ISSN: 0973-7510
E-ISSN: 2581-690X
There are various sources of microbial air pollution which are seems to be a serious public health problem all over the world. For prevention and control of air pollution caused by airborne bacteria, rapid, sensitive and reliable detection techniques are required. Therefore, our study focused on using MALDI Biotyper (MBT) for rapid recognition of various microbial air pollutants. Five hundred air samples were collected from three localities, including Qassim University (150 samples), Al-Qassim hospitals (250 samples) and poultry slaughter houses (100 samples). All air samples were collected by impactor air sampler from the indoor and outdoor environment. All samples were cultivated on nutrient and blood agar media for two days and a total of 129 isolates were purified for proteomic analysis using MALDI Biotyper (MBT) then confirmed by quantitative polymerase chain reaction (qPCR). One hundred and nineteen (92.25%) isolates were identified by MBT at the species level with a log (score) value £2. 000 whereas; 10 (7.75%) isolates were detected at the genus level with score values ranged from 1.7000 to 1.999. The MBT was able to identify 93 (72.10%) gram-positive and 36 (27.90%) gram-negative bacterial isolates. The most common genera were Staphylococcus (n = 43, 33.33%), Escherichia (n = 16, 12.40%), Enterococcus (n = 15, 11.63%) and Bacillus (n = 15, 11.63%). Staphylococcus aureus and Escherichia coli were the most frequently identified species (n = 16, 12.40% for each). In general, we detected 53 (41.10%) various bacterial species in Al-Qassim Hospitals, 41 (31.79%) in poultry slaughter houses and 35 (27.13%) in Qassim University. Throughout Al-Qassim region, the air was tainted by numerous environmental microorganisms, and the MBT was positively adjusted for their fast and accurate identification.
Airborne bacteria; MBT; Al-Qassim region; Saudi Arabia.
Air pollution is a serious problem not only in the Kingdom of Saudi Arabia (KSA) but also in many parts of the world. Albeit, this kind of contamination has different forms, some of which are unnoticeable by visualization. Pollution of air has been a grave problem since when the air we breathe carries contaminants, it can affect on our health in various approaches. Exposure to bioaerosols, holding airborne pathogens leads to various infections, including lung cancer, bronchial asthma, hypersensitivity pneumonitis, toxic reactions and various cardiac diseases1,2,3. This type of pollution can affect on people of various ages, but the incidence in elderly people and young children are frequently high. Consequently, the insufficient diagnosis and control of air quality creates significant health problems. Molds and bacteria are considered the most common microbes in indoor/outdoor areas3. The ability of microorganisms to circulate on indoor or outdoor subjects is based on the comparative moisture and the moistness content of the different objects. These may lead to obliteration, unfavorable effects on the health status 3. For that reason, it is very important to characterize indoor and outdoor airborne microorganisms.
Indoor air plays a significant role as a health contributing factor, and controlling of the indoor air quality need strategies that vary from those utilized for outdoor air4. The microbial contamination in internal and external environmental regions is caused mainly by various types of bacteria and fungi, especially molds which represents a public health problem worldwide. The temperature, wind rapidity and relative humidity are considered important as the major environmental factors which effect on the feasibility of various pathogens4,5.
Bioaerosols are airborne particles of human, animal, plant or soil origin containing microorganisms or organic compounds6,7,8. These particles become airborne when floating in the air due to wind speed, turbulence, coughing, traffic etc. Gilbert and Duchaine9, Mandal and Brandl10, Despre´s et al.11 and Fykse et al.12 evaluated the microbial contamination of bioaerosols in numerous internal and external environments and they found that the bioaerosols were contaminated by various biological agents. Moreover, Gilbert and Duchaine 9 found that the viability of the airborne microbes is based on computable aspects, for example, relative humidity, temperature and solar irradiance, as well as the particular characters of the microbes themselves, as endospore forming bacteria and pigment production. In the last few decades, the microbial concentration in bioaerosols exposure has increased due to its relationship with a wide-ranging variety of health problems and the terror of bioterrorism (terrorism involving the intended release of biological agents as bacteria, viruses, or toxins).
