ISSN: 0973-7510
E-ISSN: 2581-690X
The purpose of the research was to obtain different variants of chickpea beverages and subject them to probiotic fermentation carried out by Lactobacillus plantarum 299v in order to compare properties of received products. The study focused on the influence of the fermentation process on changes in protein profiles, antioxidative properties, pH values, and reducing power (RP). The analyzed variants of chickpea beverages exhibited diversity in terms of RP. Fermented products were characterized by significantly higher values of RP (A700) ranging from 0.1 to 0.29 than unfermented ones (value in range from 0.04 to 0.09). Whereas, pH values of fermented products were lower (from 4.46 to 5.64) to than unfermented beverages (6.68 to 7.57). The results indicate also the presence of b-conglycinin and glycinin (which are considered as one of food allergens) in analyzed chickpea-based beverages, however the probiotic fermentation contributed to strong hydrolysis of those components. Among tested variants of the chickpea products, the beverages with vanillin sugar and coconut flakes (unfermented as well as fermented) received the highest notes of general consumer acceptance in preliminary sensory assessment. The fermentation of the chickpea beverages with the probiotic strain significantly increased the reducing power of the products, revealed antioxidative-enhancing properties and contributed to hydrolysis of b-conglycinin (7S) and glycinin (11S) fractions.
antioxidants, fermented chickpea beverages, probiotics.
In intensive system of agricultural, agrochemicals that depend mainly on usage of chemical fertilizers that directly are applied to the soil to support the need of essential nutrients as growth promoting, pest and disease controlling agent. Extensive literature of inorganic fertilizers containing heavy metals results in the accumulation of potentially toxic chemicals in animals in agricultural lands1. On the other hand several industrial waste and effluents significantly contribute to heavy metal pollution on the environmental which causes hazards to human health as well as various environmental biotic and ecological set up2. The toxicity of the heavy metals like Cd++; Pb++; Zn++; Hg++; Co++; and Ni++ being soluble in nature or exchangeable elements totally rely upon their transfer rate from source to other component of ecosystem acting as soil solution, ground water plants, soil microflora and ultimately to humans through food chains1. Researchers in the past have demonstrated the harmful impact of long term usage of sewage, sludge and industrial effluents on cultivated soil due to heavy metal pollution. Major industrial effluents/waste containing heavy metals are the common source including metal finishing, nuclear and electronic industries, metal finishing operation and glass industries2. Sewage sludge utilization in agriculture is one of the most popular way of disposal and reutilizitaion. However, heavy metal content of sludge is probably the major parameter4. Heavy metals are not chemically or biologically degraded and are difficult to remove from soil or polluted sites5. Most of the heavy metals are required in low concentration in biological system in various metabolic processes but become toxic at elevated concentrations in biotic system including humans and animals6. Due to overall risk associated with heavy metals to environment and public health by heavy metal pollution, various methods have been developed to decontaminate and bioremediate waste water7. Contaminated soil, water, and other sources contained increase frequency of metal tolerant and resistant microbes. Such bacteria develop different mechanisms of tolerance and resistance to toxic metal concentration8,9. The exploitation of such metal tolerant or resistant bacteria for the removal of toxic levels of heavy metals from wastewater and contaminated soil by means of physical and chemical methods have gained increasing attention in recent times. The failure in removing low concentration of heavy metals in biotic system, conventional methods have still limited application9. The resistance determinants to metals in bacteria may be found on plasmids and being transmissible in nature and spread by horizontal transfer8. The desired resistance-genes may be possibly introduced to into more suitable hosts by means of genetic transfer techniques, for commercial production of inocula or enzymes could be exploited for bioremediation of water/soil.
Both phenotypic and molecular approaches (PCR based) have been used to investigate adaptive responses of bacteria to environmental stress and molecular mechanisms of adaptation9.
In the near future, environmental pressure is the major cause of population growth and the expectation of increased living standards2,3,10. Hill10 summarized the factors responsible for pollution and other types of environmental deterioration in many communities or societies as being caused by the combined effect of population, affluence and technology. They also stated that some of the oldest cases of environmental pollution in the world are caused by industrial discharge of heavy metal like Cu++; Pb++ and Hg++; mining, smelting and utilization by ancient civilizations. Water pollution unfavorably alter the composition of aquatic ecosystems in terms of oxygen content, presence of toxins, the living organisms and so on3,10.
The major water polluting waste includes from sewage, industrial and agricultural types10. Zhang et al. (11) reported the concentration of Cr++ Cd++, Pb++ and Hg++ in the sediments of the River Bortala in northeast China exceeded the National Standards of China. The study conducted that human activities in the vicinity of river area is the major source of metal in river sediments. Metals like Pb++, Cd++and Hg++ were found an important ecological risk factors11. Heavy metal such as Cd++, Cr++, Pb++, Ni++ and Zn++ bioaccumulation in sediment, primary producers and in aquatic animals (mollusks, fish etc) in the Tabasco Coast, Mexico was studied to evaluate the ecological potential hazard in that habitat12. Zinc showed the highest concentration in sediment (mean of 159.58 mg kg-1), in planktons (171.71 mg kg-1) and also in crabs (128.07 mg kg-1). Overall the concentration of Zn++, Ni++, Cd++ and Cr++ in river sediments exceeds the national and international threshold values12. The irrigation of agricultural soils with wastewater contaminated with heavy metals leads to an accumulation of Cu++, Zn++, Cd++, Cr++ and Pb++ in soil as well in the growing crops in that soil13. The exceeding concentration of heavy metals in the edible parts of crops grown on studied soil was above the safe limits in 90% of the vegetable samples examined. The results clearly showed that utilization of vegetable grown under metal contaminated waste water may provide a potential pathway of human exposure and toxicity13. According to study conducted in Omani standards shows that the use of treated sewage containing heavy metals represents a trend of increasing heavy metals values in soil, grassroots and snails of areas irrigated by treated sewage effluents. In a long term study conducted in India indicated that use of treated waste water of oil refinery in agricultural field indicated increasing trend of certain heavy metals compared to soil irrigated with ground water14.
In this review we have made an effort to address the recent progress in metal-microbes interactions, with special reference of toxicological impact of metals on living organisms, and the response of microbes in developing metal-tolerance and resistance mechanisms, as well as potential applications of such microbes in bioremediation of metal-polluted environments.
Effects of heavy metals on living organism
Based on their biological functions metals have been classified as; (i)- essential metals; (ii)- toxic metals and metalloids with unknown biological functions; (iii)- non-essential, non-toxic metals with no biological functions2. Heavy metals ions are chemical moieties that can exert its negative effect through various mechanisms such as oxidative damage by the production of reactive oxygen species, protein inactivation and DNA damage etc. Toxicity of copper aquatic and marine invertebrate species have been documented15. The Lethal Concentrations (LC50s) are typically less than 0.5 mg/l, but may range from 0.005 to >200 mg/l under certain conditions3,16,17. Similarly, copper exert toxic effect on aquatic plants, and most commonly used as analgicide and herbicide. Inhibition of growth generally occurs at a very low concentration18. Although it is classified as an essential element in mammalian nutritional system because of its prime importance in many enzymatic reactions .The minimum dietary requirement in humans is 0.0001 mg/day18. There are only a few reported cases of acute poisoning by copper and its salts. The chief symptoms following ingestion are epigastric burning, nausea, vomiting, and diarrhea3,18. Role of copper poisoning is known for individual suffering from Wilson’s disease however chronic copper toxicity is rarely reported6. The acute toxicity level of cadmium to aquatic invertebrates varies enormously among the different taxonomic groups. The LC50 dose is approximately 0.005 mg/l for one of the most sensitive species16,17. Toxicity of cadmium and, in its free ion are well known to plant and animal and microbial species. The chronic and acute toxicity to aquatic plants is accelerated by change in pH and concomitant effect on the presence of free cadmium ion3,17,19. Similarly, Marine algae are generally much more tolerant of cadmium than fresh water species, owing to the binding of the free cadmium ion with chloride. Potential carcinogenic effect of cadmium through inhalation has been described by USEPA and it is regulated based on renal toxic effects. Acute exposure to cadmium leads to nausea, vomiting, salivation, diarrhea, and muscular cramps20. Severe to fetal cases may show the following symptoms, liver injury, convulsion, shock, renal failure, and cardiopulmonary depression3,17. Acute toxicity of zinc to invertebrates is relatively low, with 48 to 96-h LC50 generally ranging from 0.5 to 5 mg/l17. Mortality of marine invertebrates following exposure to zinc is likely related to a progressive decrease in the ability to osmoregulation6. Toxicity of zinc to aquatic plants is highly variable, with LC50 ranging from 0.01 to 100 mg/l. This extreme variability associated with diverse physicochemical conditions on uptake and the ability of many species to adapt to high zinc levels3,17. The toxic effect of zinc is related to interference with phosphorus metabolism. The typical western diet yield 10-15 mg Zn / day in 70 kg reference man, zinc is essential in several enzymes and enzyme functions; DNA, RNA, and protein synthesis; carbohydrate metabolism and cell division and growth16,17. The minimum amount of zinc required per day ranges from 1 to 5.5 mg, depending on age, and pregnancy/lactation in females. Ingestion of 2 g Zn produce toxic symptoms: fever, diarrhea, vomiting and other gastrointestinal tract irritation, nausea, muscular weakness and pain. Symptoms associated with zinc deficiency include growth retardation, loss of taste, and hypogonadism3,17. Cobalt is moderately toxic to most aquatic species. Long term exposure of fish to cobalt exerts toxicological effects. These effects include decrease in muscle glycogen, hyperlacticemia, and necrosis of gill epithelia cells and poor oxygen uptake3,17. The total body burden of cobalt is approximately 1.5 mg in a 70 kg adult. Acute exposure to cobalt may led to a depression in iodine uptake, anorexia, nausea, vomiting and diarrhea. Neurotoxicological symptoms, including headache, peripheral neuritis and changes in reflexes have also been reported. The carcinogenic potential of copper, cadmium, zinc, and cobalt have not been evaluated by the International Agency for Research on Cancer3,17.
