This article reviews the developments related to Streptomyces chitinases regulation and their post translational modifications. Chitinases are enzymes which cleave chitin, a polymer of N-acetylglucosamine to its monomer. Bacteria produce chitinases to fulfil their nutritional needs since by-products of chitin degradation can serve as a source of carbon and nitrogen. Chitinolytic bacteria such as Streptomyces produce multiple chitinases which act synergistically to degrade crystalline form of chitin. Streptomyces are one of the major producers of chitinases in the soil. Every Streptomyces genome sequenced till date has multiple genes for chitinases. The chitinases resulting from proteolytic cleavage have different specific activities and binding efficiency. Both of the above mentioned factors contribute to complexity of the chitinolytic system. Two component systems (TCS) are the predominant signal transduction system in bacteria that regulate a wide variety of behaviours as well as fundamental processes such as metabolism and motility. Bacteria generally use two-component signal transduction pathways to couple environmental stimuli to adaptive responses. Apart from the generalized behaviours they also regulate specialised processes such as development and virulence. Thus this review focuses on the two component systems of Streptomyces, their mechanism of action, regulation of chitinases by TCS and post-translational modifications.
Streptomyces, Chitinase, Two-component systems, Glycosylation, Proteolytic cleavage
Chitinases are widely distributed in various organisms ranging from bacteria 1 to humans 2. These are enzymes that catalyze the hydrolysis of chitin, which is a b-1, 4 linked homopolymer of N-acetyl-D-glucosamine. Chitin is the second abundant polysaccharide found in nature, with the most abundant being cellulose 3, 4. Streptomyces are the main decomposers of chitin present in soil 5 since they can use chitin as a source of carbon and nitrogen, thereby playing a significant role in the turnover of chitin 6. Chitin was once considered a waste product, however, reports on applications of chitin and chitin derived products have emerged and so it attracts a special interest as a reusable material 7.
Chitinolysis is performed by three separate enzymes namely endochitinases which produce multimers of N-acetlyglucosamine (NAG), exochitinases which produce low molecular weight chitobiose that are subsequently hydrolyzed to NAG by chitobiases 8. Multiple chitinase encoding genes are distributed at different locations on the chromosomes of S. coeliocolor 9, S. avermitilis 10 and S. griseus 11. The location of various chitinase genes of three Streptomyces species is shown in table 1 A, B and C. Number of chitinase genes is quite high in S. coelicolor 9 in comparison to other chitinase producing bacteria such as Bacillus circulans 12 and Serratia marcescens 13.
Table (1):
Chitinase genes present in the genome of S. avermitilis10, S. griseus 11 and S. coelicolor 9.
| S. no | Enzyme/gene | Location on the chromosome (base) | No. of amino acid residues | |
|---|---|---|---|---|
| Start | End | |||
| 1. | Chitinase A | 2132343 | 2134028 | 561 |
| 2. | Chitinase B | 3177737 | 3179566 | 609 |
| 3. | Chitinase C precursor | 3522979 | 3524802 | 607 |
| 4. | Chitinase A | 4054020 | 4055759 | 579 |
| 5. | Endochitinase | 6815383 | 6816435 | 350 |
| 6. | Chitinase | 8252433 | 8253716 | 427 |
B
| S. no | Enzyme/gene | Location on the chromosome (base) | No. of amino acid residues | |
|---|---|---|---|---|
| Start | End | |||
| 1. | Chitinase II | 2177892 | 2179715 | 607 |
| 2. | Chitinase III | 2559778 | 2561649 | 623 |
| 3. | Putative chitinase | 2898151 | 2899548 | 465 |
| 4. | Chitinase I | 2990179 | 2991903 | 574 |
| 5. | Chitinase C | 3985104 | 3985988 | 294 |
| 6. | Putative chitinase | 3986289 | 3987110 | 273 |
| 7. | Putative chitinase | 7176841 | 7179198 | 785 |
| 8. | Putative chitinase | 7195071 | 7196342 | 423 |
| 9. | Putative chitinase | 7757174 | 7758571 | 465 |
| 10. | Putative chitinase | 8039719 | 8040786 | 355 |
C
| S. no | Enzyme/gene | Location on the chromosome (base) | No. of amino acid residues | |
|---|---|---|---|---|
| Start | End | |||
| 1. | Secreted chitinase | 5023432 | 503076 | 244 |
| 2. | Chitinase | 1524785 | 1526038 | 417 |
| 3. | Chitinase precursor | 1539609 | 1541984 | 791 |
| 4. | Chitinase precursor | 2701242 | 2702318 | 358 |
| 5. | Chitinase A precursor | 5439963 | 5441678 | 571 |
| 6. | Chitinase C | 5845252 | 5847081 | 609 |
| 7. | Secreted chitinase | 6172804 | 6174636 | 610 |
| 8. | Chitinase (Sec. protein) | 6524142 | 6526439 | 765 |
| 9. | Secreted chitinase | 6593034 | 6594557 | 507 |
| 10. | Secreted chitinase | 7003454 | 7004212 | 252 |
| 11. | Secreted chitinase | 8030688 | 8031422 | 244 |
| 12. | Chitinase | 8073282 | 8074172 | 296 |
Streptomyces chitinases display a modular structure with domains organized in following order from the N-terminus: signal peptide, substrate-binding domain, fibronectin-type III like domain and a catalytic domain 14. A signal peptide for chitinase secretion is usually present along with the catalytic domain. The catalytic domain is necessary for hydrolysis of b-1, 4 glycosidic bonds linking the N-acetyl glucosamine subunits of chitin 15. Chitinases are not the only enzymes having accessory domains in addition to catalytic domains, but several carbohydrases such as amylases, cellulases, xylanases and pectinases also possess additional domains 14. The importance of chitin binding domains has been demonstrated only in a few bacterial chitinases 16, 17.
Chitinases undergoing post-translational modifications have been reported from Streptomyces and other bacteria 12, 18. The most common post-translational modification observed in chitinases of Streptomyces is proteolytic processing. Proteolytic cleavage contributes to multiplicity of chitinases to a larger extent in Streptomyces 19 and S. marcescens 20. Glycosylation, a complex co- or post-translational modification has also been identified in the proteins of archea and bacteria. Among Streptomyces, glycosylated chitinases have been reported to occur in S. olivaceoviridis and S. griseus 21, 22. Apart from Streptomyces and Cellulomonas, glycosylated chitinases have also been identified in plants 23 and animals 24. The role of proteolytic processing of chitinases is known but a clear understanding with respect to the significance of glycosylation in chitinases in bacteria is still lacking. However, the most speculated role for glycosylation is that it protects the proteins from proteases. This property has been demonstrated for proteins such as cellulase of Cellulomonas fimi 25 and xylanase of S. lividans 26 but not in the case of chitinases.
Chitin and its importance
Chitin is an essential structural component of the fungal cell wall and is also present in the exoskeleton of arthropods and the micro-filarial sheath of nematodes. Chitin acts as a protective layer against the harsh conditions that may be endured by the pathogen or arthropod. It is also the second most abundant glycol-polymer on earth, with an estimated 1010 tonnes of chitin produced each year next only to cellulose 27. Chitin is composed largely of alternating by b- (1-4)-glycosidic bonds. Crystalline chitin occurs in three forms a-, b-, g- chitin. The variation lies in the degree of hydration, size of unit cell and number of chitin chains per unit cell 28, 29. a-chitin the most abundant form of chitin is a component of the fungal cell wall and arthropod exoskeleton, has very high tensile strength 30, 31. b-chitin occurs less frequently in nature 30. Chitin is also a component of spores produced by certain Streptomyces 31. It is structurally identical to cellulose except that chitin has acetamide groups at the C-2 position. Although both cellulose and chitin have b- (1-4)-glycosidic bonds but they differ in properties. Chitin is hydrophobic whereas cellulose is hydrophilic 32. Chitin contains 5-8% nitrogen mostly in the form of primary aliphatic amino groups 32. Chitin derived products have found immense application in field of medicine. Chitin and its derivatives have been used in wound healing and tissue regeneration 32, 33, antioxidant properties have been demonstrated and can also be used as carriers in drug delivery. Use of chitin and chitosans as immunoadjuvants and non-allergic carriers of drugs has also been shown 34. Chitin and its by-products have shown anti-microbial activity against P. aeruginosa besides applications in agriculture, detoxification and biotechnology.
