ISSN: 0973-7510
E-ISSN: 2581-690X
Lk2 and Lk3 were thermostable recombinant lipase and highly expressed in Escherichia coli BL21 (DE3). However, Lk2 and Lk3 accumulated as an inclusion body. To further characterize both recombinant lipases, the soluble enzyme must be obtained first. This study aimed to optimize the disruption of the cell membrane in order to obtain soluble and active lipases. The effects of temperature lysis, pH, and SDS concentration on lipolytic activity Lk2 and Lk3 were investigated using a three-factor Box-Behnken design response surface methods. The optimum condition for the temperature variables at 50°C, pH 8, and 0.34% SDS which gave a lipolytic activity of 0.9 U for Lk2. Meanwhile, Lk3 lipolytic activity of 0.9 U obtained at the temperature of 50°C, pH 8, and 0.1% SDS. This result showed efficient one-step membrane disruption methods using thermolysis with addition of a low concentration of detergent at pH 8. The methods used were effective and applicable in the production of active and soluble thermostable recombinant lipase.
Lipase, thermostable, recombinant, inclusion body, thermolysis
Lipases or triacylglycerol acylhydrolases catalyze carboxyl ester bond synthesis and hydrolysis. Various types of synthetic reaction catalysed by lipase, i.e.: esterification, alcoholysis, acidolysis, aminolysis, and interesterification.1,2 Through these widely reaction that could be catalysed by lipase, these enzymes were categorized as important industrial enzymes Lipase is utilized extensively in the dairy, food, flavor, biofuels, pharmaceutical, cosmetics, detergents, leather, and chemical sectors.3,4 Different sources of lipase include animals, vegetables and microbes.5-7 Another important source of lipase is from metagenome. In a metagenomics study, the lipase gene inserts into the vector plasmid and typically transforms into an Escherichia coli host. The source of the lipase gene could originate from any environment such as wastewater, soil, sea sediment, and compost.8
Lk2 and Lk3 are recombinant lipases obtained through metagenome study from domestic compost. These lipases are highly similar to the lipase of Pseudomonas stutzeri categorized as true lipase.9 Lk2 and Lk3 are strongly over expressed on Escherichia coli BL21 (DE3), were thermostable and showed hydrolytic activity on para-nitrophenyl dodecanoate.10 However, high expression level of these recombinant lipase resulting inclusion body (IBs). IBs are misfolded proteins formed in the cytosol bacterial environment. There are two types of IBs which are classic and not classic. Conventional term for recovering active proteins from IBs are made up of four steps: 1. Isolation inclusion body, 2. Solubilization through detergent or other agents, 3. Refolding and 4. Purification of the refolded proteins.11,12
Solubilization and refolding of inclusion body are crucial steps to recover functional recombinant lipase. A variety of recombinant protein solubilized using two step denaturation and refolding.13 Solubilisation using denaturant agent in various pH and activation of certained recombinant lipase by lif protein (lipase specific foldase) or chaperone discussed detailed in the literature.14,15 All of these strategies are based on the OFAT (one-factor-at-a-time) method of optimization, which keeping the other variable constant and changing one variable at a time.16 These conventional methods were time-consuming, therefore require other more efficient methods.
Optimisation employing response surface methodology (RSM) could be used to select effective methods. RSM is a set of mathematical and statistical tools for process optimization. Parameters that affect the process are called independent variables and the dependent variables are called response.17,18 For example, RSM was used to optimize the refolding of recombinant lipase from the inclusion body Escherichia coli.19 The optimization cell disruption methods using the RSM-based Box-Behnken design were done by variable temperature lysis, pH and time. Temperature at 77°C, pH 7.71 with incubation duration in 20-minute were the best values for the variables.20 However, this previous study used the thermolysis technique to disturb the Escherichia coli membrane cell.