Mechanism of Heavy metals Resistance in Bacteria
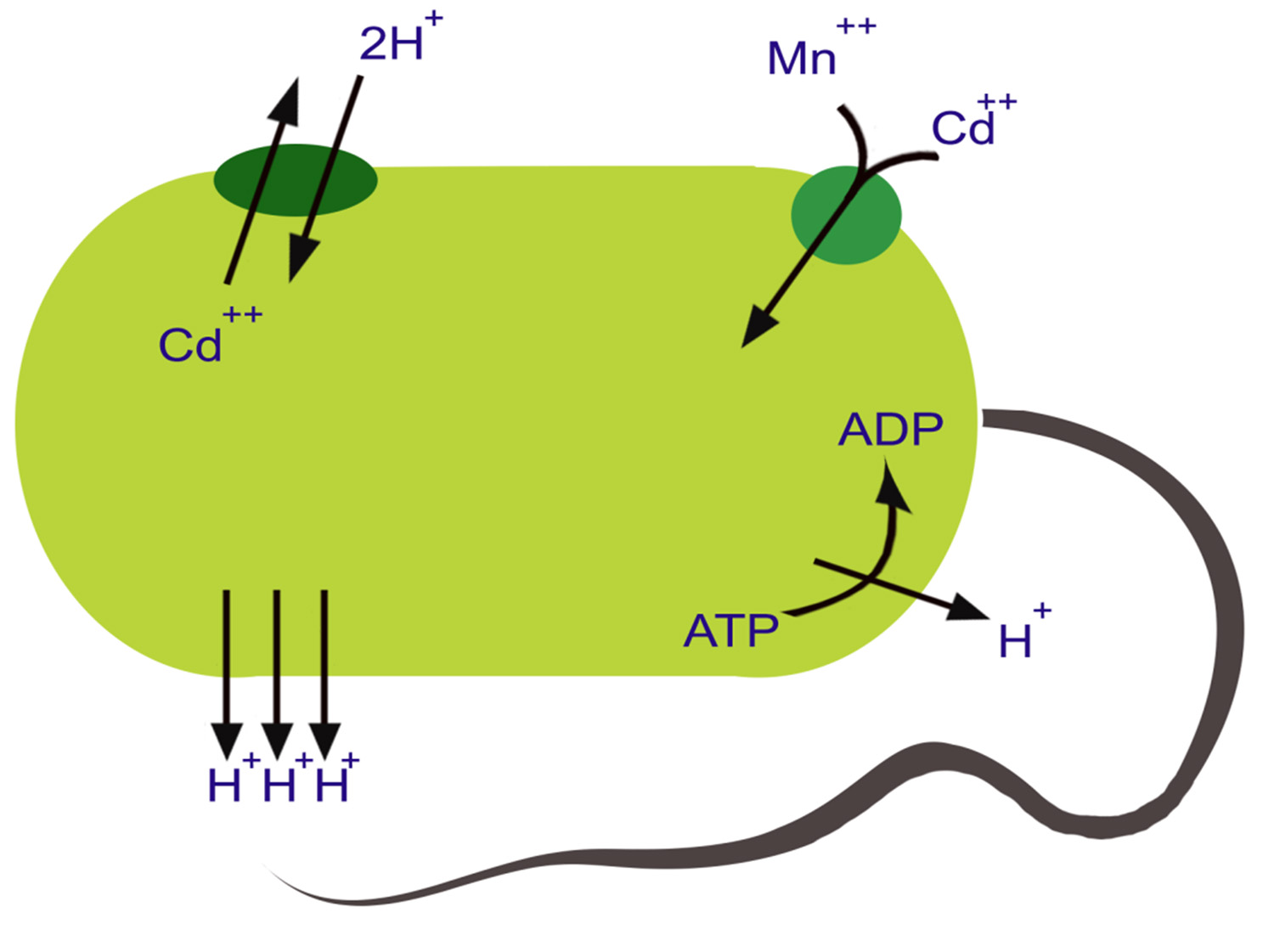
Several mechanisms have been developed by bacteria to resist and detoxify heavy metals. The response of bacteria towards heavy metals relies upon the type of the bacteria and its intrinsic ability to tolerate metal concentration. In general the response of bacteria may fall into two categories: (1) general mechanisms, which convey resistance but do not depend on metal stress for their activation, and (2) mechanisms depend on activation by specific metals (Fig. 1).
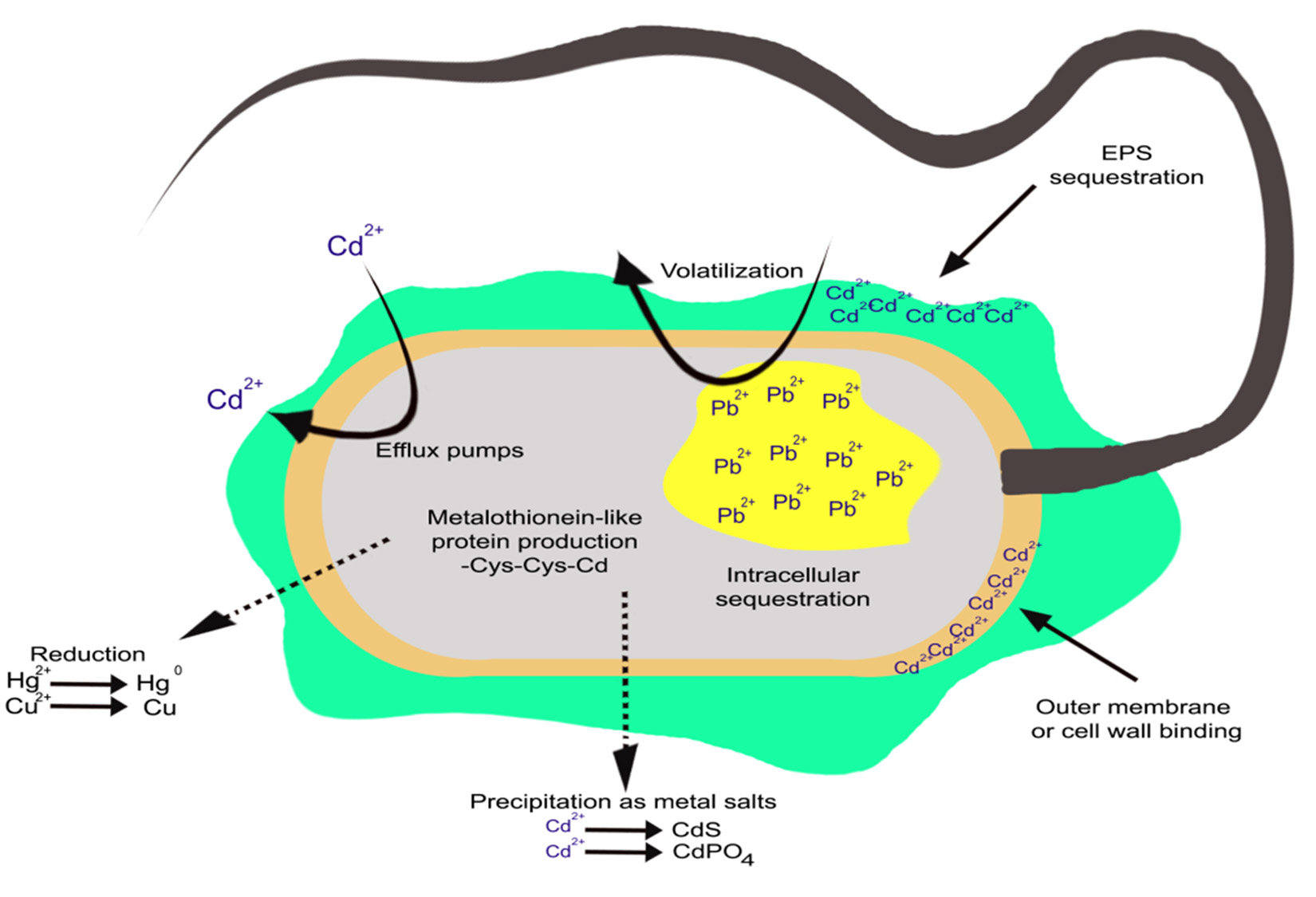
Fig. 1 . Mechanisms (intracellular and extracellular) developed by microorganisms to resist and detoxify harmful heavy metals
The production of slime layers or exopolymers around the cell, demonstrates one of the general mechanisms that do not require stimulation by metal stress. These exopolymers bind metals, as well as protecting cells against desiccation and phagocytosis. The efficiency of metal:exoplymer binding depends on pH, this is due to the negative charge, which is the functional group of these exoploymers. The exopolymers are demonstrated by Micrococcus leutus and Azotobacter spp. The production of metallo-thionein-like proteins is one of several metal-dependent mechanisms of resistance that have been demonstrated in bacteria. Metallothioneins have strong affinity with metals such as silver, zinc, cadmium, copper and mercury. The presence of metals induces synthesis of these proteins. Hence, metal detoxification is their primary function. These proteins have been found in Synechoccus spp., E. coli and Pseudomonas putida2,21.