Multiplicity of chitinases in Streptomyces
Most chitinase producing organisms produce multiple forms of chitinases. The isoforms of chitinases could result from products of multiple genes, post-translational proteolytic processing. Multiple chitinase genes have been identified in Serratia marcescens 13, Aeromonas sp. 7, P. aeruginosa 35, B. circulans 12 and Streptomyces sps 14, 36, 37. Synergy between multiple chitinases is assumed to be important for effective chitin degradation 38. Table 2 lists the Streptomyces species producing multiple chitinases. Alteromonas is a proficient degrader of chitin in the marine environments. Its capability is achieved by the production of four chitinases. All the four chitinases have varied substrate preferences and are induced in the presence of chitin. However, there was a variation in the level of transcripts relatively 38. The results of Orikoshi et al., (2005) indicated that ChiA plays a central role in chitin degradation process. Unlike Alteromonas, Pseudoalteromonas is also a marine bacterium that produces multiple chitinases showing varied preferences to substrates 39. ChiC of Pseudoalteromonas could be expressed in E. coli from its own promoter 39. S. coelicolor too has genes for multiple chitinases. It produces both family 18 and family 19 chitinases, they show diversity in the multiple domain structures and also in their sequences 15, 36.
Table (2):
Streptomyces expressing multiple chitinases.
| S. no | Streptomyces spp. | Chitinase genes | References |
|---|---|---|---|
| 1. | S. lividans | chiA | 104 |
| chiB | 14 | ||
| chiC | 131 | ||
| 2. | S. olivaceoviridis | chiO1 | 127 |
| chi92 | 127 | ||
| 3. | S. thermoviolaceus | chi40 | 113, 132, 133 |
| chi30 | |||
| chi35 | |||
| chi25 | |||
| 4. | S. griseus | chiC | 14, 128, 134 |
| chiI | |||
| chiII | |||
| chiIII | |||
| 5. | S. peucetius | chiC | 1 |
| chiA | |||
| 6. | S. coelicolor | chiA | 15, 36 |
| chiB | |||
| chiC | |||
| chiD | |||
| chiE | |||
| chiF | |||
| chiG | |||
| chiH |
Two-component signal transduction systems (TCS)
Streptomyces are soil bacteria and thus need to adapt to a wide range of environmental conditions. To accomplish this they need to monitor these conditions and respond to changes accordingly. In bacteria sensing and adapting is mediated by two-component signal transduction systems (TCS) 40, 41. Two-component systems confer the ability in microorganisms to adapt to changes in the environment by modifying the expression levels of genes in response to various stimuli 42. This adaptation can include secretion of degradative enzymes, motility, virulence and changes that result in the modification of cell wall. Two-component systems function as intracellular information processing pathways that link external stimuli to specific adaptive responses.
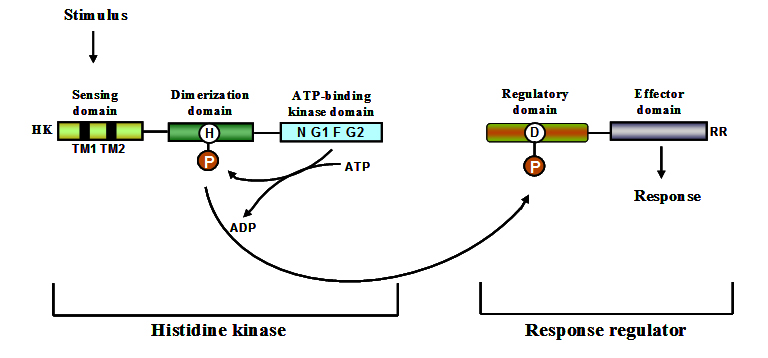
The prototypical two-component regulatory system is composed of a sensor kinase (SK) and a response regulator (RR) 43. Both sensor kinases and response regulators are modular proteins containing highly conserved modules. Sensor kinase consists of a variable N-terminal input (sensing) domain connected to a conserved C-terminal domain. A typical response regulator consists of a conserved N-terminal receiver domain and a variable C-terminal output domain. Two-component systems sense the signal through a membrane localized sensor kinase which undergoes phosphorylation at a specific histidine residue upon recognition of an appropriate signal. ATP acts as the phosphate donor in the phosphorylation process. The phosphorylated histidine residue transphosphorylates an aspartate residue located in its cognate response regulator. A scheme depicting the various steps involved in sensing and responding process is shown in figure 1. The control of longevity of the cellular response must be long enough for an effective response 44 but timely termination is necessary for the cell to adjust its behaviour as conditions change. Hydrolysis of the phosphoryl group from the response regulator resets the system to respond to additional stimuli. The process of hydrolysis can be performed by autophosphatase activity of RR 45, or phosphatase activity of SK 46, 47 or it can also be mediated aspartate phosphatases 48, 49. Through this process, the phosphate group is transferred to an aspartate of a response regulator and finally released as inorganic phosphate. Phosphorylated response regulator binds to regions upstream of genes they regulate, often leading to enhanced target expression or to a reduced expression in few cases. Complex TCS also exist in which a histidine phosphotransferase mediates the transfer of phosphate from sensor kinase to the response regulator. Functioning mechanism of a complex TCS in which a sensor kinase is located in the membrane and the phosphotransfer reaction is mediated by a histidine phosphotransferase is shown as a scheme in figure 1. Through the mechanism of phosphorylation and phosphotransfer, bacteria use TCS to translate external signals into changes in gene expression, facilitating responses to environmental stimuli.
Although auto dephosphorylation of response regulator has been reported 45, in many cases it occurs through catalysis by another protein. Some SKs possess phosphatase activity towards their response regulator. Since sensor kinase and response regulator communicate via transphosphorylation reaction, this reaction requires the formation of precise but transient complex between the phosphorylation domain of SK and phospho-acceptor domain of RR 50.
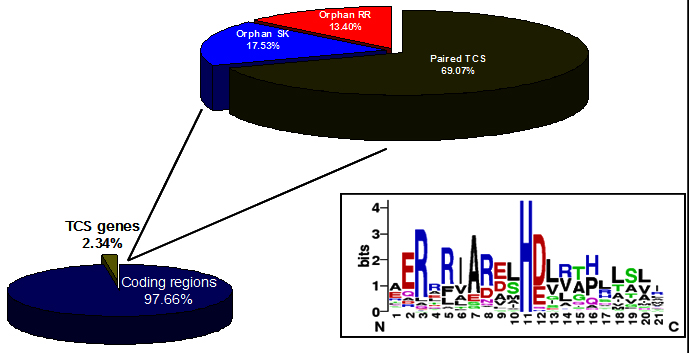
In S. coelicolor, the total number of nucleotides from TCS genes takes up approximately 2.34% of the whole chromosome (Fig. 2). Based on the previous conclusion that bacteria distributed in diverse ecological niches, tend to encode larger number of TCS than those living in limited or obligatory environments 51, 52. The total number of TCS proteins encoded by a bacterial genome, together with other signalling proteins, can be used as a measure of the adaptive potential of the organism (i.e., the bacterial intelligence quotient or “IQ”) 52. The genome sequence of S. coelicolor identified 84 genes encoding sensor kinase like proteins of which 67 are located adjacent to response regulator genes 9, 53. Except for a few proteins most of them still remain uncharacterised. The signals that activate the sensor kinases in these systems, the molecular details of phosphorelay reaction and sites in target DNA to which activated response regulators bind are defined only for a few TCS.
Fig. 1. Schematic representation of the conserved domains present in sensor kinases and re-sponse regulators 146
Fig. 2. Distribution of Two-component systems in genome of S. coelicolor. Box showing the highly conserved histidine. (http://www.p2cs.org) 147
Sensor kinases
Sensor kinases are also referred to as sensor histidine kinases. In two-component systems, they function as receptors for stimuli and as regulators that control the activity of downstream signalling components via phosphorylation. From an enzymology perspective, sensor kinases are interesting because many of them participate in three distinct but related reactions such as autophosphorylation, phosphotransfer and dephosphorylation. Sensor kinases are modular proteins with distinct structural domains playing different functional roles. Most of them have an amino terminal sensor domain which is stimulus specific and not highly conserved at the sequence level 54. The sensor domain involved in stimulus perception spans the membrane. The sensing domain is connected to a conserved cytoplasmic domain which has dimerization domain and phosphate accepting histidine (DHp). The DHp domain is in turn connected to catalytic and ATP binding domain (CA domain). A number of sensor kinases from Streptomyces have been identified and their functioning mechanism studied. Table 3 lists the identified SKs of Streptomyces and the method used for their characterization.
Table (3):
Characterized sensor kinases from Streptomyces.