The thermolysis method was done by incubating crude lysate of recombinant protein at heat temperature.21 Two previous studies successfully using thermolysis methods for partial purification of recombinant lipase from Geobacillus stearothermophilus strain AH22 and Bacillus pumilus.22,23 Thermolysis is suitable to be used in thermostable protein that can retain activity on high temperature where the other protein can denature. The present study aimed to optimizing cell disruption by using RSM methods based on three factorial models (Box-Behnken design) to recover the active soluble and thermostable recombinant lipase Lk2 and Lk3.
Materials
The LK2 and LK3 lipase genes were previously cloned in the expression vector pET-30a (+) and introduced into the host E. coli BL21(DE3).10 4-nitrophenyl dodecanoate, 2,4-nitrophenol, sodium dodecyl sulphate, TritonTM X-100, and sodium deoxycholate monohydrate were purchased from Sigma (Sigma, Chemicals, USA.). Disodium hydrogen phosphate anhydrous, NaOH, and sodium hydrogen phosphate monohydrate were pure analysis grade from Merck (Merck, Germany). All other chemicals were of the highest commercially available reagent grade.
Methods
Heterologous Expression
Each clone with kanamycin sulfate (50 g/mL) was grown in LB medium. Overexpression was achieved by inducing the cells with 1 mM IPTG and then incubating at 37°C and 150 rpm for 4 hours. Cell pellets collected by centrifugation and kept in -20°C for further methods.
Cell disruption using thermolysis with detergent added
In a 50 mM, pH 8.0 sodium phosphate buffer, the cells were resuspended. Cells expressing recombinant Lk2 and Lk3 were subsequently subjected to membrane cell solubilization by thermolysis with detergent added. This was performed by cells incubated at 50°C in 30 minutes by adding 0.1% detergents. Different detergents were used, ie. Sodium dodecyl sulfate, sodium cholate hydrate and TritonTMX-100. After incubation, the cells were centrifuged for 20 minutes at 11,000 g to separate protein supernatant and cellular debris. SDS PAGE was used to examine the protein profile of the supernatant.
SDS PAGE
SDS-PAGE was done at 110 V on a 12% running gel with SDS Running Buffer.
Protein bands were visualized by using 0.1 percent Commasie Brilliant Blue to stain the gel. The purified enzyme’s molecular mass was estimated using the peqGOLD Protein Marker III (Peqlab) molecular mass standard.
Optimisation of membrane disruption using the response surface methodology
Optimization of membrane disruption has been determined as recovery of soluble recombinant lipase expressed as lipolytic activity (U/mL). This was performed using the surface response method based on Box Benhken (BBD) design. BBD were generated by MINITAB (Trial version 19). Three independent variables included temperature, pH, and SDS concentration (Table 1.). Each experiment’s response value (lypolitics activity) is the average of three triplicates. The F-test score verified the importance of the response surface model. The coefficient of adjustment R2, the determination R2, and the lack of fit were used to assess the quadratic model’s quality. For a better understanding of the correlation between independent variables and responses, the response surface and contour plots were determined.
Table (1):
Variable levels to optimise thermolysis.
| Independent | Variable levels | ||
|---|---|---|---|
| -1 | 0 | 1 | |
| A) Temperature (ºC) | 50 | 70 | 90 |
| B) pH | 8 | 9 | 10 |
| C) SDS (%) | 0.1 | 0.55 | 1 |
Lypolitic activity assay
In a 0.9 mL acetonitrile:ethanol:buffer 1:4:95 substrate solution (v:v:v), added 0.3 mL crude enzyme and incubated at 50°C for 15 minutes. Lipolytic activity was observed by hydrolysis of 4-nitrophenyldodecanoate providing 2,4-nitrophenol which is detectable by spectrophotometry at 405 nm. The quantity of mol pNP released in one minute is one unit of enzyme activity.24
Heterologous Expression and Membrane disruption Optimization
Heterologous expression of LK2 and LK3 lipase genes in Escherichia coli BL21 (DE3) produces a large amount of recombinant lipase (Fig. 1).