Not quite, but almost all bacteria have genetic mechanisms for resistance to several metals and their ions such as Ag+, AsO2-, AsO43-, CrO42-, Co++, Cd++, Cu++, Hg++, Ni++, Pb++, Sb+++, TeO32-, Zn++ and Ti+. The mechanism of resistance is mainly through energy dependent efflux of toxic metal. These extrusion systems are ATPase type and some are chemiosmotic cation/hydrogen ion antiporters types8,22. Three general categories may includes:
- Plasmid-determined metal resistances are similar and more specific to those of antibiotic resistances. There is no general mechanism of resistance to all metal ions23.
- Plasmids encodes metal resistance system seems common among bacterial population. The general mechanism of resistance associated with plasmid and also detected on chromosomes of other bacteria24,25.
- The efflux pump mediated transport system and enzymatic detoxification of metals are common resistance mechanisms26.
Role of transport protein families in heavy metal metabolism
Metal-exporting proteins and their associated physiological functions and distribution are summarized here. CBA efflux pumps are governed by the P type ATPases, RND (Resistance Nodulation Cell Division) super family, cation diffusion facilitator and chromate efflux proteins2,27. These proteins are member of protein family that were evolved in early evolutionary history. These transport systems belongs to the ATP-Binding Cassette (ABC) family, A-type ATPases and P-type ATPases since it uses ATP as fuel for the transport process of heavy metals. In some bacteria, these Resistance-Nodulation-Cell Division (RND) exert metal cation efflux along with proton-influx. Although the actual mechanism of driving force in metal transport is not exactly known; few examples are discussed here such as the CorA proteins, HoxN, the CHR and the Cation Diffusion Facilitator (CDF) family8,22.
ATP-Binding Cassette (ABC)-Transporters
ATP-binding cassette abbreviated as ABC-Transporters, are distributed in all forms of life (prokaryotes and eukaryotes). The ABC-transporters don’t perform simultaneous uptake and efflux of compound, both of these functions occurs independently. The core structure of ABC-transporter composed of four subunits; out of which two are integral membrane proteins and two additional subunits are projecting towards the cytoplasm, hydrolyze ATP, and they might be identical or not in their structure. Therefore, the core of an ABC-transporter is a single polypeptide chain with four domains encoded by upto four genes28. Depending upon the direction of the transport the core of ABC-transporter is supported by additional proteins in gram-negative bacteria like periplasmic substrate-binding protein cooperate uptake-ABC-transporters29. While in export ABC-transporters two other proteins, outer membrane associated protein (OMF family) and a membrane fusion protein (MFP-family) are involved. The ABC core transporters are proposed to transport compounds across cytoplasmic membrane in association with the OMF and the MFP-proteins30. In bacteria such as E. coli ABC-transporters for heavy metal ions, are inducible uptake systems types and utilizes inducible substrates are Cu+, Ni2+ and Ag+29,31.
P-type ATPases
P-type ATPases formed by single polypeptide chain utilize single transmembrane channel of the transporter and the ATP-binding domain where hydrolysis takes place. However, in certain cases additional subunits may be used by transporters. Similar to ABC-transporters, P-type ATPases can transport in unidirectional way as an efflux or an uptake system8. P-type ATPases can transport inorganic metal ions both divalent and monovalent as well as protons. For example Salmonella typhimurium harbors two P-type ATPases that are inducible in nature responsible for influx of MgtA MgtB and Mg32. E. coli has 4 well known P-type ATPases encoded by its genome8. One of the transporter is Mgt A (now designated AtmA, o898, gb#2367363) and causes uptake of Mg2+ (and Ca2+?). Another transport K+ (AtkB, gb#1786914), Cu2+ (AtcU, gb#1786691) or efflux Zn2+ (AtzN, gb#1789879)33. In addition to the P type driven efflux system for heavy metal, previously efflux system for Cd in Staphylococcus aureus, Cad A has been reported34. CadA belongs to eucaryotic P-type ATPases required for mono and divalent inorganic ions and protons transport. However Cad A subfamily also includes other members which involved in metal transport. In addition to this, two P-type ATPases are found, responsible for concentration dependant influx and efflux of copper in Enterococcus hirae33.
F-type, V-type and A-type ATPAses
A-type ATPases are a distributed in mitochondrial inner membranes and chloroplast thylakoid membranes and bacterial membranes, F-ATPase together with V-ATPase and A-ATPase, belong to superfamily of related ATP synthases, which differ in terms of function35.
In E. coli and S. aureus, ArsA subunit is not responsible for the export and it is solely attributed by the chemiosmotic gradient, while in other cases, efflux is additionally mediated by hydrolysis of ATP. In each of the case, arsenate is not exported directly firstly it is reduced to arsenite, then it might be required to distinguish structurally similar phosphate form that cannot be further reduced and arsenate form36,37.
Resistance-Nodulation-Cell Division (RND) transporters
RND (resistance-nodulation-cell division) type transporters are high (31000 amino acid residues) proteins mainly distributed in Gram-negative bacteria. Billions of years ago they are evolved from a common ancestor through gene duplication and comprises of two related halves, one is hydrophobic membrane part and other is hydrophilic domain38. It has been hypothesized that RND proteins are proton-driven export systems where one half is substrate channel and other half-protein is a proton-channel, cooperating as a proton-substrate-antiporter22,24,38. RND-pumps in connection with a OMF (outer membrane factor) protein and a MFP (membrane fusion protein) makes a trans envelope channel that allow extrusion of substrate into the extracellular lumen from the cytoplasm22,38. In each case, RND-systems are multidrug resistance factors and common substrates for RND-driven efflux system are organic compounds as and heavy metal cations as well. As the emergence of antibiotic resistance has become a greatest threat and hamper our ability to tackle the infections associated pathogenic bacterial species. RND-driven transporter family of proteins are major contributor involved in multiple drug resistance in bacteria29,38,39. These transporters also have the ability to detoxify beta lactamase inhibitors, efforts have not been made in understanding the features particularly the members of this efflux pump transporters. RND transporters have diversity in substrate utilization and most commonly utilize heavy metal ions. The CzcCBA efflux pump transporter (where CzcA is RND protein, CzcC is OMF protein and CzcB is MFP protein) responsible for detoxification of Zn2+, Co2+ and Cd2+ 38,40,41. This plasmid-encoded pump is found on a megaplasmid of A. eutrophus. This bacterium also harbors CnrCBA efflux system, which extrudes Co2+ and Ni2+. In close relation with this, A. xylosoxidans, harbors NccCBA-system acquired for resistance to Cd2+, Co2+ and Ni2+ (38), in fluctuating copper concentrations and other metals RND systems ensuring the Myxococcus bacterium life cycle completion41.