SK |
Organism |
Approaches used to characterize |
Function regulated |
References |
|---|---|---|---|---|
AbsA1 |
S. coelicolor |
Genetic, Biochemical |
Antibiotic production |
135-137 |
SenS |
S. reticuli |
Genetic, Biochemical |
Production of catalase-peroxidase |
138, 139 |
ChiS |
S. coelicolor, S. thermoviolaceus |
Genetic |
Chitinase |
113, 114 |
EcrA1 |
S. coelicolor |
Genetic |
Antibiotic production |
140 |
VanS |
S. coelicolor |
Genetic, Biochemical |
Resistance to vancomycin |
141 |
PhoR |
S. lividans |
Genetic |
Alkaline phosphatase and antibiotics |
142 |
RapA1 |
S. coelicolor |
Genomic, Proteomic |
Antibiotic production |
143 |
CseC |
S. coelicolor |
Genetic |
Sigma factor |
144 |
AfsQ1 |
S. coelicolor S. lividans |
Genetic |
Secondary metabolites |
145 |
Sensor kinases can be categorised into two major groups. One group comprises the SKs of classical two-component signalling systems in which the kinase domain is at the carboxyl terminus of the protein. The other group comprises of hybrid sensor kinases which also contain a response regulator domain along with the kinase domains. Sensor kinases function as dimeric proteins that undergo autophosphorylation on a conserved histidine residue in response to specific stimuli 55. They consist of an ATP binding kinase domain and a motif H-box containing the histidine residue which gets phosphorylated. The kinase domain consists of three conserved motifs namely, N, F, and G boxes 56. Modular nature of sensor kinases and response regulators is shown in figure 1.
Based on the organisation of H-box and kinase domains as well as on differences in the amino acid sequences, five types of SKs were identified. Type I SKs predominate in bacteria where as in archea type II SKs predominate. Type III SKs predominate in gram-positive bacteria. Type IV were a minor type found in bacteria. Chemosensory sensor kinases belonged to type V 57. All bacterial genomes sequenced till date contain SKs with mycoplasma being the only exception 58. The number of SKs seemed to increase as the genome size increases 59. Generally free living bacteria possess larger genomes when compared to pathogenic bacteria. Thus, the number of SKs was more in free living bacteria in comparison to the pathogenic forms 57.
Response regulators
Most of the signal transduction systems in bacteria are based on the central phosphate transfer mechanism involving two-components, a sensor kinase and a response regulator 41. A response regulator is a two domain protein having N-terminal conserved receiver domain and a variable effector domain at the C-terminal end 60. Even though sequence conservation in response regulators exists it cannot be used as an indicator of functional similarity, for example DivK of Caulobacter crescentus and Spo0F of Bacillus subtilis, share 30% identity but have completely different functions. Similar situation exists even for response regulators such as OmpR and PhoB where they share 37% identity but have unrelated functions 61. This leads to problems in assigning function to RRs based on sequence similarity. Response regulators primarily exist in two different conformations, i.e. the active/inactive form. Phosphorylation of receiver domain generates the activated form of response regulator in most cases 62, 63. Domain rearrangements on phosphorylation have been identified in response regulators by solving the crystal structures of phosphorylated and non-phosphorylated forms 64. In prokaryotes, response regulators are the terminal component in pathways functioning as phosphorylation activated switches to effect the response 41, 65. They have the capability to catalyze phosphotransfer from histidine to aspartate and can also catalyze their phosphorylation from small molecule phosphodonors independently of SK 66-68. Effector domains in RRs vary in function and some RRs even lack the effector domain 69. The effector domain can have varied functions such as DNA binding, RNA binding 70, enzyme activity 40, 71 and also protein-protein interaction domain 72. Modular nature of response regulators is shown in figure 1.
Orphan sensor kinases and response regulators
Classical two-component regulatory systems are naturally encoded in locus that includes both sensor kinase (SK) and response regulator (RR) genes 73. The organization of SKs and RRs in a locus favors co-expression of the corresponding proteins and decreases the chances of cross-talk between non-cognate SKs and RRs. However, genomes of bacteria also encode for solitary SKs and RRs also referred to as orphan sensor kinases and response regulators. Even though there is hardly any bacterial genome which does not have orphan sensor kinases and response regulators, there is a considerable variation in the number of orphan SKs and RRs. Two out of 32 sensor kinases in E. coli genome are categorized as orphan SKs 74. This number is quite less when compared to Caulobacter crescentus genome in which 57% TCS genes exist as orphans 75. Interestingly of the 84 sensor kinase genes identified in the S. coelicolor genome 67 are located in pair with their cognate response regulator gene and 17 genes were identified as orphan 53. This shows that 20% sensor kinases in S. coelicolor are orphan sensor kinases. This number is quite less in comparison to C. crescentus. Few orphan sensor kinases specifically phoshorylate/interact with a particular response regulator which has been identified by in vitro phosphotransfer experiments using recombinant proteins. Interestingly, though orphan sensor kinases and response regulators lack their prototypical partners they function in very unorthodox ways to modify gene expression. Orphan Sensor kinases sometimes regulate primary functions mediated by a non-orphan TCS. This was observed in the case of GacS and GacA TCS of Pseudomonas aeruginosa where in RetS and GacS reciprocally control the expression of virulence factors responsible for acute and chronic infections 76. The orphan RetS protein binds to GacS protein thereby, inhibiting its ability to autophosphorylate using ATP and also stimulating the dephosphorylation of GacS~P 76. Non-binding of RetS to PilS proved its specificity towards GacS 77.
Annotation of the 3.3 Mb genome sequence of Lactobacillus plantarum WCFS1 revealed the presence of 13 paired TCS genes, and one orphan SK and RR 78. SKs and RRs that belong to a common two-component signalling pathway are often encoded by genes that are organized as a locus on the bacterial chromosome. TarC, response regulator, shares no apparent association with a cognate HK. This was indicated by RT-PCR analysis, which reveals a single monocistronic mRNA that is derived from a promoter located immediately upstream of tarC. While it is unclear how orphan response regulators like TarC modulate gene expression, one could envision their involvement as intermediaries of in vivo cross talk between otherwise independent signal transduction systems. In fact, a recent report supports phosphoryl transfer from a sensory kinase to a non-cognate response regulator in E. coli 79, and cross talk has been suggested in the regulation of S. mutans acid tolerance by Li and co-workers 80.
Signal integration in TCS
To respond to diverse environmental changes with greater sensitivity, information is also conveyed between TCSs to form a complex signal transduction network 81. In a classical TCS sensor kinase autophosphorylates and transfers the phosphate to its cognate response regulator. The phosphorylation of response regulator triggers conformational changes, due to which it is able to perform its designated function. However, complex TCS do exist, in bacteria these systems are designated as phosphorelays. In a typical phosphorelay the SK autophosphorylates and transfers the phosphate to a stand-alone RR, the phosphorylated RR serves as a phosphate donor for a SK which in turn transphosphorylates a RR which eventually performs its designated function 82. There also exist a group of proteins whose primary function is to connect two TCS and they most often serve to connect two independent two-component systems. The connector proteins employ variety of strategies to perform their roles, the most important being inhibiting autophosphorylation of SK 83, promoting dephosphorylation of RR 48, inhibiting dephosphorylation of RR 84, activating a sensor kinase 85, inhibiting recruitment of RNA polymerase 86, 87 and sometimes by sequestering proteins which in turn promote protein degradation 88. The connector proteins display distinct quantitative and kinetic properties that determine the timing and intensity of the response output. The genes which are regulated at the transcriptional level by connector proteins often display an increased mRNA induction when compared to directly regulated genes.
Mathematical modeling demonstrated that PhoP/PhoQ, PmrA/PmrB of Salmonella enterica which are connected by PmrD protein 85 showed increased level of mRNA induction 84, 89, 90. The connector proteins are also known to expand the spectrum of signals that influence the activity of RR 81. Interaction of connector proteins and their targets is a highly specific reaction such that the connector proteins do not interact even with their closest homologs as observed in the case of PmrD were it seldom interacts with YgiX RR a close homolog of PmrA 91, 92.
The interaction between aspartate phosphatases RapA, RapB, RapE and Spo0E proteins is also a highly specific reaction as demonstrated 93. Even though target specificity is predominant among connector proteins, few connector proteins also display dual functions such as in the case of RapH which it promotes dephosphorylation of Spo0F~P and also inhibits the DNA binding activity of response regulator ComA. Thus RapH controls both competence as well as sporulation 94.