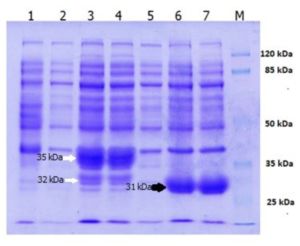
Fig. 1. SDS-PAGE of over expressed Lk2 and Lk3 in E. coli BL21(DE3): [1] pET-30a vector; [2] Lk2 without IPTG induction; [3]&[4] Lk2 with IPTG induction; [5] Lk3 without IPTG induction; [6]&[7] Lk3 with IPTG induction. Lk2 protein bands are indicated by white arrows. Lk3 protein bands are indicated by a black arrow.
The LK2 and LK3 lipase genes have a high degree of homology with the Pseudomonas stutzeri gene according to family 1.1, ranging from 96 to100% 10. The presence of peptide signals in the LK2 gene sequence results in two proteins (Fig. 1, line 3 and 4) that are overexpressed. Based on the in silico analysis, the approximate weight of the fusion protein is 35 kDa that still contains a putative signal peptide, when the region was cleaved resulting 277 amino acid residues corresponded to the protein with molecular weight of 32 kDa. The LK3 gene sequence produces a single protein band of 31 kDa in size.
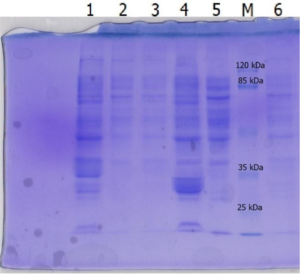
Isolation of recombinant lipase Lk2 or Lk3 by routine ultrasonication method could not overcome the problem of inclusion bodies, this showed by large amount of protein bands in the cell debris (Fig. 2, line 1 and 4). So we did thermolysis with detergent added to get soluble recombinant lipase.
Fig. 2. SDS-PAGE of lysis cell using ultrasonication method; [1] Lk2 cell debris, [2]&[3] Lk2 cell supernatant; [4] Lk3 cell debris, [5]&[6] Lk3 cell supernatant.
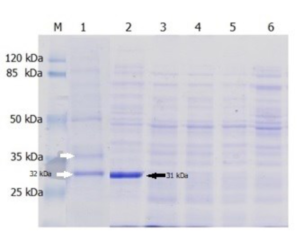
Thermolysis with a 0.1% added detergent was carried out with varies; sodium cholate hydrate, triton X-100 or SDS. The result was only thermolyzing by adding SDS can be used to obtain soluble Lk2 and Lk3 (Fig. 3). Based on this, thermolysis optimization was done using SDS added. SDS classified as strong ionic detergents are capable of lysing the cell within a second.25 However, after the soluble recombinant lipase was obtained, SDS binding protein was precipitated using 30 mM K2HPO4 to prevent denaturation effect.26
Fig. 3. The lysate’s profile on SDS-PAGE: [M] Protein Ladder, [1] LK2 from thermolysis+0.1% SDS at 50 °C, [2] LK3 from thermolysis+0.1% SDS at 50 °C, [3] LK2 from thermolysis+0.1% sodium cholate hydrate at 50 °C, [4] LK3 from thermolysis+0.1% sodium cholate hydrate at 50 °C, [5] LK2 from thermolysis+0.1% triton x-100 at 50 °C, [6] LK3 from thermolysis+0.1% triton x-100 at 50 °C.
Thermolysis has been classified as a nonmechanical (physical) cell lysis method.27 This method was first performed by28 in which, at 80°C, Escherichia coli would release recombinant thermostable esterase. At elevated temperatures, the membrane did not explode completely, but became readily permeable. In this situation, the detergent easily attaches itself with a membrane component like lipopolysaccharides and proteins. The disruption of the cell membrane is therefore efficient.
Membrane disruption optimization based on response surface methodology
To study the optimization variable within the recovery of recombinant lipases Lk2 and Lk3, temperature, pH and SDS concentration were studied to assess their effect on the recovery of soluble recombinant lipase using rsm based on a three level BBD factorial design. A total 15 run experiments and the resulting enzyme activity (U/mL) are presented in Table 2.