In hospitals, it could be imperative to examine the air for the reason that it is a grave and extensive hazard that patients acquire nosocomial infections through the bioaerosols during staying in the hospital8,13. Therefore, examining the air at common places could assist in reducing the amount of possible bioaerosols assaults. To diminish the assault of infection, initial recognition of biological agents is compulsory. Frequent checking of the airborne for identification of definite microbes is potential13. Therefore, study the bioaerosol environment is very crucial as indicated by the National Research Council14.
In addition, airborne bacteria can be established in several surroundings, nevertheless in many conditions, they don’t exist a public health danger to the unprotected persons. Therefore, the intensive care of present levels of out-of-doors airborne biological agents is compulsory to assess the possible dangers affecting the health of individuals15.
Airborne fungi are also of public health importance, since they may cause serious diseases such as allergy and respiratory diseases 16. On the other hand, Rotz et al.17 indicated that bacteria, virus, fungi and toxins are considered the most common biological agents of bio-terrorism attack in accordance with several microorganisms that developed from the centers for prevention and monitoring of various diseases. Valen8 found that bioaerosols can be established in both indoor and outdoor environment, and they frequently hold various microorganisms like bacteria, virus and fungi, which perhaps pathogenic or saprophytic. It has been recorded by previous study that few numbers (1%) of different microbes assembled from the surroundings can be cultured in various test centers by traditional methods. Nevertheless, molecular methods can identify approximately 99% of the remaining non-cultivable microbes by analysis of their nucleotide sequences and protein fingerprinting. Consequently, rapid and accurate identification of airborne biological agents using mass spectrometry technology has revealed a well-known method nearly in all clinical laboratories18. Since the genetic methods used for recognition of various types of pathogens are time consuming and highly expensive, an alternative method using Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-TOF MS) has positively been utilized for recognition of various microorganisms recovered from various sources19,20. This method needs simple tools and comparatively rapid machinist training. It was noticed that using of the mass spectrometry technology represented by MALDI-TOF-MS needs small quantity of biological materials and comprises simple procedures without preliminary measurement, for instance gram staining, when compared with DNA analysis based technologies21,22,23.
To the best of our knowledge, there are few published researches related to the use of the MALDI Biotyper for identification of airborne pathogens12,24. From the previously mentioned data, our study aimed to rapid and precise identification of airborne pathogens isolated from the indoor and outdoor environment in certain areas in Al-Qassim region using mass spectrometry technology.
Location and time of sampling
Five hundred air samples were collected from three localities. The 1st location (150 samples) was at the Qassim University (QU) classrooms and offices of the university which are located in Almulyda, North of Prince Nayef Airport (Al-Qassim region), next to the station for distribution of petroleum products (Aramco). The 2nd sampling location (250 samples) was hospitals in Al-Qassim region, including King Fahad Specialist Hospital Buraidah (50 samples), Buraidah Central Hospital (50 samples), Qassim National Hospital Buraydah (50 samples), King Saud Hospital Unaizah (50 samples), Al Bukayriyah General Hospital (50 samples). The 3rd sampling location was poultry slaughter houses (100 samples). All microbial air samples were collected by impactor air sampler from indoor and outdoor air in six replicates for each sample. The collection of air samples was carried out in the winter season of 2017, at Al-Qassim region, KSA. Throughout this season, there was an extensive utilization of environment insulation, which most probable rises the number of biological agents in the indoor and outdoor air, corresponding to high levels of breathing distress. The samples were collected by a six-stage impactor air sampler, therefore, the air was impacted in prepared plates with specific media for bacteria. The air sampler was stayed throughout the collection time, about two meters above floor level.
Isolation of bacteria
Tryptose Soy agar plus Benlate media was used for bacterial isolation. The petri dishes were incubated for 72 h at 28-30°C, after inoculation of the air samples in specific plates. The colonies of bacteria were then estimated after the 1st and 3rd day of incubation and the total of colony forming unit (CFU) m-3 air was also recorded. Moreover, the distinct colonies considered were isolated and compared with each other again. The isolated strains were stored at -70°C in glycerin 20% for further investigation. For studying of the morphological characters, the isolates were stained by particular staining technique and observed microscopically and morphological properties of the bacterial cells were demonstrated. Brain heart infusion agar (BHI) was used in our study as a suitable medium for growing of different types of microorganisms. Samples were collected using BHI agar for the enumeration of bacterial colony forming unit (CFU).