Sensor kinases which can activate multiple RRs also known to exist, thus can feed multiple signals into a particular pathway. This phenomenon has been studied in great detail in the chemotaxis TCS of E. coli where swimming behavior is modified in response to changes in the concentration of different substrates. CheW a membrane protein senses the stimuli and alters the phosphorylated state of CheA (sensor kinase) which results in the phosphorylation of CheB and CheY (response regulators) 95. The phosphorylation state of either CheY or CheB dictates the swimming behavior which can be smooth or tumble. The change in swimming pattern is modulated by the interaction of RRs with flagellar motor protein. E. coli has five sensory receptors which are localized at the bacterial poles. These proteins form sensory complexes by teaming up with CheA and CheW thus enabling processing of multiple signals at one time 96. Marine bacterium Vibrio harveyi responds to two types of auto-inducer (AI) molecules known as AI-1 and AI-2. AI-1 is produced specifically by V. harveyi and AI-2 is a product of metabolism from wide variety of bacteria. The response to the presence of AI-1 and AI-2 is mediated by LuxN and LuxQ which acts via a periplasmic protein LuxP. All these sensors converge in phosphotransfer domain containing protein LuxU which in turn phosphorylates the LuxO a response regulator 97. Phosphorylated RR activates a repressor which turns off the genes for bioluminescence. The convergence of signal originating from two sensors onto a single RR help V. harveyi in responding to its own cell density as well as from other bacteria 96.
TCS regulating extracellular enzymes
Bacteria possess multiple signal transduction pathways to adapt to changes in the environment. The changes in the environment which bacteria sense and respond can be of either biotic or abiotic in nature. The capacity to utilize variety of nutrients is highly developed in actinomycetes. Bacillus subtilis produces a variety of degradative enzymes which enable the bacterium to grow on many different substrates. These enzymes are a-amylase, levansucrase, b-glucanase, xylanase and proteases. The production of all these enzymes is regulated by a TCS designated as DegS/DegU 98 where in degS encodes a sensor kinase and degU encodes for a response regulator. Deletion of degS and degU genes did not lead to variation in the phenotype. However, deletion led to the reduced production of these degradative enzymes 98. A different class of mutations were also identified in degS and degU which led to the hyper production of degradative enzymes 99. DegU which had threonine 98 mutated to isoleucine, valine 131 mutated to leucine displayed strong phosphorylation signals when compared to wild type DegU protein 100. This indicated that DegU also has autophosphatase property where amino acid residue threonine 98 and valine 131 play a very significant role.
Proteins, apart from sensor kinases and response regulators, are known to be involved in the signal transduction processes 84 where a small protein protects the dephosphorylation of response regulator. Interestingly in the case of DegU there also exists a protein designated DegR which stabilizes the phosphorylated DegU. DegR protects dephosphorylation of DegU however the exact mechanism still remains unclear 101. TCS involved in extracellular protease production was also identified in Staphylococcus aureus. ArlS/ArlR TCS which not only regulates protease production, but also plays a significant role in virulence mechanisms of S. aureus. ArlS/ArlR TCS also functions as a regulator of peptidoglycan hydrolase activity as well as in bio-film formation 102.
Regulation of chitinases
Chitinase production in bacteria is regulated by a repressor/inducer system in which chitin or products of chitin degradation act as inducers. Experiments on cultures without carbon source to prevent catabolite repression revealed that N-acetylglucosamine is the best inducer of chitinase 103. Induction of chitinases is substrate specific and not induced by pectin, xylan or cellulose. Streptomyces have a remarkable ability to utilize chitin as a source of carbon by producing multiple chitinases. Regulation of chitinases in S. lividans happens at the transcriptional level, chitin induces the production of chitinases and combination of chitin and glucose represses their production 104.
Catabolite repression of chitinase and other genes involved in utilization of carbon sources was identified in ccrA1 mutant of S. coelicolor. Glucose repression of chitinase (chi63) production was abolished in a ccrA1 mutant of S. coelicolor indicating its role in regulating chi63 production 105. The role of glucose kinase in glucose repression of chitinases has been established in S. lividans by introducing glkA gene from S. coelicolor into S. lividans G015 mutant which lacked glucose repression capabilities 106. Interestingly, glucose repression of chitinase chi63 promoter is independent of glucose kinase glkA 107. DNA binding protein Cpb1 which had affinity towards chitinase promoters was purified from intracellular proteins of S. lividans by affinity purification 108. This protein exhibited ability to bind chitinase promoters indicating its possible role in regulation. Disruption of cpb1 gene provided a partial relief from glucose repression of chitinase production 108.
Reg1, a DNA binding protein identified in S. lividans has a helix-turn-helix motif (HTH) in the N-terminus. Disruption of reg1 gene relieved the carbon catabolite repression of both ±-amylases and chitinases indicating the involvement of reg1 109. Streptomyces in which reg1 gene was disrupted lost the capability of chitin mediated induction of chitinases 109 indicating the role of reg1 in induction of chitinases. Above mentioned studies indicated that more than one mechanism of glucose repression operates in Streptomyces spp. 107. Presence of direct repeats has been reported in the promoters of chitinase genes from many Streptomyces spp. 1, 15, 110. Partial characterization of S. plicatus chi63 and chi35 promoters identified that a single base pair substitution resulted in a strain which produced chitinase even in the presence of glucose. This mutant produced chitinase constitutively even in the absence of chitin 110. Further work on chi63 promoter has identified regions which influence both glucose repression as well as chitin induction 111. Chitinase (chiC) regulated by a quorum-sensing system was identified for the first time in the case of opportunistic pathogen P. aeruginosa 112.
Chitinases regulated by TCS
Chitinases are enzymes which degrade chitin which is a homopolymer of b-1, 4-N-Acetyl-D-glucosamine, one of the most abundant biopolymers on earth 112. Chitinase production is constitutive and their production is enhanced by the presence of chitin. N-Acetyl glucosamine, the product of chitin breakdown is utilized by the bacterium as a source of carbon and nitrogen 113.
Expression of chitinases is regulated in the producing bacteria by means of catabolite repression and substrate induction. Two-component systems regulating chitinases have been reported from few Streptomyces sp. 113-116. A hybrid TCS sensor kinase regulates chitinase production in Vibrios 117. Two-component system involved in regulation of chitinase was first identified in S. thermoviolaceus and subsequently in S. coelicolor based on sequence homology to TCS from S. thermoviolaceus. Both these TCS not just share a high sequence homology but also share functional similarity with each other 53. S. peucetius, well know producer of anti-cancer drugs doxorubicin and daunorubicin can degrade chitin and utilize for its growth effectively1. Chitinase production in S. peucetius is negatively regulated by ChiS/ChiR two-component system 118.
Allosamidin a family 18 chitinase inhibitor is produced by Streptomyces sp. AJ9463. Unlike other Streptomyces, this bacterium also produces chitinases. Streptomyces sp. AJ9463 chi65 is regulated by two-component system which functions in very unique manner. Allosamidin can activate the transcription of chi65 mediated by Chi65S/Chi65R TCS. Although allosamidin is involved in the transcriptional regulation it cannot activate on its own and it requires the presence of N, N”-diacetylchitobiose 116, 119. Hybrid sensor kinase involved in regulating the production of chitinase was first reported in Vibrio furnissi and Vibrio cholerae. This hybrid sensor kinase (ChiS) controls the expression of approximately 50 genes of which many are involved in degradation of chitin 117.
Post translational modification of Chitinases
Glycosylation
Glycosylation, the post-translational modification of proteins by carbohydrates has long been recognized as a key strategy to influence structure and function of proteins in eukaryotes 120. For long it was believed that glycosylation occurs exclusively in eukaryotes, however this has been challenged by the identification of glyco-proteins from prokaryotes 121. The surface layer (S-layer) glycoprotein of Halobacterium halobium (Salinarum) was the first prokaryotic glycoprotein to be identified 121. Identification of glycoproteins is most often based on their aberrant migration in SDS-PAGE. However, glycosylation of proteins can also be detected by oxidation of carbohydrates mediated by periodic acid and condensation of generated aldehyde by Schiff’s base with a chromogen or an indicator enzyme. Oxidized sugar could also be detected by digoxigenin (DIG), DIG in turn is detected by Anti-DIG antibodies 122. Lectin based identification of glycoprotein’s is also a feasible option. The enzymatic removal of glycan from the protein can identify whether glycosylation is N- linked or O-linked 2, 123.
Tunicamycin, an inhibitor of N-linked glycosylation has been used to study the functional significance of glycosylation in Sf9 (Spodographa frugiperda) cells expressing a chitinase from a tick Haemaphysalis longicornis 24. Tunicamycin along with enzymatic removal of glycans can be used to identify the nature of glycosylation 2. Identification of glycans by mass spectrometry involves the separation of glycan either by enzymatic or by chemical methods 120. N-linked glycans are removed by N-glycosidases such as PNGaseF. O-linked glycosylation can be removed both by chemical method such as reductive alkaline b-elimination and by enzymatic method using O-glycanases 2, 123. A few chitinases that have been found to be glycosylated are ChiA from Cellulomonas uda 18, ChiC from S. griseus 21, CHT1 from Haemaphysalis longicornis 24 and (human analog of chitinases) chitotriosidase produced by macrophages in humans 2. The functional significance of glycosylation in chitinases is not known however, in the case of cellulases glycosylation plays a significant role in binding to crystalline substrate and also protects the enzyme from cleavage by proteases 25.