Role of other proteins
High-Affinity Nickel Transport Protein (hoxN) family
In Ralstonia eutropha formerly known as A. eutrophus is the first member of another protein family found. This protein plays an important role in a chemolithoautotrophic life style for the uptake of Ni2+ as it is required by hydrogenases that carry assimilation of molecular hydrogen42. It has been found that Streptomyces utilizes HoxN for high-affinity nickel uptake whereas in case of Gram negative bacteria an ABC-transporter proteins are required for the same43. HoxN transporters favours the uptake of divalent cations by the chemiosmotoc gradient and it has no ATPases activity. Recently, there are various members of the HoxN-family transporters proteins are found which are either involved in Co2+ or Ni2+ uptake, but HoxN-proteins are restricted to one of the two cations44.
Chromate ion transporter (CHR) family
The majority members of this small family proteins carry efflux of chromate, in prokaryotes including archea. It is unclear that how transport is mediated; however, anion efflux is an energy dependant phenomenon in bacteria. Members of the ChrA-family of transporters utilize sulfate or chromate45.
Metal Inorganic Transport (MIT) family
The CorA protein of MIT family of transporters in S. typhimurium carry nonspecific and fast uptake for large number of divalent ions where Mg2+ comes in the form of leading molecule. CorA-transport proteins have been localized in the genome of many prokaryotic and eukaryotes. This family of protein has been abbreviated as MIT for “metal inorganic transport”46,47.
Cation Diffusion Facilitator (CDF) family
CDF is the last family of heavy metal transport in microorganisms found in archaea, bacteria, worms, yeasts, mammals including plants27,48,49. Experimental evidence suggest that the members of this group transport Cd2+ or Co2+ and Zn2+ 50,51. The first two members of this family were CzcD that recognizes periplasmic cations as a sensor for control of Czc system of the RND-transporter while other one is ZRC1p responsible for the homoeostasis of zinc/cadmium and synthesis of glutathione in S. cerevisiae, baker’s yeast51. Mostly CDF transporters poses six transmembrane alpha helices with histidine packed large hydrophilic domain; that are localized on the N-terminal of the first span, and/or between spanning four and five or sometimes in carboxy-terminal portion of the protein51,52 .
Copper Resistance mechanism
Plasmid encoded Copper resistance in Gram-negative bacteria
The detailed description of increased level of copper resistance given with plasmids in Xanthomonas53, Pseudomonas53,54,55, and E.coli53, 56. In Pseudomonas, CopR and CopS are two regulatory genes and four are structural genes copABCD53. The comparable E.coli genes are called pcoABCD and PcoRS where PcoR and PcoS are transcriptionaly regulated by a classical regulatory system56. The PcoS, localized in the membrane having autokinase activity that can be labeled at a specific conserved regions of histidine residue and senses 32P from [d-32P] ATP. PcoR act as a DNA-binding responder which gets trans-phosphorylated on a specific aspartate amino acid residue through 32P-labeled PcoS56.
The four structural proteins are CopD, two periplasmic proteins CopA and CopC, are blue copper proteins that poses 11 and 1 Cu2+, respectively and other is outer membrane protein CopB56. It has considered that accumulation of excess copper in the periplasm provide protection to the cell from toxic effect of copper ions. However the role of CopB and CopD are involved in stirring of copper across the transmembrane is not exactly clear. In a cop operon of mutant that contains only CopD not other genes provides hyper-accumulation and hyper-sensitivity of cellular copper53, indicating copD importance in uptake system of copper by the cell. In some cases there are problems in identifying uptake and efflux of copper. Copper-resistant Pseudomonas tura cells appears bright blue when allowed to grow in high concentration of copper enriched media, where as those of E.coli and Xanthomonas turns brown representing no storage of periplasmic copper53. However, there is scanty of evidence for copper efflux (not uptake) associated with the E. coli copper resistance system. Resolution of this problem still needs further research. In Pseudomonas syringae and E. coli53,56 certain chromosomal genes named as cut responsible for copper movement across the membrane and its resistance. The for these gene functions showed by three hypothetical chromosomal gene product functions are obtained through tentative models such as CutB for attachment to the intracellular domain, CutU responsible for influx and CutE for efflux for proposed membrane. All of these models are under the control of cellular uptake-efflux of copper-binding proteins protecting the cytoplasm from damage associated with copper-related redox56.
Chromosomal Copper resistance in Gram-positive bacteria
At present time the best studied copper resistance and transport system is of Gram-positive bacteria Streptococcus pneumoniae53,59 and Enterococcus faecalis53,57,58. Notably, the two genes such as CopA, for uptake CopB, that is makes efflux-type ATPase respectively, are located on a single operon57. The system is under the regulation of both copper-limited (when the CopA uptake ATPases is required) and copper excess (where the copB efflux ATPase is needed53,57,58 (Fig. 2). CopA and CopB proteins in Enterococcus may correspond to CutU and CutE, although their names are different because cop names have been employed for the plasmid genes of Pseudomonas that mediate copper resistance. Mutants of E. hirae lack CopA uptake ATPase become somewhat copper resistant and require increased copper concentration on growth medium. While the bacterial mutants that lack the CopB efflux ATPase have gained increased sensitivity to copper53,58. Cop operon is regulated through two genes products, CopY and CopZ where CopY act as a apo-repressor and CopZ as a antirepressor. The CopY apo-repressor become active only in presence intracellular Cu+ bind to the operator/promotor DNA in the operon. CopY convert into a DNA-binding repressor by the moderate level of intracellular Cu+ 53,57. At elevated concentration of intracellular Cu+, Cu+ binds to CopZ antirepressor which then binds to CopY-Cu+, making an inactive complex. The co-induction and synthesis of each type of ATPase through 20 µM Ag+ or 2 mM Cu2+ could be explained by tentative model53,58.
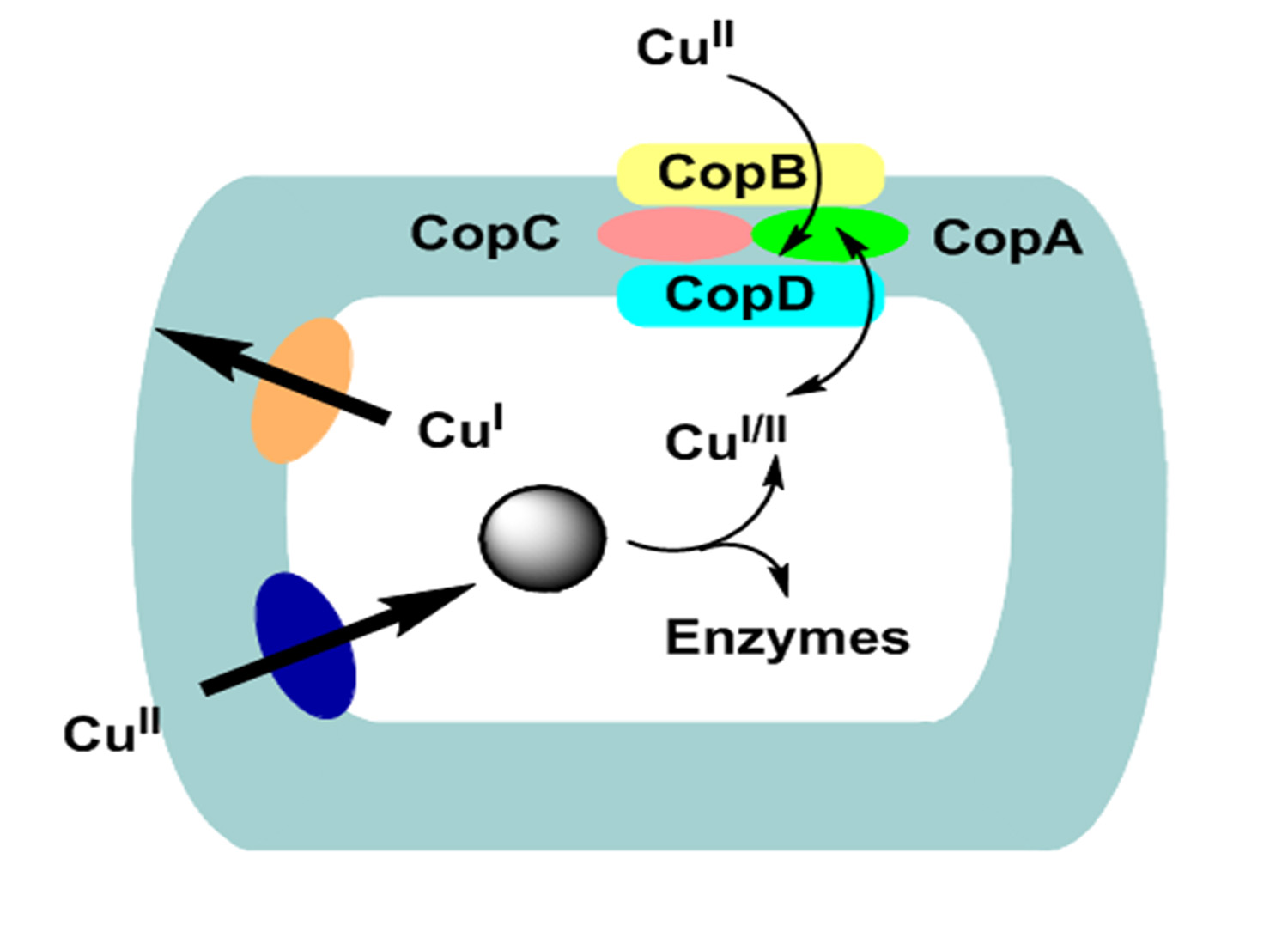
Fig. 2. Copper transport, hypothesized CutU (uptake) and CutE (efflux) P-type ATPases model in Gram-positive bacteria
Significance of P-type ATPase for cadmium resistance in Gram-positive bacteria
The ATPase for Cd2+ efflux composed of 727-aa found in Staphylococcal plasmid PI 258 was the first system widely distributed among Gram-positive bacteria at present time60,61. The Cd2+ ATPase are also present in Bacillus subtilis as well as Bacillus cereus62,63. The protein structure of this transporter is typical of P-type ATPases that poses metal- binding motif, with a vicinal Cys pair. This motif shows similarity with cadmium-, copper- and mercury-binding regions located on efflux ATPase as well as other proteins (Fig 3). The membranous ATPase region closely shows homologous relationship with other P-type of ATPase of microorganisms, animals and plants60,61,64,65.