Table (2):
Independent variables and response values in a Box-Behnken design.
| Std Order | Run Order | Pt Type | Blocks | Temperature (°C) | pH | SDS (%) | Activity (U/mL) | |
|---|---|---|---|---|---|---|---|---|
| Lk2 | Lk3 | |||||||
| 1 | 1 | 2 | 1 | 50 | 8 | 0,55 | 0.90 | 0.89 |
| 2 | 2 | 2 | 1 | 90 | 8 | 0.55 | 0.54 | 0.63 |
| 3 | 3 | 2 | 1 | 50 | 10 | 0.55 | 0.67 | 0.80 |
| 4 | 4 | 2 | 1 | 90 | 10 | 0.55 | 0.30 | 0.53 |
| 5 | 5 | 2 | 1 | 50 | 9 | 0.1 | 0.82 | 0.86 |
| 6 | 6 | 2 | 1 | 90 | 9 | 0.1 | 0.49 | 0.59 |
| 7 | 7 | 2 | 1 | 50 | 9 | 1 | 0.69 | 0.84 |
| 8 | 8 | 2 | 1 | 90 | 9 | 1 | 0.39 | 0.57 |
| 9 | 9 | 2 | 1 | 70 | 8 | 01 | 0.57 | 0.76 |
| 10 | 10 | 2 | 1 | 70 | 10 | 0.1 | 0.38 | 0.69 |
| 11 | 11 | 2 | 1 | 70 | 8 | 1 | 0.55 | 0.73 |
| 12 | 12 | 2 | 1 | 70 | 10 | 1 | 0.35 | 0.66 |
| 13 | 13 | 0 | 1 | 70 | 9 | 0.55 | 0.52 | 0.70 |
| 14 | 14 | 0 | 1 | 70 | 9 | 0.55 | 0.55 | 0.70 |
| 15 | 15 | 0 | 1 | 70 | 9 | 0.55 | 0.52 | 0.65 |
Multiple regression was used to evaluate the data in Table 2 and was modified to the second order (quadratic) regression model for enzymatic activity, the following equation:
Uncoded Units Regression Equation for Lk2
Activity = 0,85 – 0.04383 Temperature + 0.441 pH + 0.1255 SDS + 0.000252 Temperature*Temperature – 0.0304 pH*pH – 0.1877 SDS*SDS
Uncoded Units Regression Equation for Lk3
Activity = 1,6945 – 0,01067 Temperature – 0,04098 pH – 0,02800 SDS + 0,000029 Temperature*Temperature
The significance of the model for thermolysis optimization Lk2 and Lk3 was confirmed by an analysis of variance (ANOVA) as shown in Tables 3 and 4.
Table (3):
ANOVA for quadratic model of Lk2.
Source |
DF |
Adj SS |
Adj MS |
F-Value |
P-Value |
|---|---|---|---|---|---|
Model |
6 |
0.383704 |
0.063951 |
72.78 |
0.000 |
Linear |
3 |
0.333816 |
0.111272 |
126.63 |
0.000 |
Temperature |
1 |
0.232721 |
0.232721 |
264.84 |
0.000 |
pH |
1 |
0.090488 |
0.090488 |
102.98 |
0.000 |
SDS |
1 |
0.010608 |
0.010608 |
12.07 |
0.008 |
Square |
3 |
0.049888 |
0.016629 |
18.92 |
0.001 |
Temperature*Temperature |
1 |
0.037568 |
0.037568 |
42.75 |
0.000 |
pH*pH |
1 |
0.003409 |
0.003409 |
3.88 |
0.084 |
SDS*SDS |
1 |
0.005333 |
0.005333 |
6.07 |
0.039 |
Error |
8 |
0.007030 |
0.000879 |
||
Lack-of-Fit |
6 |
0.006607 |
0.001101 |
5.21 |
0.170 |
Pure Error |
2 |
0.000422 |
0.000211 |
||
Total |
14 |
0.390734 |
S=0.0296430, R2=98.20%, adjusted R2=96.85%, predicted R2=92.99%
Table (4):
ANOVA for quadratic model of Lk3.