The hemolytic activity of isolated colonies
The isolated strains were examined for their degree of hemolysis. Blood agar plates supplemented with 5% sheep blood (Oxoid, UK) were used for this purpose. From each isolate, a fresh colony was inoculated onto agar medium and then preserved in 28-30°C for two successive days. After that, the colonies were categorized as a-hemolytic, b-hemolytic or g-hemolytic.
Identification of airborne pathogens using protein fingerprinting (Microflex LT)
All air samples were analyzed using the MBT compass software (Bruker Daltonics, Germany). According to the instructions of the company, identification scores of ³2.000 will be revealed as identification at the species level, whereas, the scores ranged from 1.700 to 1.999 will be indicated as identification at the genus levels. In contrast, the scores of 1.700 or less will be recognized as misidentification25,26.
Ethanol/formic acid extraction protocol was carried out according to the instructions provided by the Bruker Daltonics company. Briefly, one fresh colony was moved into sterilized tube containing 300 µl of highly purified water then mixed thoroughly. 900 ul of absolute ethanol was then added and then properly centrifuged at 13000 g for 2 min. The residue of the tube was left at room temperature for a couple of minutes. After that 50µl of 70% formic acid was added to the tube and strongly mixed. Fifty microliters of 70% acetonitrile were then added and centrifugation was carried out at 13000 rpm for a couple of min. One microliter of aliquot, was directly inoculated onto stainless steel target plate. Subsequently, 1µl of matrix solution (Sigma Aldrich, USA) was added to each hole. The target plate was consecutively placed in the MBT device for direct microbial identification. All examined samples were triplicated for precise identification.
Molecular assays
The commonly identified isolates were Staphylococcus aureus, Enterococcus faecalis, Bacillus subtilis, Acinetobacter baumanni and Escherichia coli. Therefore, we studied their virulence by real time PCR (RT-PCR, Applied Biosystem, USA). Firstly, DNA of identified isolates was extracted using a QIAamp tissue kit (Qiagen, Hilden, Germany) which was performed based on the manufacturer’s recommendations. The designated primers were shown in Table 1.
Table (1):
Primer sequences, target genes and amplicon sizes of certain virulence genes of the most frequently isolated airborne bacteria
| Species | Target gene | Primer sequence (5’−3’) | Size (bp) | References |
|---|---|---|---|---|
| Staphylococcus aureus | nuc | GCG ATT GAT GGT GAT ACG GTT | 270 | Zhang et al. 27 |
| AGC CAA GCC TTG ACG AAC TAA AGC | ||||
| Enterococcus faecalis | gelE | TAAGAAAGAACATCACCACGA | 213 | Asmat et al. 28 |
| TATGACAATGCTTTTTGGGAT | ||||
| Bacillus subtilis | bsub | AAGTCGAGCGGACAGATGG | 595 | Wattiau et al. 29 |
| CCAGTTTCCAATGACCCTCCCC | ||||
| Acinetobacter baumanni | recA | CCTGAATCTTCYGGTAAAAC | 452 | AL-Jubori et al. 30 |
| GTTTCTGGGCTGCCAAACATTAC | ||||
| Escherichia coli | iss | ATGTTATTTTCTGCCGCTCTG
CTATTGTGAGCAATATACCC |
266 | Yaguchi et al. 31 |
A total of 1,900 isolates were grown on the air sample nutrient agar plates after one day of incubation. One hundred twenty-nine purified colonies were obtained after sub-culturing for the next investigation. As shown in Table 2, 36 (27.90%) of these isolates were from Qassim University, 26 (20.16%) were from Buraidah Hospitals, 10 (7.75%) were from King Saud Hospital in Unaizah, 17 (13.18%) were from Al Bukayriyah General Hospital and 40 (31%) were from a poultry slaughterhouse in Buraidah. Throughout the first day of air sampling, the temperature was 37°C and the humidity was 70%. While in the 2nd and 3rd day of sampling, the temperature was 39°C and the humidity was 50%.