Proteolytic processing
Streptomyces are the major producers of chitinases in soil. Multiple chitinase genes have been identified in the genome sequences of S. coelicolor 9, S. avermitilis 10 and S. griseus 11. Several isoforms of chitinases have also been identified and they could have been the products of different genes or the result of post-translational proteolytic processing. This assumption was first made for the multiple chitinases in Bacillus circulans 12. Post-translational proteolytic processing has also been observed in the case of chitinases of Serratia marcescens 124, 125 and also in the case of cellulases of S. reticuli 126. Among Streptomyces, proteolytic cleavage of chitinases was first observed in S. olivaceoviridis where in 70 kDa chitinase served as a precursor for 30 kDa and 20.5 kDa chitinase. Chitinase of 20.5 kDa is derived from 30 kDa or 70 kDa chitinase upon proteolytic cleavage 19. Even though both 30 kDa and 20.5 kDa chitinases had the same active site 20.5 kDa chitinase shows reduced specific activity 19.
Chitinases which have originated from the same precursor by proteolytic cleavage also show differences in binding and substrate specificities 127. In the case of S. marcescens chitinase both precursor and mature chitinase show similar specific activities and optimal reaction temperature 125. Family 19 chitinase C-1 identified in S. griseus was also derived from Chitinase C by proteolytic cleavage 128.
Origin of multiple chitinases from the same precursor by proteolytic cleavage is not just limited to bacteria. Chitinases of plants such as Phaseolus vulgaris L. cv Saxa namely PvChiE, PvChiF and PvChiG were all derived from PvChi4 by differential proteolytic cleavage 129. Human analogue of chitinases namely chitotriosidase secreted by macrophages exhibits multiple forms of chitotriosidases both by alternative splicing of mRNA and also by proteolytic cleavage 2. S. olivaceoviridis produces an autocatalytic chitinase 130 which has a lysine C-endoproteinase in the C-terminus. This protein remains as a 92 kDa protein in the presence of protease inhibitors. However, 70 kDa and 22 kDa fragments are produced on removal of the protease inhibitors. The resultant 22 kDa fragment has proteolytic activity 130.
Chitinases- importance and application
Chitinases are enzymes which cleave the glycosidic linkages of chitin to generate low molecular weight oligosaccharides. Based on the mode of cleavage chitinases can be broadly grouped into two categories. The endochitinases which cleave randomly and generate chito-oligosaccharides of various sizes and exochitinases cleave from the non-reducing end of chitin microfibril 29. Based on the sequence similarity chitinolytic enzymes can be grouped into families 18 and19. Family 18 chitinases are the most diverse of the three families and are found in bacteria, fungi, viruses, animals and plants. Family 19 chitinases are abundant in plants 129 and a few Streptomyces 3. Family 18 and family 19 chitinase do not share sequence similarity, have different structures and molecular mechanisms indicating that these enzymes are likely to have evolved from different ancestors 20.
- Vetrivel KS, Pandian SK, Chaudhary U and Dharmalingam K. Purification, cloning, and DNA sequence analysis of a chitinase from an overproducing mutant of Streptomyces peucetius defective in daunorubicin biosynthesis. Can J Microbiol. 2001; 47(3):179-187.
- Renkema GH, Boot RG, Strijland A, Donker-Koopman WE, van den Berg M, Muijsers AO and Aerts JM. Synthesis, sorting, and processing into distinct isoforms of human macrophage chitotriosidase. Eur J Biochem. 1997; 244(2):279-285.
- Watanabe T, Kanai R, Kawase T, Tanabe T, Mitsutomi M, Sakuda S and Miyashita K. Family 19 chitinases of Streptomyces species: characterization and distribution. Microbiology. 1999; 145(12):3353-3363.
- Metcalfe AC, Krsek M, Gooday GW, Prosser JI and Wellington EM. Molecular analysis of a bacterial chitinolytic community in an upland pasture. Appl Environ Microbiol. 2002; 68(10):5042-5050.
- Okazaki K, Yamashita Y, Noda M, Sueyoshi N, Kameshita I and Hayakawa S. Molecular cloning and expression of the gene encoding family 19 chitinase from Streptomyces sp. J-13-3. Biosci Biotechnol Biochem. 2004; 68(2):341-351.
- Saito A, Miyashita K, Biukovic G and Schrempf H. Characteristics of a Streptomyces coelicolor A3(2) extracellular protein targeting chitin and chitosan. Appl Environ Microbiol. 2001; 67(3):1268-1273.
- Ueda M, Kojima M, Yoshikawa T, Mitsuda N, Araki K, Kawaguchi T, Miyatake K, Arai M and Fukamizo T. A novel type of family 19 chitinase from Aeromonas sp. No.10S-24. Cloning, sequence, expression, and the enzymatic properties. Eur J Biochem. 2003; 270(11):2513-2520.
- Gomes RC, Semedo LT, Soares RM, Linhares LF, Ulhoa CJ, Alviano CS and Coelho RR. Purification of a thermostable endochitinase from Streptomyces RC1071 isolated from a cerrado soil and its antagonism against phytopathogenic fungi. J Appl Microbiol. 2001; 90(4):653-661.
- Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature. 2002; 417(6885):141-147.
- Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Shinose M, Takahashi Y, Horikawa H, Nakazawa H, Osonoe T, Kikuchi H, Shiba T, Sakaki Y and Hattori M. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc Natl Acad Sci U S A. 2001; 98(21):12215-12220.
- Ohnishi Y, Ishikawa J, Hara H, Suzuki H, Ikenoya M, Ikeda H, Yamashita A, Hattori M and Horinouchi S. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J Bacteriol. 2008; 190(11):4050-4060.
- Watanabe T, Oyanagi W, Suzuki K and Tanaka H. Chitinase system of Bacillus circulans WL-12 and importance of chitinase A1 in chitin degradation. J Bacteriol. 1990; 172(7):4017-4022.
- Suzuki K, Sugawara N, Suzuki M, Uchiyama T, Katouno F, Nikaidou N and Watanabe T. Chitinases A, B, and C1 of Serratia marcescens 2170 produced by recombinant Escherichia coli: enzymatic properties and synergism on chitin degradation. Biosci Biotechnol Biochem. 2002; 66(5):1075-1083.
- Saito A, Fujii T and Miyashita K. Distribution and evolution of chitinase genes in Streptomyces species: involvement of gene-duplication and domain-deletion. Antonie Van Leeuwenhoek. 2003; 84(1):7-15.
- Saito A, Ishizaka M, Francisco PB, Jr., Fujii T and Miyashita K. Transcriptional co-regulation of five chitinase genes scattered on the Streptomyces coelicolor A3(2) chromosome. Microbiology. 2000; 146(11):2937-2946.
- Howard MB, Ekborg NA, Taylor LE, Weiner RM and Hutcheson SW. Genomic analysis and initial characterization of the chitinolytic system of Microbulbifer degradans strain 2-40. J Bacteriol. 2003; 185(11):3352-3360.
- Chuang HH, Lin HY and Lin FP. Biochemical characteristics of C-terminal region of recombinant chitinase from Bacillus licheniformis: implication of necessity for enzyme properties. FEBS J. 2008; 275(9):2240-2254.
- Reguera G and Leschine SB. Biochemical and genetic characterization of ChiA, the major enzyme component for the solubilization of chitin by Cellulomonas uda. Arch Microbiol. 2003; 180(6):434-443.
- Romaguera A, Menge U, Breves R and Diekmann H. Chitinases of Streptomyces olivaceoviridis and significance of processing for multiplicity. J Bacteriol. 1992; 174(11):3450-3454.
- Suzuki K, Taiyoji M, Sugawara N, Nikaidou N, Henrissat B and Watanabe T. The third chitinase gene (chiC) of Serratia marcescens 2170 and the relationship of its product to other bacterial chitinases. Biochem J. 1999; 343 Pt 3:587-596.
- Itoh Y, Takahashi K, Takizawa H, Nikaidou N, Tanaka H, Nishihashi H, Watanabe T and Nishizawa Y. Family 19 chitinase of Streptomyces griseus HUT6037 increases plant resistance to the fungal disease. Biosci Biotechnol Biochem. 2003; 67(4):847-855.