Cadmium, Zinc and Cobalt resistance in Gram-negative bacteria result from a three polypeptide Chemiosmotic Antiporter
The three-component Czc (Cd2+, Zn2+, Co2+) is an efflux system that functions to transport divalent cation/proton antiporter which is comprises of outer membrane (CzcC), inner membrane (CzcA) and membrane-spacing (CzcB) proteins transporting cations from the cytoplasm across the periplasmic space of cell to the extracellular environment cooperatively38,66. Among the four proteins, CzcA (1064 aa) is the largest and essential for cation transport and plays a central role in the efflux protein complex. Only CzcA forms most of the part of transmembrane a-helices and is thus capable of making a tunnel like structure. Deletions of CzcB or CzcC genes confer cobalt resistance in strains while CzcA alone is probably able to catalyze a slow efflux of Co, In the primary sequence of CzcA no putative ATP-binding site is present in contrast to CadA protein of S. aureus. Thus, it is hypothesized that the underlying mechanism for the Czc-encoded efflux protein action is mediated through cation-proton antiport38,66,67.
Secondary structure of CzcA protein reveals that it poses four domains two hydrophilic and two hydrophobic domains. The N-terminal membrane region comprises of transmembrane a-helices having charged amino acid residues (both positive and negative) spanning in the center of membrane, contrary to the hydrophobic region located in the middle of two predicted cytoplasmic domains. Therefore, N-terminal domain might act as a proton “tunnel” transport protons across the cytoplasmic membrane through a charge-relay-system27,38. Accordingly, hydrophobic domain that does not have charged residues in the middle of the membrane might act as a cation tunnel. The CzcA protein contain limited amount of cysteine and histidine residues and thus decreased possible metal binding sites. Metal cation may be required by CzcB protein ( composed of 521 aa) second largest protein of czc. CzcB contains two possible metal binding sites each site contain four histidine residues. Both of the sites show homology with each other; in coordination with each other the CzcA and the CzcB proteins function as Zn2+ efflux pump. In the proposed initial cation-binding site of CzcAB no cysteine residues are present, in contrast to CadA (Cd2+ pump in the first place)27,38. The CzcC protein (346 aa) third protein of efflux complex thought to be function as a modificator switching. The transporter substrate specificity starts from Zn2+ only to Cd2+, Zn2+, and Co2+. CzcC protein which does not have cysteines or histidines, is solely dependent on the CzcB protein for their function. Czc operpon is induced by a novel two-component regulatory system comprises of the soluble regulatory protein CzcR and a membrane-bound sensor CzcD (Fig 4)27,38. These proteins do not show any resemblance from the two component trans-phosphorylating sensor/transducer regulatory proteins and thus, the underlying biochemical mechanism to sense and regulate might be entirely different67.
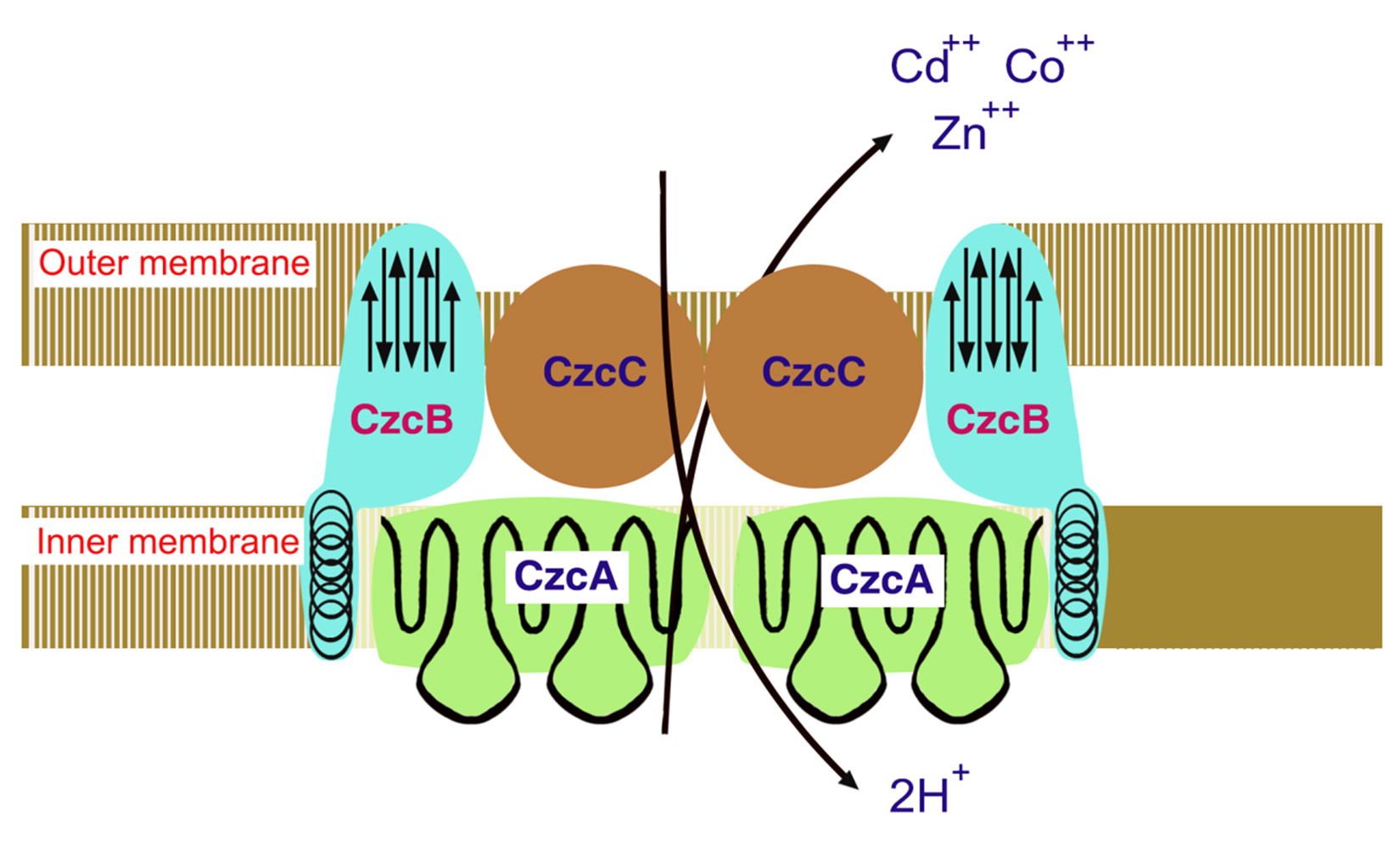 Fig. 4. Czc model for Cd2+, Zn2+, and Co2+ efflux system functioning as proton/cationantiporter consisting of inner membrane (CzcA), outer membrane (CzcC) and membrane fusion (CzcB) proteins functioning as a dimer
Fig. 4. Czc model for Cd2+, Zn2+, and Co2+ efflux system functioning as proton/cationantiporter consisting of inner membrane (CzcA), outer membrane (CzcC) and membrane fusion (CzcB) proteins functioning as a dimerBiological treatment technologies for metals remediation
Biological treatment technologies for metals remediation are in continuous development. Metals and their salts have the ability to inhibit biological activity. Although some metals are essential micronutrients, they become toxic to animal, humans and plants. The storage of heavy metals in the ecosystem, particularly within the food chain, could lethal to human health. Microorganisms have advanced mechanism to protect themselves from the lethal doses of metals such as adsorption, oxidation/reduction, or methylation. These mechanisms can be adopted with some manipulation in treatment strategies for the cleanup of metal polluted environmental68. Natural bacterial populations play an important role in metabolism of major elemental as well as organic compounds69. The pollutants in the environmental pose a greatest risk to existing natural bacterial populations, their adaptive responses in polluted environments have been studied extensively. Such findings suggest that bacteria which survive and flourish in metal polluted environments have acquired genetic systems that counteract the effect of elevated metal ion concentrations8. Microbial metabolism processes are extensively studied for metal remediation such as biotransformation (involves microbial oxidation and reduction of metals). Bioprecipitation (by precipitating metal ions at the cell surface through microbial mechanisms) such as cation efflux to change pH or using a cell-bound phosphatase to release phosphate at the cell surface, and biosorption using natural microorganisms21,68,69 or using recombinant microbial biomass to adsorb metal ions8. Bioaccumulation involves the transfer of a metal from a contaminated matrix to biomass. Microbial biomass has been shown to adsorb inorganic as well as organic compounds from aqueous phase. Metals can be accumulated by selected living organisms or onto inactivated nonliving biomass. The mechanisms to remove heavy metals from water can include two distinct pathways depending on whether the cells are living or dead. Biomass has been shown to be as efficient as many ion exchange resins for this type of removal. The process can be employed to concentrate and recovering metals, primarily from water solutions69,70,71,72. Selected microorganisms through oxidation or reduction of metals remediate various types of metal contaminates in surrounding environment. The redox reactions can accomplished directly by the organism or may be as a consequence of a reducing agents released by them69,70,71,72. Biological methylation refers to the process in which organisms attach a methyl group (-CH3) to an inorganic form of metal. Methylation directly affects the mobility of the metal. Among metals known to be methylated or demethylated by microorganisms are mercury, arsenic, cadmium, and lead69,70,71,72. Biosorption refers to the process of being taken up or hold by either adsorption or absorption of a molecule by an organism. Biosorption has been extensively studied process of metal