Table (2):
The tested areas and airborne bacteria recognized by MALDI Biotyper Compass Software
| Species | Qassim University | Al-Qassim Hospitals |
Poultry slaughter house |
Total no. of isolates | |||||
|---|---|---|---|---|---|---|---|---|---|
| Buraidah | Unaizah | Al Bukayriyah | |||||||
| Class rooms | offices | King Fahad Specialist Hospital | Buraidah Central Hospital | Qassim National Hospital | King Saud Hospital | Al Bukayriyah General Hospital | |||
| Acinetobacter baumannii | 3 | 1 | 1 | 1 | 0 | 1 | 2 | 0 | 9 |
| Acinetobacter lwoffii | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Aeromonas enteropelogenes | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 2 |
| Arthrobacter gandavensis | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 |
| Bacillus megatorium | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| Bacillus pumilus | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 5 |
| Bacillus subtilis | 0 | 2 | 1 | 0 | 1 | 0 | 1 | 2 | 7 |
| Citrobacter freundii | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 5 |
| Corynebacterium amycolatum | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| Corynebacterium durum | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Corynebacterium simulans | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| Corynebacterium spp. | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Enterococcus casseliflavus | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 |
| Enterococcus faecalis | 1 | 0 | 1 | 2 | 1 | 0 | 3 | 4 | 12 |
| Enterococcus faecium | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 |
| Enterococcus gallinarum | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| Escherichia coli | 2 | 1 | 1 | 1 | 0 | 2 | 2 | 7 | 16 |
| Gemella haemolysans | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| Kocuria rosea | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| Pantoea calida | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 |
| Proteus vulgaris | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 |
| Pseudomonas putida | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Shewanella putrefaciens | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 |
| Serratia liquefaciens | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 6 |
| Staphylococcus aureus | 2 | 3 | 1 | 2 | 1 | 2 | 2 | 3 | 16 |
| Staphylococcus capitis | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| Staphylococcus epidermidis | 1 | 1 | 0 | 2 | 2 | 3 | 1 | 3 | 13 |
| Staphylococcus haemolyticus | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 2 | 5 |
| Staphylococcus hominis | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 3 |
| Staphylococcus simulans | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 4 |
| Total | 21 | 15 | 7 | 12 | 7 | 10 | 17 | 40 | 129 |
Table (3):
Log (score) values of airborne pathogens identified by MALDI Biotyper
| Species | Total no. of isolates | Log (score) values | |||
|---|---|---|---|---|---|
| 0.000-1.699 | 1.7000-1.999 | 2.000-2.299 | 2.300-3.000 | ||
| Acinetobacter baumannii | 9 | 0 | 1 | 5 | 3 |
| Acinetobacter lwoffii | 1 | 0 | 0 | 0 | 1 |
| Aeromonas enteropelogenes | 2 | 0 | 0 | 2 | 0 |
| Arthrobacter gandavensis | 2 | 0 | 0 | 1 | 1 |
| Bacillus megatorium | 1 | 0 | 0 | 1 | 0 |
| Bacillus pumilus | 5 | 0 | 1 | 1 | 3 |
| Bacillus subtilis | 7 | 0 | 1 | 3 | 3 |
| Citrobacter freundii | 5 | 0 | 0 | 2 | 3 |
| Corynebacterium amycolatum | 2 | 0 | 0 | 2 | 0 |
| Corynebacterium durum | 1 | 0 | 0 | `1 | 0 |
| Corynebacterium simulans | 1 | 0 | 0 | 0 | 1 |
| Corynebacterium spp. | 1 | 0 | 0 | 1 | 0 |