- Hassan F, Meens J, Jacobsen HJ and Kiesecker H. A family 19 chitinase (Chit30) from Streptomyces olivaceoviridis ATCC 11238 expressed in transgenic pea affects the development of T. harzianum in vitro. J Biotechnol. 2009; 143(4):302-308.
- Nielsen KK, Bojsen K, Roepstorff P and Mikkelsen JD. A hydroxyproline-containing class IV chitinase of sugar beet is glycosylated with xylose. Plant Mol Biol. 1994; 25(2):241-257.
- You M, Xuan X, Tsuji N, Kamio T, Taylor D, Suzuki N and Fujisaki K. Identification and molecular characterization of a chitinase from the hard tick Haemaphysalis longicornis. J Biol Chem. 2003; 278(10):8556-8563.
- Langsford ML, Gilkes NR, Singh B, Moser B, Miller RC, Jr., Warren RA and Kilburn DG. Glycosylation of bacterial cellulases prevents proteolytic cleavage between functional domains. FEBS Lett. 1987; 225(1-2):163-167.
- Kluepfel D, Vats-Mehta S, Aumont F, Shareck F and Morosoli R. Purification and characterization of a new xylanase (xylanase B) produced by Streptomyces lividans 66. Biochem J. 1990; 267(1):45-50.
- Sutherland TE, Maizels RM and Allen JE. Chitinases and chitinase-like proteins: potential therapeutic targets for the treatment of T-helper type 2 allergies. Clin Exp Allergy. 2009; 39(7):943-955.
- Merzendorfer H and Zimoch L. Chitin metabolism in insects: structure, function and regulation of chitin synthases and chitinases. J Exp Biol. 2003; 206(Pt 24):4393-4412.
- Dahiya N, Tewari R and Hoondal GS. Biotechnological aspects of chitinolytic enzymes: a review. Appl Microbiol Biotechnol. 2006; 71(6):773-782.
- Aam BB, Heggset EB, Norberg AL, Sorlie M, Varum KM and Eijsink VG. Production of chitooligosaccharides and their potential applications in medicine. Mar Drugs. 2010; 8(5):1482-1517.
- Chater KF, Biro S, Lee KJ, Palmer T and Schrempf H. The complex extracellular biology of Streptomyces. FEMS Microbiol Rev. 2010; 34(2):171-198.
- Khoushab F and Yamabhai M. Chitin research revisited. Mar Drugs. 2010; 8(7):1988-2012.
- Zhang J, Xia W, Liu P, Cheng Q, Tahirou T, Gu W and Li B. Chitosan modification and pharmaceutical/biomedical applications. Mar Drugs. 2010; 8(7):1962-1987.
- Muzzarelli RA. Chitins and chitosans as immunoadjuvants and non-allergenic drug carriers. Mar Drugs. 2010; 8(2):292-312.
- Wang SL and Chang WT. Purification and characterization of two bifunctional chitinases/lysozymes extracellularly produced by Pseudomonas aeruginosa K-187 in a shrimp and crab shell powder medium. Appl Environ Microbiol. 1997; 63(2):380-386.
- Saito A, Fujii T, Yoneyama T, Redenbach M, Ohno T, Watanabe T and Miyashita K. High-multiplicity of chitinase genes in Streptomyces coelicolor A3(2). Biosci Biotechnol Biochem. 1999; 63(4):710-718.
- Sato M, Machida K, Arikado E, Saito H, Kakegawa T and Kobayashi H. Expression of outer membrane proteins in Escherichia coli growing at acid pH. Appl Environ Microbiol. 2000; 66(3):943-947.
- Orikoshi H, Nakayama S, Miyamoto K, Hanato C, Yasuda M, Inamori Y and Tsujibo H. Roles of four chitinases (ChiA, ChiB, ChiC, and ChiD) in the chitin degradation system of marine bacterium Alteromonas sp. strain O-7. Appl Environ Microbiol. 2005; 71(4):1811-1815.
- Techkarnjanaruk S and Goodman AE. Multiple genes involved in chitin degradation from the marine bacterium Pseudoalteromonas sp. strain S91. Microbiology. 1999; 145(4):925-934.
- Stock JB, Ninfa AJ and Stock AM. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989; 53(4):450-490.
- Stock AM, Robinson VL and Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000; 69:183-215.
- Cheung J and Hendrickson WA. Sensor domains of two-component regulatory systems. Curr Opin Microbiol. 2010; 13(2):116-123.
- Kenney LJ. How important is the phosphatase activity of sensor kinases? Curr Opin Microbiol. 2010; 13(2):168-176.
- Silversmith RE. Auxiliary phosphatases in two-component signal transduction. Curr Opin Microbiol. 2010; 13(2):177-183.
- Wolanin PM, Webre DJ and Stock JB. Mechanism of phosphatase activity in the chemotaxis response regulator CheY. Biochemistry. 2003; 42(47):14075-14082.
- Hsing W, Russo FD, Bernd KK and Silhavy TJ. Mutations that alter the kinase and phosphatase activities of the two-component sensor EnvZ. J Bacteriol. 1998; 180(17):4538-4546.
- Dutta R, Yoshida T and Inouye M. The critical role of the conserved Thr247 residue in the functioning of the osmosensor EnvZ, a histidine Kinase/Phosphatase, in Escherichia coli. J Biol Chem. 2000; 275(49):38645-38653.
- Perego M, Glaser P and Hoch JA. Aspartyl-phosphate phosphatases deactivate the response regulator components of the sporulation signal transduction system in Bacillus subtilis. Mol Microbiol. 1996; 19(6):1151-1157.
- Ishikawa S, Core L and Perego M. Biochemical characterization of aspartyl phosphate phosphatase interaction with a phosphorylated response regulator and its inhibition by a pentapeptide. J Biol Chem. 2002; 277(23):20483-20489.
- Casino P, Rubio V and Marina A. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell. 2009; 139(2):325-336.
- Rodrigue A, Quentin Y, Lazdunski A, Mejean V and Foglino M. Two-component systems in Pseudomonas aeruginosa: why so many? Trends Microbiol. 2000; 8(11):498-504.
- Galperin MY. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 2005; 5:35.
- Hutchings MI, Hoskisson PA, Chandra G and Buttner MJ. Sensing and responding to diverse extracellular signals? Analysis of the sensor kinases and response regulators of Streptomyces coelicolor A3(2). Microbiology. 2004; 150(9):2795-2806.
- Krell T, Lacal J, Busch A, Silva-Jimenez H, Guazzaroni ME and Ramos JL. Bacterial sensor kinases: diversity in the recognition of environmental signals. Annu Rev Microbiol. 2010; 64:539-559.
- Dutta R, Qin L and Inouye M. Histidine kinases: diversity of domain organization. Mol Microbiol. 1999; 34(4):633-640.
- Stock A, Chen T, Welsh D and Stock J. CheA protein, a central regulator of bacterial chemotaxis, belongs to a family of proteins that control gene expression in response to changing environmental conditions. Proc Natl Acad Sci U S A. 1988; 85(5):1403-1407.
- Kim D and Forst S. Genomic analysis of the histidine kinase family in bacteria and archaea. Microbiology. 2001; 147(5):1197-1212.
- Kiil K, Ferchaud JB, David C, Binnewies TT, Wu H, Sicheritz-Ponten T, Willenbrock H and Ussery DW. Genome update: distribution of two-component transduction systems in 250 bacterial genomes. Microbiology. 2005; 151(11):3447-3452.
- Whitworth DE and Cock PJ. Two-component systems of the myxobacteria: structure, diversity and evolutionary relationships. Microbiology. 2008; 154(2):360-372.
- Jenal U and Galperin MY. Single domain response regulators: molecular switches with emerging roles in cell organization and dynamics. Curr Opin Microbiol. 2009; 12(2):152-160.
- Galperin MY. Diversity of structure and function of response regulator output domains. Curr Opin Microbiol. 2010; 13(2):150-159.
- Dyer CM and Dahlquist FW. Switched or not?: the structure of unphosphorylated CheY bound to the N terminus of FliM. J Bacteriol. 2006; 188(21):7354-7363.
- Stock AM and Guhaniyogi J. A new perspective on response regulator activation. J Bacteriol. 2006; 188(21):7328-7330.
- Bachhawat P and Stock AM. Crystal structures of the receiver domain of the response regulator PhoP from Escherichia coli in the absence and presence of the phosphoryl analog beryllofluoride. J Bacteriol. 2007; 189(16):5987-5995.
- Stock J and Da Re S. Signal transduction: response regulators on and off. Curr Biol. 2000; 10(11):R420-424.