Source |
DF |
Adj SS |
Adj MS |
F-Value |
P-Value |
|---|---|---|---|---|---|
Model |
4 |
0.157647 |
0.039412 |
821.06 |
0.000 |
Linear |
3 |
0.157160 |
0.052387 |
1091.37 |
0.000 |
Temperature |
1 |
0.142453 |
0.142453 |
2967.72 |
0.000 |
pH |
1 |
0.013438 |
0.013438 |
279.95 |
0.000 |
SDS |
1 |
0.001270 |
0.001270 |
26.45 |
0.000 |
Square |
1 |
0.000486 |
0.000486 |
10.13 |
0.010 |
Temperature*Temperature |
1 |
0.000486 |
0.000486 |
10.13 |
0.010 |
Error |
10 |
0.000480 |
0.000048 |
||
Lack-of-Fit |
8 |
0.000454 |
0.000057 |
4.45 |
0.196 |
Pure Error |
2 |
0.000026 |
0.000013 |
||
Total |
14 |
0.158127 |
S=0.0069283, R2=99.70%, adjusted R2=99.58%, predicted R2=99.27%
The high F-value of Lk2 (72.78) and Lk3 (821.06) and the small P-value (0.000) confirmed that both models were significant. Table 3 and Table 4 indicated that the linear terms (temperature, pH and SDS) were important for the activity response. This means that each of the independent variables gives effect to the cell disruption and can be observed based on recombinant lipase activity. The terms cross-interaction between Temperature*pH, Temperature*SDS and pH*SDS have no significant effects and are therefore not included in the final equation of the model. For square interaction in the Lk3 quadratic regression model, the terms pH*pH and SDS*SDS were eliminated as they have no significant effect.
The coefficient of determination (R2) is a goodness-of-fit test used in ANOVA to show how well a statistical model fits a set of data. The range value between 0-1 and the best model closest to 1. According to R2 value for each quadratic model, 98.20% of the total of variance can be explained by the model for Lk2 and 99.70% for Lk3, respectively. The high adjusted R2 also confirm significance of the model.29 The Lk2 and Lk3 regression models have a non-significant lack of fit when compared to the pure error, according to the ANOVA table. This indicates that the model adequately describes the experimental data.30
A final test of model significance was obtained by comparing the experimental response to the predicted response (Fig. 4).
Fig. 4, shows less difference between the clustering of the experimental run and the predicted values. These plots show the validation of the models.
The effect of factors on membrane disruption was studied using response surface analysis
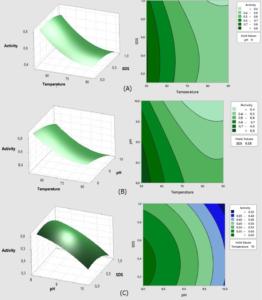
To visualize the interaction effects of the covariates on the response, the amended regression equation was shown as a three-dimensional response area and two-dimensional contour plot. Fig. 5 and 6 indicate the impact of temperature, pH, and SDS concentrations on enzymatic activity response, as well as their reciprocal effects.
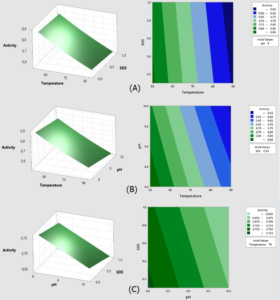
Fig. 5. Surface and contour graphs of the response Lk2; (A) At pH 9, the effect of temperature and SDS content on lipase activity was investigated.; (B) At a concentration of 0.55 percent SDS, the effect of temperature and pH on lipase activity was investigated.; (C) Effect of pH and SDS concentration at temperature 70°C.