| Enterococcus casseliflavus | 2 | 0 | 0 | 2 | 0 |
| Enterococcus faecalis | 12 | 0 | 1 | 6 | 5 |
| Enterococcus faecium | 2 | 0 | 1 | 1 | 0 |
| Enterococcus gallinarum | 1 | 0 | 0 | 0 | 1 |
| Escherichia coli | 16 | 0 | 0 | 9 | 7 |
| Gemella haemolysans | 1 | 0 | 0 | 1 | 0 |
| Kocuria rosea | 2 | 0 | 0 | 0 | 2 |
| Pantoea calida | 2 | 0 | 0 | 1 | 1 |
| Proteus vulgaris | 2 | 0 | 0 | 0 | 2 |
| Pseudomonas putida | 1 | 0 | 0 | 1 | 0 |
| Shewanella putrefaciens | 2 | 0 | 1 | 1 | 0 |
| Serratia liquefaciens | 6 | 0 | 1 | 4 | 1 |
| Staphylococcus aureus | 16 | 0 | 0 | 7 | 9 |
| Staphylococcus capitis | 2 | 0 | 1 | 1 | 0 |
| Staphylococcus epidermidis | 13 | 0 | 2 | 6 | 5 |
| Staphylococcus haemolyticus | 5 | 0 | 0 | 3 | 2 |
| Staphylococcus hominis | 3 | 0 | 0 | 2 | 1 |
| Staphylococcus simulans | 4 | 0 | 0 | 3 | 1 |
| Total | 129 | 0 | 10 | 67 | 52 |
Amongst the 129 colonies examined by MBT Compass software, respectable recognition of various types of airborne pathogens on the species levels was detected in 119 (92.25%) isolates with a score value ³2.000 (Table 3). Whereas; 10 (7.75%) isolates were identified by score values ranged from 1.7000 to 1.999 (Table 3). The MBT was able to identify 93 (72.10%) gram-positive bacterial and 36 (27.90%) gram-negative bacterial air isolates (Table 4). The most common identified gram-positive genera were Staphylococcus (n = 43, 33.33%), Enterococcus (n = 15, 11.63%) and Bacillus (n = 15, 11.63%). The most common identified gram-negative genera were Escherichia (n = 16, 12.40%). As can be seen in Table 4, we identified 8 various gram-positive bacterial species by MBT including 43 different Staphylococcus sp., 15 Bacillus sp., 15 Enterococcus sp., 10 Acinetobacter sp., 5 Corynebacterium sp., 2 Arthrobacter sp., 2 Kocuria sp. and 1 Gemella sp. Moreover, 8 gram-negative bacterial species were identified including 16 Escherichia sp., 6 Serratia sp., 5 Citrobacter sp., 2 for each Aeromonas, Pantoea, Proteeus, Shewanella sp. and 1 Pseudomonas sp.
Table (4):
Incidence of airborne bacteria isolated from examining air samples of different localities
| Genus | Qassim University | Al-Qassim Hospitals | Poultry house slaughter houses | Total (129) | |
|---|---|---|---|---|---|
| Gram positive bacteria | |||||
| Acinetobacter | 5 | 5 | 0 | 10 | |
| Arthrobacter | 0 | 0 | 2 | 2 | |
| Bacillus | 5 | 4 | 6 | 15 | |
| Corynebacterium | 3 | 2 | 0 | 5 | |
| Enterococcus | 1 | 9 | 5 | 15 | |
| Gemella | 0 | 0 | 1 | 1 | |
| Kocuria | 2 | 0 | 0 | 2 | |
| Staphylococcus | 11 | 18 | 14 | 43 | |
| Total | 27 | 38 | 28 | 93 | |
| Gram negative bacteria | |||||
| Aeromonas | 0 | 1 | 1 | 2 | |
| Citrobacter | 2 | 2 | 1 | 5 | |
| Escherichia | 3 | 6 | 7 | 16 | |
| Pantoea | 1 | 1 | 0 | 2 | |
| Proteus | 0 | 0 | 2 | 2 | |
| Pseudomonas | 0 | 1 | 0 | 1 | |
| Shewanella | 0 | 0 | 2 | 2 | |
| Serratia | 2 | 4 | 0 | 6 | |
| Total | 8 | 15 | 13 | 36 | |
| Total no. of gram +ve and –ve bacteria | 35 | 53 | 41 | 129 | |
As shown in Fig. 1, Staphylococcus aureus and Escherichia coli were the most frequently identified species (n = 16, 12.40% for each), followed by Staphylococcus epidermidis (n = 13, 10.10%), Enterococcus faecalis (n = 12, 9.30%) and Acinetobacter baumanni (n = 9, 6.98%). In general, we detected 53 (41.10%) various bacterial species in Al-Qassim Hospitals, 41 (31.79%) in poultry slaughterhouse and 35 (27.13%) in Qassim University (Fig. 2). We found a significant variation between the total numbers of airborne pathogens recovered from the Al-Qassim Hospitals compared with Qassim University and poultry slaughter house.