- Lukat GS, McCleary WR, Stock AM and Stock JB. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci U S A. 1992; 89(2):718-722.
- McCleary WR and Stock JB. Acetyl phosphate and the activation of two-component response regulators. J Biol Chem. 1994; 269(50):31567-31572.
- Wolfe AJ. Physiologically relevant small phosphodonors link metabolism to signal transduction. Curr Opin Microbiol. 2010; 13(2):204-209.
- Qian W, Han ZJ and He C. Two-component signal transduction systems of Xanthomonas spp.: a lesson from genomics. Mol Plant Microbe Interact. 2008; 21(2):151-161.
- Shu CJ and Zhulin IB. ANTAR: an RNA-binding domain in transcription antitermination regulatory proteins. Trends Biochem Sci. 2002; 27(1):3-5.
- Simms SA, Keane MG and Stock J. Multiple forms of the CheB methylesterase in bacterial chemosensing. J Biol Chem. 1985; 260(18):10161-10168.
- Francis NR, Wolanin PM, Stock JB, Derosier DJ and Thomas DR. Three-dimensional structure and organization of a receptor/signaling complex. Proc Natl Acad Sci U S A. 2004; 101(50):17480-17485.
- Lavin JL, Kiil K, Resano O, Ussery DW and Oguiza JA. Comparative genomic analysis of two-component regulatory proteins in Pseudomonas syringae. BMC Genomics. 2007; 8: 397.
- Blattner FR, Plunkett G, 3rd, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997; 277(5331):1453-1462.
- Skerker JM, Prasol MS, Perchuk BS, Biondi EG and Laub MT. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 2005; 3(10):e334.
- Goodman AL, Merighi M, Hyodo M, Ventre I, Filloux A and Lory S. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 2009; 23(2):249-259.
- Raghavan V and Groisman EA. Orphan and hybrid two-component system proteins in health and disease. Curr Opin Microbiol. 2010; 13(2): 226-231.
- Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R, Tarchini R, Peters SA, Sandbrink HM, Fiers MW, Stiekema W, Lankhorst RM, Bron PA, Hoffer SM, Groot MN, Kerkhoven R, et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc Natl Acad Sci U S A. 2003; 100(4):1990-1995.
- Verhamme DT, Arents JC, Postma PW, Crielaard W and Hellingwerf KJ. Investigation of in vivo cross-talk between key two-component systems of Escherichia coli. Microbiology. 2002; 148(1):69-78.
- Li YH, Lau PC, Tang N, Svensater G, Ellen RP and Cvitkovitch DG. Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J Bacteriol. 2002; 184(22):6333-6342.
- Mitrophanov AY and Groisman EA. Signal integration in bacterial two-component regulatory systems. Genes Dev. 2008; 22(19):2601-2611.
- Perraud AL, Weiss V and Gross R. Signalling pathways in two-component phosphorelay systems. Trends Microbiol. 1999; 7(3):115-120.
- Wang L, Grau R, Perego M and Hoch JA. A novel histidine kinase inhibitor regulating development in Bacillus subtilis. Genes Dev. 1997; 11(19):2569-2579.
- Kato A and Groisman EA. Connecting two-component regulatory systems by a protein that protects a response regulator from dephosphorylation by its cognate sensor. Genes Dev. 2004; 18(18):2302-2313.
- Eguchi Y and Utsumi R. A novel mechanism for connecting bacterial two-component signal-transduction systems. Trends Biochem Sci. 2005; 30(2):70-72.
- Ansaldi M and Dubnau D. Diversifying selection at the Bacillus quorum-sensing locus and determinants of modification specificity during synthesis of the ComX pheromone. J Bacteriol. 2004; 186(1):15-21.
- Ansaldi M, Theraulaz L and Mejean V. TorI, a response regulator inhibitor of phage origin in Escherichia coli. Proc Natl Acad Sci U S A. 2004; 101(25): 9423-9428.
- Bougdour A, Cunning C, Baptiste PJ, Elliott T and Gottesman S. Multiple pathways for regulation of sigmaS (RpoS) stability in Escherichia coli via the action of multiple anti-adaptors. Mol Microbiol. 2008; 68(2):298-313.
- Kox LF, Wosten MM and Groisman EA. A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. 2000; 19(8):1861-1872.
- Kato A, Latifi T and Groisman EA. Closing the loop: the PmrA/PmrB two-component system negatively controls expression of its posttranscriptional activator PmrD. Proc Natl Acad Sci U S A. 2003; 100(8):4706-4711.
- Soncini FC, Garcia Vescovi E, Solomon F and Groisman EA. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J Bacteriol. 1996; 178(17):5092-5099.
- Soncini FC and Groisman EA. Two-component regulatory systems can interact to process multiple environmental signals. J Bacteriol. 1996; 178(23):6796-6801.
- Perego M and Brannigan JA. Pentapeptide regulation of aspartyl-phosphate phosphatases. Peptides. 2001; 22(10):1541-1547.
- Smits WK, Bongiorni C, Veening JW, Hamoen LW, Kuipers OP and Perego M. Temporal separation of distinct differentiation pathways by a dual specificity Rap-Phr system in Bacillus subtilis. Mol Microbiol. 2007; 65(1):103-120.
- Bourret RB and Stock AM. Molecular information processing: lessons from bacterial chemotaxis. J Biol Chem. 2002; 277(12):9625-9628.
- Bijlsma JJ and Groisman EA. Making informed decisions: regulatory interactions between two-component systems. Trends Microbiol. 2003; 11(8):359-366.
- Bassler BL. Small talk. Cell-to-cell communication in bacteria. Cell. 2002; 109(4):421-424.
- Msadek T, Kunst F, Henner D, Klier A, Rapoport G and Dedonder R. Signal transduction pathway controlling synthesis of a class of degradative enzymes in Bacillus subtilis: expression of the regulatory genes and analysis of mutations in degS and degU. J Bacteriol. 1990; 172(2):824-834.
- Tanaka T, Kawata M and Mukai K. Altered phosphorylation of Bacillus subtilis DegU caused by single amino acid changes in DegS. J Bacteriol. 1991; 173(17):5507-5515.
- Dahl MK, Msadek T, Kunst F and Rapoport G. Mutational analysis of the Bacillus subtilis DegU regulator and its phosphorylation by the DegS protein kinase. J Bacteriol. 1991; 173(8):2539-2547.
- Mukai K, Kawata-Mukai M and Tanaka T. Stabilization of phosphorylated Bacillus subtilis DegU by DegR. J Bacteriol. 1992; 174(24):7954-7962.
- Fournier B and Hooper DC. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J Bacteriol. 2000; 182(14):3955-3964.
- Felse PA and Panda T. Regulation and cloning of microbial chitinase genes. Appl Microbiol Biotechnol. 1999; 51(2):141-151.
- Miyashita K, Fujii T and Saito A. Induction and repression of a Streptomyces lividans chitinase gene promoter in response to various carbon sources. Biosci Biotechnol Biochem. 2000; 64(1):39-43.
- Ingram C, Delic I and Westpheling J. ccrA1: a mutation in Streptomyces coelicolor that affects the control of catabolite repression. J Bacteriol. 1995; 177(12):3579-3586.
- Saito A, Fujii T, Yoneyama T and Miyashita K. glkA is involved in glucose repression of chitinase production in Streptomyces lividans. J Bacteriol. 1998; 180(11):2911-2914.
- Ingram C and Westpheling J. The glucose kinase gene of Streptomyces coelicolor is not required for glucose repression of the chi63 promoter. J Bacteriol. 1995; 177(12):3587-3588.
- Fujii T, Miyashita K, Ohtomo R and Saito A. DNA-binding protein involved in the regulation of chitinase production in Streptomyces lividans. Biosci Biotechnol Biochem. 2005; 69(4):790-799.
- Nguyen J, Francou F, Virolle MJ and Guerineau M. Amylase and chitinase genes in Streptomyces lividans are regulated by reg1, a pleiotropic regulatory gene. J Bacteriol. 1997; 179(20):6383-6390.
- Delic I, Robbins P and Westpheling J. Direct repeat sequences are implicated in the regulation of two Streptomyces chitinase promoters that are subject to carbon catabolite control. Proc Natl Acad Sci U S A. 1992; 89(5):1885-1889.
- Ni X and Westpheling J. Direct repeat sequences in the Streptomyces chitinase-63 promoter direct both glucose repression and chitin induction. Proc Natl Acad Sci U S A. 1997; 94(24):13116-13121.
- Folders J, Algra J, Roelofs MS, van Loon LC, Tommassen J and Bitter W. Characterization of Pseudomonas aeruginosa chitinase, a gradually secreted protein. J Bacteriol. 2001; 183(24):7044-7052.