Fig. 6. Surface and contour graphs of the response Lk3; (A) At pH 9, the effect of temperature and SDS content on lipase activity was investigated.; (B) At a concentration of 0.55 percent SDS, the effect of temperature and pH on lipase activity was investigated.; (C) Effect of pH and SDS concentration at temperature 70°C.
Fig. 5 demonstrates how raising the temperature range from 50°C to 90°C and the pH from 8 to 10 reduces Lk2 activity. Within the SDS range of 0.1 to 0.6 percent and pH range of 8 to 9 at 50°C, high activity (>0.8 U) is visible.
An Lk3 regression model produced the response surface and contour plot for the interaction variable and activity illustrated in Fig. 4.
The surface and contour graphs in Fig. 4 demonstrate this, the higher the temperature, pH and SDS concentration, the lower the activity of Lipase Lk3. High activity >0.85 U observed at SDS concentrations from 0.1 to 0.2%, pH 8 at 50°C.
The predicted optimal conditions can be calculated statistically from the regression model which was performed. The response optimizer predictions were calculated using Minitab based on the Lk2 and Lk3 regression models. This can be done once we know that the model was valid and significant according to the above result. Response optimization was set to be maximized to achieve optimum activity. Lk2 optimum activity predicted at 0.90 U with independent variable as follows; temperature at 50°C, pH 8 and 0.34% SDS. Meanwhile, for Lk3, the optimum activity was expected to be 0.90 U with temperature variables at 50°C, pH 8 and 0.1% SDS.
The two recombinant lipase Lk2 and Lk3 showed the best lysis condition (lipase activity) at medium temperature at 50°C. Optimization of thermolysis as well as activity measurements were also performed from thermostable cellulase FnCe15A,20 thermostable laccase,31 hyperthermophilic esterase were expressed in E. coli.21 The temperature of these thermolysis studies varied from 60 to 96°C. In the previous reported methods, thermolysis with addition of 0.2% triton-X detergent successfully isolating recombinant steryl glucosidase (SGs) from E. coli host.32 The detergent may bind the hydrophobic and hydrophilic site of the bacterial cell membrane. Detergent can disrupting the lipid-lipid, lipid-protein, and also protein-protein interaction.33 Combination of low concentration of detergent (0.05–1%) with a 0.5–1 mol urea effectively solubilized inclusion body of recombinant enzyme and enhanced the activity.34
This study demonstrated that the optimal pH for lysis cells was at pH 8. In the previous thermolysis study, pH 9 showed an optimal condition for release of recombinant and indigenous Escherichia coli proteins. The pH has affected the charged bacterial cell wall molecule as well as the inclusion of body proteins.35 When the charged species is established, anionic detergent such as SDS attaches easily. This pH and detergent collaborate to permeate cell membrane and solubilized inclusion body.
These results demonstrate the importance of optimizing the thermolysis method with activity determination to achieve soluble and active recombinant thermostable lipase.
In summary, two recombinant thermostable lipases Lk2 and Lk3 can be easily isolated by thermolysis using low concentration detergent. The present study is based on a three-factor Box-Behnken factorial design-response surface methodology to find the best thermolysis condition or maximal activity. By statistical analysis, the optimal conditions of temperature and pH were determined at 50°C and pH 8, where SDS concentration 0.34% for Lk2 and 0.1% for Lk3, respectively. The maximum lipolytic activity of 0.9 U for Lk2 and Lk3 can be achieved under the abovementioned conditions. Isolation and solubilization of recombinant thermostable lipase as an inclusion body could easily be achieved by thermolysis with the addition of a low concentration of SDS, which could be directly applied in biocatalytic or purified first.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
All authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication. All authors read and approved the final manuscript for publication.