remediation by microorganisms69,71,72,73. When the metals ions are strongly adhered on soil or sediment or in complex forms, ions exchange technology as well as biosorption unable to remove and recover metal ions like in case of Hg++. The treatment through adsorption and bioprecipitation process shows increased sensitivity to conditions, e.g.: change in pH and the presence of various inorganic and organic compounds69,70,71,72. The effectiveness pre-cipitation and ion exchange is limited in treating metal-polluted wastes, particularly for dilute metal wastes in contrast to commonly employed strategies69,72. The studied mechanism of heavy metals biosorption mediated through pure microbial species reveals that one of microbial group which have great capacity of metal sorption are majority of Gram-positive bacteria69,72,74. Biosorption has been considered as an alternative technology in treatment of industrial wastewater by means of removing the metal residues from aqueous solution69,72,73. Table 1, shows a list of microorganisms that have been successfully used to remove heavy metals or their ions from contaminated sites. Recently, the technology of phytoremediation (utilization plants to detoxify heavy metal ions) have been applied extensively using natural plants such as, with the aid of soil natural bacterial communities has shown great potential making a biological tool for remediation of heavy metals or their ions68,72,75. Another promising new tool that can be used together with the aid of microorganisms in heavy metal cleanup from contaminated soil and wastewater is the nano-particle technology21.
Table (1):
Microorganisms species that have successfully used to remove heavy metals from contaminated environments
| Microorganism type | Species | Heavy metal removed | Environment type |
|---|---|---|---|
| Bacteria | Bacillus cereus strain XMCr-6 | Cr (VI) | Soil |
| Kocuriaflave | Cu | Water | |
| Bacillus cereus | Cr (VI) | Water | |
| Sporosarcinaginsengisoli | As (III) | Water and soil | |
| Pseudomonas veronii | Cd, Zn, Cu | Water | |
| Pseudomonas putida | Cr (VI) | Soil | |
| Enterobacter cloacae B2-DHA | Cr (VI) | Soil | |
| Bacillus subtilis | Cr (VI) | Soil | |
| Filamentous Fungi | Aspergillusversicolor | Ni, Cu | Water |
| Aspergillus fumigates | Pb | water | |
| Gloeophyllumsepiarium | Cr (VI) | Soil | |
| Rhizopusoryzae | Cr (VI) | soil | |
| Yeast | Saccharomyces cerevisiae | Pb, Cd | Water |
| Algae | Spirogyra and Cladophora | Pb (II), Cu (II) | Water |
| Spirulina and Spirogyra | Cr, Cu, Fe, Mn, Zn | Water | |
| Hydrodictylon | As | Water | |
| Oedogonium | As | Water |
Table adapted with modification from (72)
Bioremediation
Biologically-based pollution treatment, or bioremediation, is a technology that has standard engineering approaches. Currently bioremediation is attracting considerable attention as a potential tool for degradation of troublesome pollutants by employing competent microorganisms for the restoring the health of contaminated soil and/or water18,76,77,78.
Concepts of bioremediation have evolved from treatment and management of industrial and municipal wastewater including solid wastes. From the beginning of late 9th century, involvement of soil bacteria and fungi in decontamination processes have continued to add newer methods of land treatment for the purpose biodegradation of particular type of toxic components76,77,79. Developments in wastewater treatment technologies have been shifted in controlling and managing the contamination of ground waters78,80. Many naturally occurring bacteria and fungi detoxify organic compounds and heavy metals. Microbial bioremediation may therefore provide a means for on-site treatment of organic pollutants that circumvents excavation or harsh chemical treatments. In addition, a number of metal-resistant bacteria and fungi have been described at the biochemical and genetic levels18,78, 81. Various researchers have suggested the feasibility and benefits of introducing new metabolic activities into a particular microbial environment using one type of microbial species36,78,82,83. The use of microbes to treat industrial waste streams in treatment tanks or immobilized filers will allow the recovery and disposal of metal pollutants. The new biocatalytic activity can be achieved at inoculated site either through the growth and metabolism of introduced microorganisms or by the exchange of genetic material responsible for degradation21,79. The only possible remediation methods of heavy metals are based on concentration and subsequent degradation. Afterwards, the residual end products can be damped in a controlled manner or recycled for recovery of metal. Metals, which are ready available in industrial or municipal wastewater can be removed via biologically, precipitation, induced adsorption, and transformation or complexation processes78. At low concentration transition metals such as nickel, cobalt, zinc and copper are essential for various cellular and metabolic processes of microbial species. However, elevated levels of these transition metals can be cytotoxic. Low concentration of heavy metal contamination above threshold levels are most effectively can be removed through bisorption technology72. Microbes are known to significantly alter the metal distribution in the surrounding environments. Metal removal from aqueous and soil system using one or more combinations of microorganisms can be accomplished by involving in situ and ex situ techniques. Bacteria are more efficient in metal transformation, biosorption, bioaccumulation and biotransformation. This clearly holds s promise for effective economical and eco-friendly metal bioremediation72.
Metal microbe interaction is complex and can be understood through its surface phenomenon, biosorption using dead biomass and uptake by living biomass. Presence of various resistance mechanisms in bacteria is an excellent survival strategy of bacteria to transform metal in the presence of even elevated concentration in aqueous and soil system. However for biosorption technology, innovative, economically feasible and biomass regeneration and conversion of recovered metal into usable form are the best option. Further, bioremediation of metal from contaminated soil required more integrated approach applying ex situ and in situ techniques. Advance innovation and developing most feasible technology is required to address this complex environmental problem where microbial technology seems to be the solution of the problem considering the physiological and genetic basis of metal-microbes interactions.
