among carbapenem‑resistant Klebsiella pneumoniae strains isolated from patients in Turkey. Ann Lab Med. 2015;35:595‑601.
Crossref - Hu L, Zhong Q, Shang Y, et al. The prevalence of carbapenemase genes and plasmid‑mediated quinolone resistance determinants in carbapenem‑ resistant Enterobacteriaceae from five teaching hospitals in central China. Epidemiol Infect. 2014;142:1972‑1977.
Crossref - Dortet L, Poirel L, Al Yaqoubi F, Nordmann P. NDM-1, OXA-48 and OXA-181 carbapenemase-producing Enterobacteriaceae in Sultanate of Oman. Clin Microbiol Infect. 2012;18(5):E144‑148.
Crossref - Antunes NT, Lamoureaux TL, Toth M, Stewart NK, Frase H, Vakulenko SB. Class D β-lactamases: are they all carbapenemases?. Antimicrobial agents and chemotherapy. 2014;58(4):2119-25.
Crossref - Potron A, Poirel L, Rondinaud E, Nordmann P. Intercontinental spread of OXA-48 beta-lactamase-producing Enterobacteriaceae over a 11-year period, 2001 to 2011. Eurosurveillance. 2013;18(31):20549.
- Poirel L, Naas T, Nordmann P. Diversity, epidemiology, and genetics of class D β-lactamases. Antimicrob Agents Chemother. 2010;54(1):24-38.
Crossref - Logan LK, Weinstein RA. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. The J Infect Dis. 2017;215(1-15):S28-S36.
Crossref - Queenan AM, Bush K. Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev. 2007;20(3):440-458.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.