FUNDING
This work is supported by SIMLITABMAS research project program, Ministry of Research, Technology and Higher Education with grant No. 2/E1/KP.PTNBH/2021 and scholarship to TH from SMARTD Indonesian Agency for Agricultural Research and Development, DIPA No. SP DIPA- 018.09.1.411971/2016.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Rahman R, Salleh AB, Basri M. Molecular and structural biology of new lipases and proteases. 2013:1-212.
- Sharma R, Chisti Y, Banerjee UC. Production, purification, characterization, and applications of lipases. 2001;19(8):627-662.
Crossref - Javed S, Azeem F, Hussain S, et al. Bacterial lipases: A review on purification and characterization. Prog Biophys Mol Biol. 2018;132:23-34.
Crossref - Mohamed SA, Abdel-Mageed HM, Tayel SA, El-Nabrawi MA, Fahmy AS. Characterization of Mucor racemosus lipase with potential application for the treatment of cellulite. Process Biochem. 2011;46(3):642-648.
Crossref - Chandra P, Enespa, Singh R, Arora PK. Microbial lipases and their industrial applications: A comprehensive review. Microbial Cell Factories BioMed Central. 2020;19:1-42.
Crossref - Adetunji AI, Olaniran AO. Production strategies and biotechnological relevance of microbial lipases: a review. Brazilian J Microbiol. 2021;52(3):1257-1269.
Crossref - Fahmy AS, Abo-Zeid AZ, Mohamed TM, Ghanem HM, Borai IH, Mohamed SA. Characterization of esterases from Cucurbita pepo cv.”Eskandrani.” Bioresour Technol. 2008;99(2):437-443.
Crossref - Almeida JM, Alnoch RC, Souza EM, Mitchell DA, Krieger N. Metagenomics: Is it a powerful tool to obtain lipases for application in biocatalysis? Biochim Biophys Acta – Proteins Proteomics. 2020;1868(2):140320.
Crossref - Nurhasanah N, Nurbaiti S, Madayanti F, Akhmaloka A. Diversity Of Gene Encoding Thermostable Lipasefrom Compost Based On Metagenome Analysis. 2015.
- Nurhasanah, Nurbaiti S, Madayanti F, Akhmaloka. Heterologous expression of gene encoded thermostable lipase and lipolytic activity. J Pure Appl Microbiol. 2017;11(1):135-139.
Crossref - Pontrelli S, Chiu T-Y, Lan EI, Chen FY-H, Chang P, Liao JC. Escherichia coli as a host for metabolic engineering. Metab Eng. 2018;50:16-46.
Crossref - Singh A, Upadhyay V, Upadhyay AK, Singh SM, Panda AK. Protein recovery from inclusion bodies of Escherichia coli using mild solubilization process. Microb Cell Fact. 2015;14(1):1-10.
Crossref - Yang Z, Zhang L, Zhang Y, et al. Highly Efficient Production of Soluble Proteins from Insoluble Inclusion Bodies by a Two-Step-Denaturing and Refolding Method. PLoS One. 2011;6(7):e22981.
Crossref - Haddad L, Babaeipour V, Mofid MR. The effect of cell disruption techniques and chaotropic agents on the downstream purification process of mecasermin produced as inclusion body in E. coli. Res Pharm Sci. 2015;10(6):553-561.
Crossref - Ogino H, Inoue S, Akagi R, Yasuda M, Doukyu N, Ishimi K. Refolding of a recombinant organic solvent-stable lipase, which is overexpressed and forms an inclusion body, and activation with lipase-specific foldase. Biochem Eng J. 2008;40(3):507-511.
Crossref - Packiam KAR, Ramanan RN, Ooi CW, Krishnaswamy L, Tey BT. Stepwise optimization of recombinant protein production in Escherichia coli utilizing computational and experimental approaches. Appl Microbiol Biotechnol. 2020;104(8):3253-3266.
Crossref - Ko B, Kaymak-Ertekin F. Response surface methodology and food processing applications. 2010.