None
The author declares that there are no conflict of interest.
- Inceer H, Ayaz S, Beyazaנolu O, Emine S. Cytogenetic effects of copper chloride on the root tip cells of Helianthus annuns.Turk. J. of Biol., 2003; 27: 43-46.
- Pepper IL, Gerba CP, Gentry TJ. Environmental Microbiology 3rd edition. 2015 Academic Press, San Diego, USA.
- Jan AT, Azam M, Siddiqui K, Ali A, Choi I, Rizwanul Haq QM. Heavy metals and human health: mechanistic insights into toxicity and counter defense system of antioxidants. Int. J. Molec. Sci., 2015; 16: 29592-29630.
- Aralp LC, Erdincler A,Onay TT. Heavy metal removal from wastewater and leachate co-treatment sludge by sulfur oxidizing bacteria. Water Sci. Techno., 2001; 44: 53-58.
- Mejare M, Bulow L. Metal-binding proteins and peptides in bioremediation and phytoreme-diation of heavy metals. Trends Biotechnol., 2001; 19: 67-73.
- Bozhkov A, Padalko V, Dlubovskaya V,Menzianova N. Resistance to heavy metal toxicity in organisms under chronic exposure. Ind. J. Experimen. Biol., 2010;48: 679-696.
- Valls M, de Lorenzo V. Exploiting the genetic and biochemical capacities of bacteria for the bioremediation of heavy metal pollution. FEMS Microbiol. Rev., 2002; 26:327-338.
- Issazadeh K, Jahanpour N, Pourghorbanali F, Raeisi G, Faekhondeh J. Heavy metal resistance by bacterial strains. Ann. Biol. Res., 2013; 4: 60-63.
- Yang T, Ming-Li C,Jian-Hua W. Genetic and chemical modification of cells for selective separation and analysis of heavy metals of biological and environmental significance. Trends Anal. Chem., 2015; 66: 90-102.
- HillMK. Understanding Environmental Pollution 3rd edition. 2010, Cambridge University Press, Cambridge, UK.
- Zhang Z, Juying L, Mamat Z, Qing Fu Y. Source Identification and pollution evaluation of heavy metals in the surface of sediments of the Bortala River, northeast China. Exotoxicol. Environ. Saf., 2016;126: 94-101.
- Mendoza-Carranza M, Sepתlveda-Lozada A, Diaz-Ferreira C, Giessen V. Distribution and bio-concentration of heavy metals in a tropical aquatic food web: a case study of a tropical estuarine in SE Mexico. Environ. Poll., 2016; 210: 155-165.
- Balkhair KS, Ashraf MA. Field accumulation risk of heavy metals in soil and vegetable crop irrigated with sewage water in western region of Saudi Arabia. Saudi J. Biol. Sci., 2016; 23: S32-S44.
- Al-Musharafi SK, Mahmoud IY, Al-Bahry SN. Heavy metal pollution from treated sewage effluent. APCBEE Procedia, 2013; 5: 344-348.
- 15. El Assal FM, Abdel-Meguid ZA. Impact of heavy metal pollution of Procambarus clarkia (Crustacea: Decapoda) from Egypt. J. Water Resour., 2017; 7; e270.
- Sharma RK, Agarwal M. Biological effects of heavy metals: an overview. J. Environ. Biol., 2005; 26: 301-313.
- 17. Jaishankar M, Tseten T, Anbalagan N, Mathew BB, Beedegowda KN. Toxicity, mechanism and health effects of some heavy metals. Toxicol., 2014; 7: 60-72.
- Oves M, Khan SM, Qari HA, Felemban NM, Almeelbi T. Heavy metals: Biological importance and detoxification strategies. J. Bioremed. Biodegrad., 2016; 7: e334.
- Amari T, Ghnaya T, Abdelly C. Nickel, cadmium and lead phytotoxicity and potential of halophytic plants in heavymetal extraction. South Afri. J. Botany, 2017; 111: 99-110.
- USEPA. Exposure Factors Handbook, edition (final report). Office of Research and Development, U.S. Environmental Protection Agency, 2011 .
- Gupta A, Joia J, Sood A, Sood R, Sidhu C, Gaganjot K. Microbes as potential tool for remediation of heavy metals: A review. J.Microb. Chem. Biotechnol., 2016; 8: 364-372.
- Long F, Su C-C, Lie H-T, Bolla JR, Do SV, Yu EW. Structure and mechanism of the tripartite CusCBA heavy-metal efflux complex. Phil. Trans. Royal Soc., B.2012; 367: 1047-1058.
- Banu H, Prasad KP. Role of plasmid in microbiology. J.Aquacult. Res. Develop., 2017; 8: 1-8.
- Ahmed M.. Implications of bacterial resistance against heavy metals in bioremediation: a review. IIOAB Journal, 2012; 3: 39-46.
- Ozturk A. The use of Streptomyces coelicolor in the removal of heavy metals.Adv. Tech. Biol. Med., 2016; 4: e168.
- Canovas D, Cases I, de Lorenzo V. Heavy metal tolerance and metal homeostasis in Pseudomonas putida as revealed by complete genome analysis. Environ. Microbiol., 2003; 5: 1242-1256.
- Nies DH. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev., 2003; 27: 313-339.
- Ponte-Sucre A. ABC Transporters in Micro-organisms, Caister Academic Press, UK, 2009.
- Delmar JA, Su C-C. Yu EW. Structural mechanisms of heavy-metal extrusion by the Cus efflux system.Biometals, 2013; 24: 593-607.
- Davidson A.L, Chen J. ATP binding cassette transporters in bacteria. Ann. Rev. Biochem., 2004; 73: 241-268.
- Su C-C, Long F, Zimmermann MT, Rajashnkar KR, Jernigan RL, Yu EW.Crystal structure of the CusBA heavy-metal efflux complex of the Escherichia coli. Nature., 2011; 470: 558-563.
- Palmgren MG,Niseen P. P-type ATPases. Ann. Rev. Biophys., 2011; 40: 243-266.
- Arguello JM, Raimunda A, Padilla-Benavides T. Mechanisms in copper homeostasis in bacteria. Front. Microbiol., 2013; 3: Article 73.
- Chan H, Babayan V, Blyumin E, Gandhi C, Hak K, Harake D, Kumar K, Lee P, Li TT, Liu HY, Lo TCT, Meyer CJ, Stanford S, Zamora KS, Saier Jr. MH. The P-type ATPases superfamily. J. Molec. Microbiol. Biotechnol., 2010; 19: 5-104.
- Gruber G, Wieczorek H, Harvey W, Muller V. Structure-function relationships of A-, F- and V-ATPases. The J.Experimen. Biol., 2001; 204: 2597-2605.
- Satyapal GK, Rani S, Kumar M, Kumar N. Potential role of arsenic resistant bacteria in bio-remediation: current status and future prospect. J. Microb. Biochem. Technol., 2016; 8: 256-258.
- Deckers-Hebestreit G, Greie JC, Stalz WD, Altendorf K. The ATP synthase of Escherichia coli: structure and function of F0 subunits. Biochim. Biophys. Acta., 2000; 1458: 364-373.
- Nies DH. RND efflux pump for metal cations. In: Microbial Efflux Pump (Yu, E. W., Zhang, Q. and Brown, M. H. Eds.). Pp. 79-122, Caister Academic Press, Norfolk, UK, 2013.
- Anes J, McCusker MP, Fanning S, Martins M. The ins and outs of RND efflux pumps in Escherichia coli. Front. Microbiol., 2015; 6: 587.
- Moraleda-Munoz A, Perez J, Extremera AL, Munoz-Dorado J. Differential regulation of six heavy metal efflux systems in the response of Myxococcus Xanthus to copper. App. Environ. Microbiol., 2010; 76: 6069-6076.
- Valencia EY, Braz VS, Guzzo C, Marques M. Two RND proteins in heavy metal efflux in Caulo-bactercrescentus belong to separate clusters within proteobacteria. BMC Microbiol., 2013; 13: 79.
- Eitinger T, Mandrand-Berthelot MA. Nickel transport systems in microorganisms. Arch. Microbiol., 2000; 173: 1-9.
- Amoroso MJ, Schubert D, Mitscherlich P, Schumann P, Kothe E. Evidence for high affinity nickel transport genes in heavy metal resistant Streptomyces spec. J. Basic Microbiol., 2000;40: 295-301.
- Eitinger T. Secondary transporters for nickel and cobalt ions: theme and variations. Biometals., 2005; 18: 399-405.