- Tsujibo H, Hatano N, Okamoto T, Endo H, Miyamoto K and Inamori Y. Synthesis of chitinase in Streptomyces thermoviolaceus is regulated by a two-component sensor-regulator system. FEMS Microbiol Lett. 1999; 181(1):83-90.
- Kormanec J, Sevcikova B and Homerova D. Cloning of a two-component regulatory system probably involved in the regulation of chitinase in Streptomyces coelicolor A3(2). Folia Microbiol (Praha). 2000; 45(5):397-406.
- Homerova D, Knirschova R and Kormanec J. Response regulator ChiR regulates expression of chitinase gene, chiC, in Streptomyces coelicolor. Folia Microbiol (Praha). 2002; 47(5):499-505.
- Suzuki S, Nakanishi E, Ohira T, Kawachi R, Ohnishi Y, Horinouchi S, Nagasawa H and Sakuda S. Chitinase inhibitor allosamidin is a signal molecule for chitinase production in its producing Streptomyces II. Mechanism for regulation of chitinase production by allosamidin through a two-component regulatory system. J Antibiot (Tokyo). 2006; 59(7):410-417.
- Li X and Roseman S. The chitinolytic cascade in Vibrios is regulated by chitin oligosaccharides and a two-component chitin catabolic sensor/kinase. Proc Natl Acad Sci U S A. 2004; 101(2):627-631.
- Rabbind Singh A, Senthamaraikannan P, Thangavel C, Danda R, Pandian SK and Dharmalingam K. ChiS histidine kinase negatively regulates the production of chitinase ChiC in Streptomyces peucetius. Microbiol Res. 169(2-3):155-162.
- Suzuki S, Nakanishi E, Ohira T, Kawachi R, Nagasawa H and Sakuda S. Chitinase inhibitor allosamidin is a signal molecule for chitinase production in its producing Streptomyces I. Analysis of the chitinase whose production is promoted by allosamidin and growth accelerating activity of allosamidin. J Antibiot (Tokyo). 2006; 59(7):402-409.
- Hitchen PG and Dell A. Bacterial glycoproteomics. Microbiology. 2006; 152(6):1575-1580.
- Benz I and Schmidt MA. Never say never again: protein glycosylation in pathogenic bacteria. Mol Microbiol. 2002; 45(2):267-276.
- Broadway RM, Williams DL, Kain WC, Harman GE, Lorito M and Labeda DP. Partial characterization of chitinolytic enzymes from Streptomyces albidoflavus. Lett Appl Microbiol. 1995; 20(5):271-276.
- Morelle W and Michalski JC. Analysis of protein glycosylation by mass spectrometry. Nat Protoc. 2007; 2(7):1585-1602.
- Gal SW, Choi JY, Kim CY, Cheong YH, Choi YJ, Bahk JD, Lee SY and Cho MJ. Isolation and characterization of the 54-kDa and 22-kDa chitinase genes of Serratia marcescens KCTC2172. FEMS Microbiol Lett. 1997; 151(2):197-204.
- Gal SW, Choi JY, Kim CY, Cheong YH, Choi YJ, Lee SY, Bahk JD and Cho MJ. Cloning of the 52-kDa chitinase gene from Serratia marcescens KCTC2172 and its proteolytic cleavage into an active 35-kDa enzyme. FEMS Microbiol Lett. 1998; 160(1):151-158.
- Moormann M, Schlochtermeier A and Schrempf H. Biochemical Characterization of a Protease Involved in the Processing of a Streptomyces reticuli Cellulase (Avicelase). Appl Environ Microbiol. 1993; 59(5):1573-1578.
- Blaak H and Schrempf H. Binding and substrate specificities of a Streptomyces olivaceoviridis chitinase in comparison with its proteolytically processed form. Eur J Biochem. 1995; 229(1):132-139.
- Ohno T, Armand S, Hata T, Nikaidou N, Henrissat B, Mitsutomi M and Watanabe T. A modular family 19 chitinase found in the prokaryotic organism Streptomyces griseus HUT 6037. J Bacteriol. 1996; 178(17):5065-5070.
- Lange J, Mohr U, Wiemken A, Boller T and Vogeli-Lange R. Proteolytic processing of class IV chitinase in the compatible interaction of bean roots with Fusarium solani. Plant Physiol. 1996; 111(4):1135-1144.
- Radwan HH, Plattner HJ, Menge U and Diekmann H. The 92-kDa chitinase from Streptomyces olivaceoviridis contains a lysine-C endoproteinase at its N-terminus. FEMS Microbiol Lett. 1994; 120(1-2):31-35.
- Fujii T and Miyashita K. Multiple domain structure in a chitinase gene (chiC) of Streptomyces lividans. J Gen Microbiol. 1993; 139(4):677-686.
- Tsujibo H, Minoura K, Miyamoto K, Endo H, Moriwaki M and Inamori Y. Purification and properties of a thermostable chitinase from Streptomyces thermoviolaceus OPC-520. Appl Environ Microbiol. 1993; 59(2):620-622.
- Tsujibo H, Okamoto T, Hatano N, Miyamoto K, Watanabe T, Mitsutomi M and Inamori Y. Family 19 chitinases from Streptomyces thermoviolaceus OPC-520: molecular cloning and characterization. Biosci Biotechnol Biochem. 2000; 64(11):2445-2453.
- Akagi K, Watanabe J, Hara M, Kezuka Y, Chikaishi E, Yamaguchi T, Akutsu H, Nonaka T, Watanabe T and Ikegami T. Identification of the substrate interaction region of the chitin-binding domain of Streptomyces griseus chitinase C. J Biochem. 2006; 139(3):483-493.
- Anderson TB, Brian P and Champness WC. Genetic and transcriptional analysis of absA, an antibiotic gene cluster-linked two-component system that regulates multiple antibiotics in Streptomyces coelicolor. Mol Microbiol. 2001; 39(3):553-566.
- Sheeler NL, MacMillan SV and Nodwell JR. Biochemical activities of the absA two-component system of Streptomyces coelicolor. J Bacteriol. 2005; 187(2):687-696.
- McKenzie NL and Nodwell JR. Phosphorylated AbsA2 negatively regulates antibiotic production in Streptomyces coelicolor through interactions with pathway-specific regulatory gene promoters. J Bacteriol. 2007; 189(14):5284-5292.
- Ortiz de Orue Lucana D, Zou P, Nierhaus M and Schrempf H. Identification of a novel two-component system SenS/SenR modulating the production of the catalase-peroxidase CpeB and the haem-binding protein HbpS in Streptomyces reticuli. Microbiology. 2005; 151(11):3603-3614.
- Bogel G, Schrempf H and Ortiz de Orue Lucana D. DNA-binding characteristics of the regulator SenR in response to phosphorylation by the sensor histidine autokinase SenS from Streptomyces reticuli. FEBS J. 2007; 274(15):3900-3913.
- Li YQ, Chen PL, Chen SF, Wu D and Zheng J. A pair of two-component regulatory genes ecrA1/A2 in S. coelicolor. J Zhejiang Univ Sci. 2004; 5(2):173-179.
- Hutchings MI, Hong HJ and Buttner MJ. The vancomycin resistance VanRS two-component signal transduction system of Streptomyces coelicolor. Mol Microbiol. 2006; 59(3):923-935.
- Sola-Landa A, Moura RS and Martin JF. The two-component PhoR-PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans. Proc Natl Acad Sci U S A. 2003; 100(10):6133-6138.
- Lu Y, Wang W, Shu D, Zhang W, Chen L, Qin Z, Yang S and Jiang W. Characterization of a novel two-component regulatory system involved in the regulation of both actinorhodin and a type I polyketide in Streptomyces coelicolor. Appl Microbiol Biotechnol. 2007; 77(3):625-635.
- Paget MS, Leibovitz E and Buttner MJ. A putative two-component signal transduction system regulates sigmaE, a sigma factor required for normal cell wall integrity in Streptomyces coelicolor A3(2). Mol Microbiol. 1999; 33(1):97-107.
- Ishizuka H, Horinouchi S, Kieser HM, Hopwood DA and Beppu T. A putative two-component regulatory system involved in secondary metabolism in Streptomyces spp. J Bacteriol. 1992; 174(23):7585-7594.
- West AH and Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001; 26(6):369-376.
- Barakat M, Ortet P, Jourlin-Castelli C, Ansaldi M, Mejean V and Whitworth DE. P2CS: a two-component system resource for prokaryotic signal transduction research. BMC Genomics. 2009; 10:315.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.