- Montgomery DC. Design and analysis of experiments [Internet]. Second edition. New York : Wiley. [1984] ©1984; 2005. https://search.library.wisc.edu/catalog/999529863402121
- Akbari N, Khajeh K, Ghaemi N, Salemi Z. Efficient refolding of recombinant lipase from Escherichia coli inclusion bodies by response surface methodology. Protein Expr Purif. 2010;70(2):254-259.
Crossref - Mohammad SF, Feng Y, Yang G. Optimization of cell culture and cell disruption processes to enhance the production of thermophilic cellulase FnCel5A in E.coli using response surface methodology. PLoS One. 2019;14(1):e0210595.
Crossref - Ren X, Yu D, Han S, Feng Y. Thermolysis of recombinant Escherichia coli for recovering a thermostable enzyme. Biochem Eng J – Biochem ENG J. 2007;33(1):94-98.
Crossref - Ekinci AP, Dincer B, Baltas N, Adıguzel A. Partial purification and characterization of lipase from Geobacillus stearothermophilus AH22. J Enzyme Inhib Med Chem. 2016;31(2):325-331.
Crossref - Laachari F, El Bergadi F, Sayari A, et al. Bacillus pumilus susundan elde edilen yeni termostabil lipazın biyokimyasal karakterizasyonu. Turkish J Biochem. 2015;40(1):8-14.
Crossref - Lee DW, Koh YS, Kim KJ, et al. Isolation and characterization of a thermophilic lipase from Bacillus thermoleovorans ID-1. FEMS Microbiol Lett. 1999;179(2):393-400.
Crossref - Brown RB, Audet J. Current techniques for single-cell lysis. J R Soc Interface. 2008;5 Suppl (Suppl 2):S131-S138.
Crossref - Zilionis A. Removal of sodium dodecyl sulfate from protein samples. Chemija. 2018;29(4).
Crossref - Geciova J, Bury D, Jelen P. Methods for disruption of microbial cells for potential use in the dairy industry – A review. Int Dairy J. 2002;12(6):541-553.
Crossref - Ren X, Yu D, Yu L, Gao G, Han S, Feng Y. A new study of cell disruption to release recombinant thermostable enzyme from Escherichia coli by thermolysis.
J Biotechnol. 2007;129(4):668–73.
Crossref - Jin ML, Wang YM, Huang M, Lu ZQ, Wang YZ. Optimization of culture medium for exopolysaccharide production by enterobacter cloacae Z0206 using response surface methodology. Asian J Chem. 2011;23(9):3799-802.
- Ekpenyong M, Antai S, Asitok A, Ekpo B. Response surface modeling and optimization of major medium variables for glycolipopeptide production. Biocatal Agric Biotechnol. 2017;10:113-21.
Crossref - Koschorreck K, Wahrendorff F, Biemann S, Jesse A, Urlacher VB. Cell thermolysis – A simple and fast approach for isolation of bacterial laccases with potential to decolorize industrial dyes. Process Biochem. 2017;56:171-176.
Crossref - Eberhardt F, Aguirre A, Paoletti L, et al. Pilot-scale process development for low-cost production of a thermostable biodiesel refining enzyme in Escherichia coli. Bioprocess Biosyst Eng. 2018;41(4):555-564.
Crossref - Shehadul Islam M, Aryasomayajula A, Selvaganapathy PR. A Review on Macroscale and Microscale Cell Lysis Methods. Micromachines. 2017;8(3):83.
Crossref - Mohammadian A, Kaghazian H, Kavianpour A, Jalalirad R. Solubilization of inclusion body proteins using low and very low concentrations of chemicals: implications of novel combined chemical treatment designs in enhancement of post-solubilization target protein purity and biological activity. J Chem Technol Biotechnol. 2018;93(6):1579-1587.
Crossref - Falconer RJ, O’Neill BK, Middelberg AP. Chemical treatment of Escherichia coli. II. Direct extraction of recombinant protein from cytoplasmic inclusion bodies in intact cells. Biotechnol Bioeng. 1998;57(4):381-386.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.