- Wu W, Huang H, Ling Z, Yu Z, Jiang Y, Liu P, Li X. Genome sequencing reveals mechanisms for heavy metals resistance and polycyclic aromatic hydrocarbon degradation in Delftialacustris strain LZ-C. Ecotoxicol., 2016; 25: 234-247.
- Kalia K, Patel PC. Mechanisms for arsenic resistance in microbial cells. J. Cell Tissue Res., 2005; 5: 369-380.
- Kirsten A, Herzberg M, Voigt A, Seravalli J, Grass G, Scherer J, Nies DH. Contribution of five secondary metal uptake systems to metal homeostasis of Cupriavidusmetallidurans CH34. J. Bacteriol., 2011; 193: 4652-4663.
- Williams LE, Pittman JK, Hall JL. Emerging mechanisms for heavy metal transport in plants. Biochem.Biophys.Acta – Biomembrans., 2000; 1465: 104-126.
- Chiang HC, Lo JC, Yeh KC. Genes associated with heavy metal tolerance and accumulation in Zn/Cd hyperaccumulatorArabidopsis halleri: A genomic survey with cDNA microarray. Environ. Sci. Technol., 2006; 40: 6792-6798.
- Montanini B, Blaudez D, Jeandroz S, Sanders D. Phylogenetic and functional analysis of the cation diffusion facilitator (CDF) family: improved signature and prediction of substrate specificity. BMC Genom., 2007; 8: 107.
- Kolaj-Rbin O, Russell D, Hayes KA, Pembroke JT, Soulimane T. Cation Diffusion family structure and function. FEBS Lett., 2015; 589: 1283-1295.
- Haney CJ, Grass G, Franke S, Rensing C. New developments in the understanding of the cation diffusion facilitator family. J. Indust. Microbiol. Biotechnol., 2005; 32: 215-226.
- Rademacher C,Masepohl B. Copper-responsive gene regulation in bacteria. Microbiol., 2012; 158: 2451-2464.
- Mihdhir AA, Assaeedi ASA, Abulreesh HH, Osman GEH.Detection, identification and characterization of Some Heavy Metals Tolerant Bacteria. J. Microb. Biochem. Technol., 2016; 8: 226-230.
- Mihdir AA, Assaeedi ASA, Abulreesh HH, Osman GEH Detection of heavy metal resistance genes in an environmental Pseudomonas aeruginosa isolate. Brit. Microbiol. Res. J., 2016;17: 1-9; Article no.BMRJ.28655.
- Bondarczuk K, Piotrowska-Seget Z. Molecular basis of active copper resistance mechanisms in Gram-negative bacteria. Cell Biol.Toxicol., 2013; 29: 397-405.
- Zhang A, Wang D, Wang Y, Hasman H, Aarestrup FM, Alwathnani H A, Zhu YG, Rensing C. Genome sequence of copper resistance and sensitive Enterococcus faecalis strains isolated from copper-fed pigs in Denmark. Stand. Genom. Sci., 2015; 10: 35.
- Solioz M, Abicht HK, Mermod M, Mancini S. Response of Gram-positive bacteria to copper stress. J. Biol. Inorganic Chem. 2010; 15: 3-14.
- Fu Y, Tsui H-CT, Bruce KE, Sham L-T, Higgins KA, Lisher JP, Kazmierczak KM, Maroney MJ, Dann III, CE, Winkler ME, Gierdoc DP. A new structural paradigm in copper resistance in Streptococcus pneumoniae.Nature Chem. Biol., 2013; 9: 177-183.
- Endo G, Silver S. CadC, the transcriptional regulatory protein of the cadmium resistance system of Staphylococcus aureus plasmid pI258. J. Bacteriol., 1995; 177: 4437-4441.
- Crupper SS, Worrell V, Stewart GC, Iandolo JJ. Cloning and expression of cadD, a new cadmium resistance gene in Staphylococcus aureus. J. Bacteriol.,1999; 181: 4071-4075.
- Solovieva IM, Entian KD. Metalloregulation in Bacillus subtilis: the copZ chromosomal gene in involved in cadmium resistance. FEMS Microbiol. Lett., 2004; 236: 115-122.
- Huang F, Guo CL, Lu GN, Yi X., Zhu LD, Dang Z. (2014) Bioaccumulation characterizationof cadmium by growing Bacillus cereus RC-1 and its mechanisms. Chemosphere, 2014; 109: 134-142.
- Song WY, Martinoia E, Lee J, Kim D, Kim DY, Vogt E, Shim D, Choi KS, Hwang I, Lee Y. A novel family of Cys-rich membrane proteins mediates cadmium resistance in Arabidopsis. Plant Physiol., 2004; 135: 1027-1039.
- Sheng XF, Xia JJ. Improvement of rape (Brassica napus) plant growth and cadmium uptake by cadmium-resistant bacteria.Chemosphere, 2006; 64: 1036-1042.
- Liu P, Chen X, Huang Q, Chen W. The role of CzcRS two-component systems in the heavy metal resistance of Pseudomonas putida X4.Int. J. Molec. Sci., 2015; 16: 17005-17017.
- Prasad MNV. Metals in the Environment: Analysis and Biodiversity. 2001; Marcel Dekker Inc., New York.
- Hashim MA, Soumyadeep M, Sahu JN, Sengupta B. Remediation technologies for heavy metal contaminated ground water. J. Environ. Manag., 2011; 92: 2355-2388.
- Igiri BE, Okoduwa SIR, Idoko GO, Akabuogu EP, Adeyi AO, Ejiogu IK. Toxicity and bioremediation of heavy metals contaminated ecosystems from tannery wastewater: A review. J. Toxicol., 2018; 2018: Article ID 2568028, 16 pages.
- Choudhary M, Kumar R, Datta A, Nehra V, Garg N. Bioremediation of heavy metals by microbes. In: Bioremediation of Salt Affected Soils: An Indian Perspective. Arora S., Snigh A. K., Snigh Y. P. (Eds.). Pp. 233-255, 2017; Springer International Publishing, Switzerland.
- Mishra GK. Microbes in heavy metal remediation: A review on current trends and patents. Recent Pat. Biotechnol., 2017; 11:188-196.
- Ojuederie OB, Babablola OO. Microbial and plant-assisted bioremediation of heavy metal polluted environments: A review. Int. J. Environ. Res. Public Health, 2017; 14: 1504
- Pagenellia F, Esposito A, Toro L,Veglio F. Metal speciation and pH effect on Pb, Cu, Zn, and Cd biosorption onto Sphaerotilusnatans: Langmuir-type empirical model. Water Res., 2003; 37: 627-633.
- Yilmaz EI. Metal tolerance and biosorption capacity of Bacillus circulansstrain EB1.Res. Microbiol., 2003; 154: 409-415.
- Wood JL, Liu W, Tang C, Franks AE. Microorganisms in heavy metal bioremediation: strategies for applying microbial-community engineering to remediate soils. AIMS Bioeng., 2016; 3: 211-229.
- Philp, JC, Atlas RM. Bioremediation of contaminated soils and aquifers. In: Bio-remediation: Applied Microbial Solutions for Real-World Environmental Cleanup, Atlas, R. M. and Philp J. C. (eds.). Pp. 139-236, 2005, ASM Press, Washington DC, USA.
- Uqab B, Mudasir S, Sheikh AQ, Nazir R. Bioremediation: A management tool. J. Bioremed. Biodegrad., 2016; 7: e331
- Shah MP. Bioremediation-waste water treatment. J. Bioremed. Biodegred., 2018; 9: 1-10.
- Shah MP. Prokaryotes: a promising agents in environmental bioremediation. Adv.Recycl.Waste Managt., 2017; 2: e103.
- Prommer H, Barry A. Modeling bioremediation of contaminated ground water. In: Bioremediation: Applied Microbial Solutions for Real-World Environmental Cleanup, Atlas, R. M. and Philp J. C. (eds.). Pp. 108-138, 2005; ASM Press, Washington DC, USA.
- Siddiquee S, Rovina K, Al Azad S, Naher L, Suryani S, Chaikaew P. Heavy metals contaminants removal from wastewater using the potential filamentous fungi biomass: A review. Microb. Chem. Biotechnol., 2015; 7: 384-393.
- Xenia ME, Refugio RV. Microorganisms metabolism during bioremediation of oil contaminated soil. J. Bioremed. Biodegrad., 2016; 7: e340
- Shah MP. Microorganisms in bioremediation. J.Bioremed. Biodegrad., 2016; 7: e176.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.