 Fig. 1. Genera of gram-positive and negative airborne bacteria recovered from three different localities in Al-Qassim region
Fig. 1. Genera of gram-positive and negative airborne bacteria recovered from three different localities in Al-Qassim region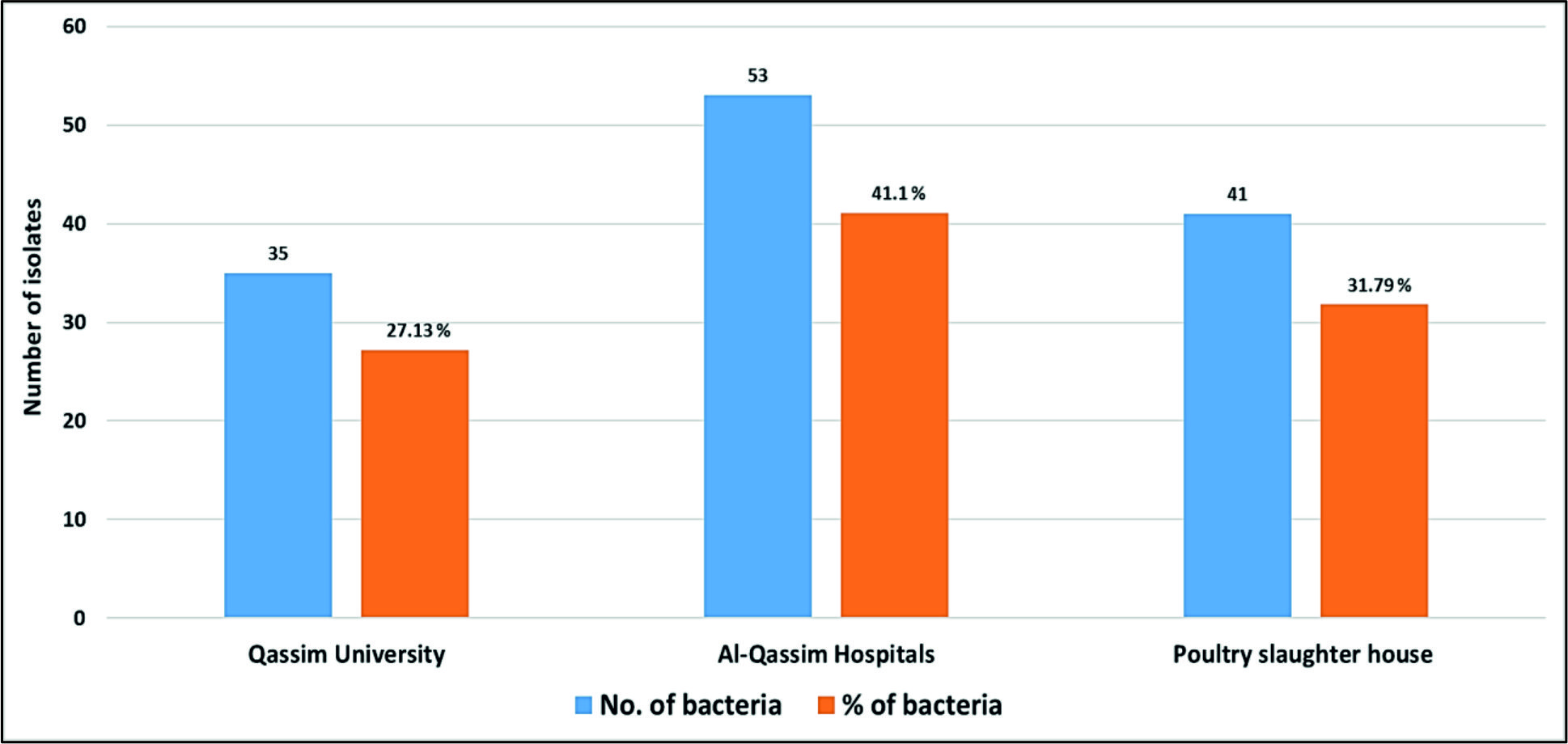 Fig. 2. Numbers and percentages of various airborne pathogens isolated from three different areas in Al-Qassim region
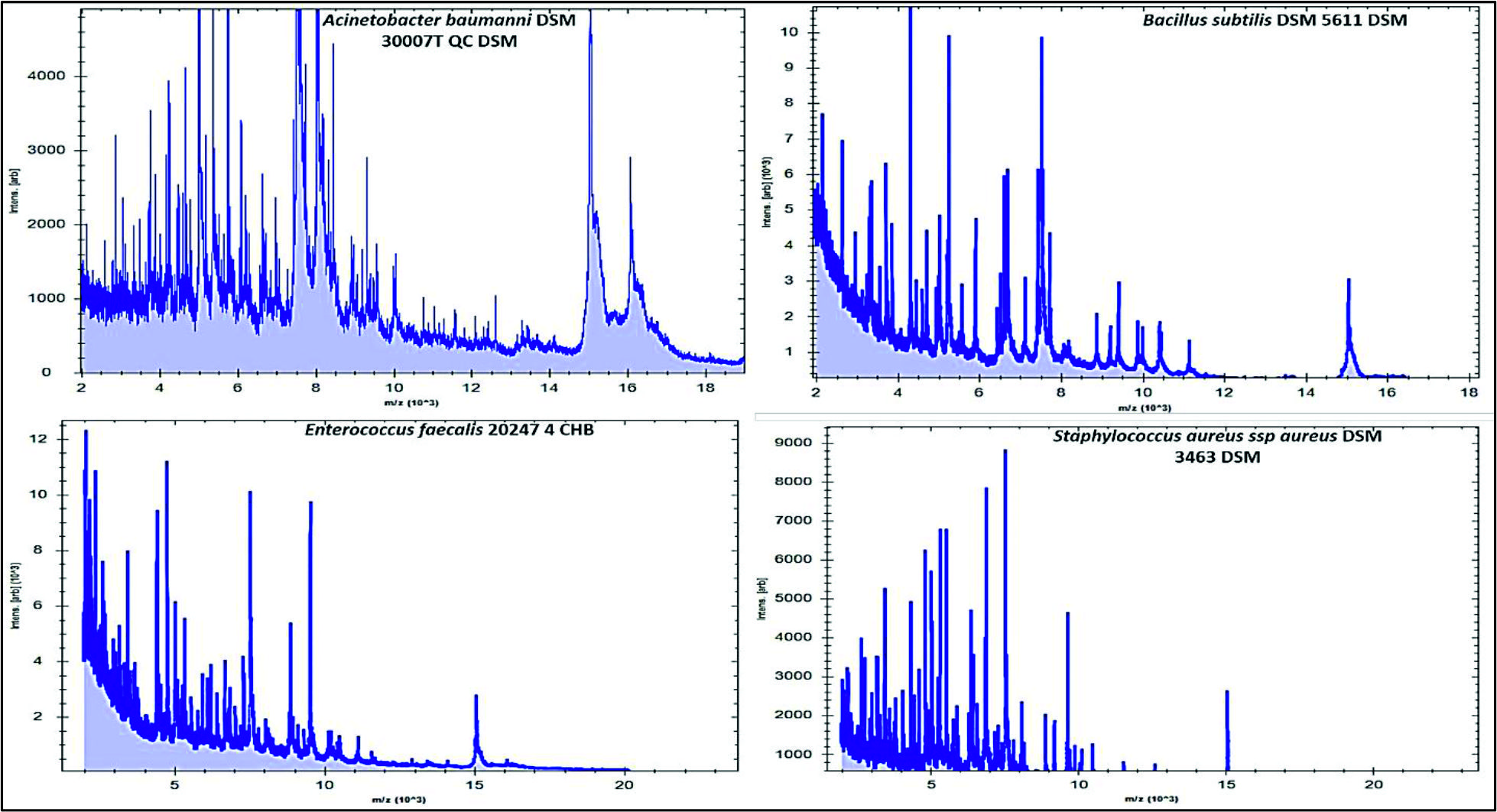
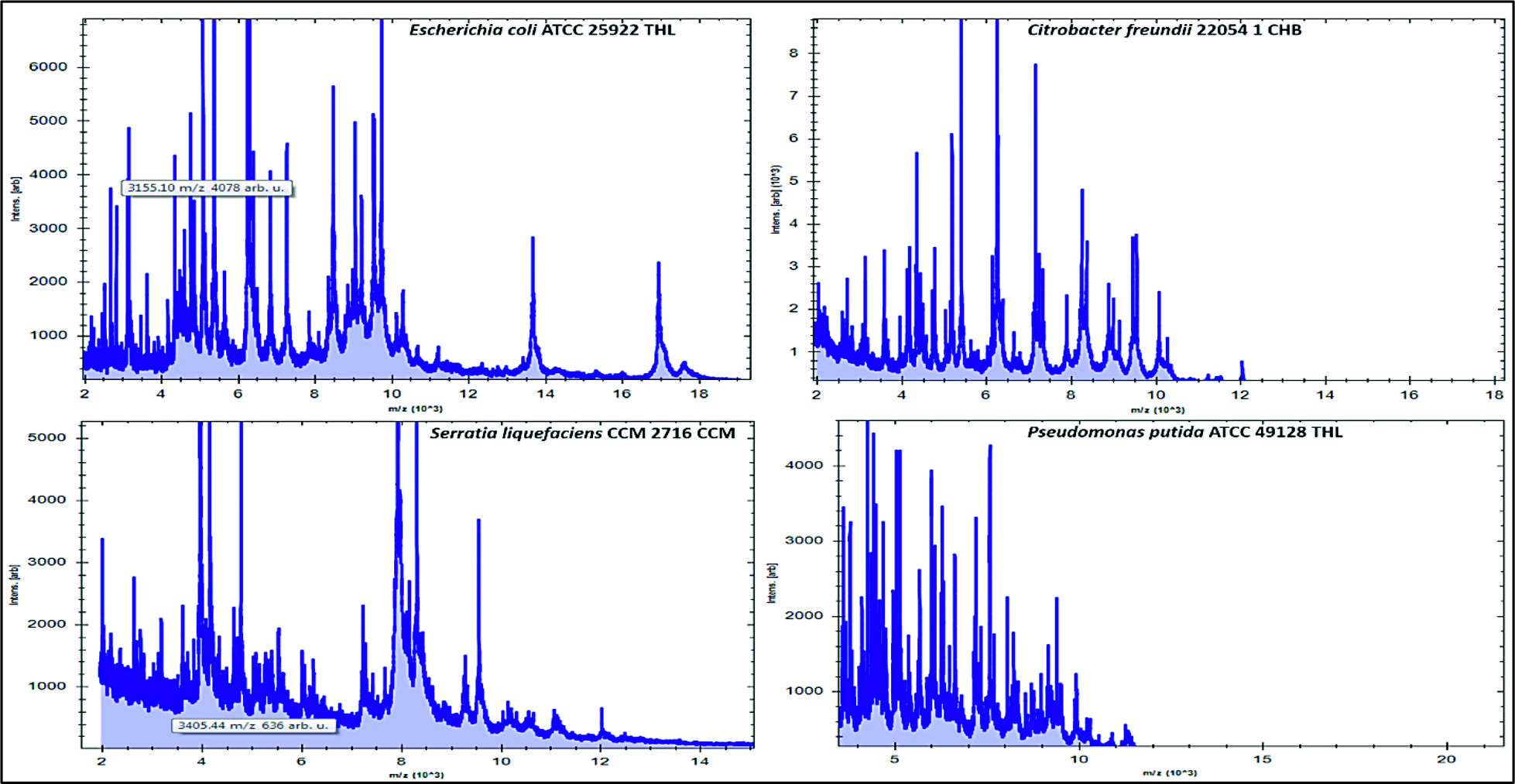
Fig. 2. Numbers and percentages of various airborne pathogens isolated from three different areas in Al-Qassim regionAs can be seen in Fig. 3, the data analysis of MBT illustrated that numerous spectra for the most commonly isolated gram positive bacteria as Bacillus subtilis, Enterococcus faecalis and Staphylococcus aureus were scattered within the range from 2,000 to 15,000 m/z, and from 2,000 to 16,000 m/z for Acinetobacter baumanni with higher peak intensities were determined between 4,000 and 8,000 m/z for all identified gram positive bacteria. Likewise, for the commonly isolated gram negative bacteria as Citrobacter freundii, Srratia liquefaciens and Pseudomonas putida, several spectra were scattered within the range from 2,000 to 10,000 m/z and from 2,000 to 17,000 m/z for Escherichia coli with higher peak intensities were detected between 4,000 to 8,000 for all identified gram-negative bacteria (Fig. 4).
As community awareness of possible hazards arising from bioaerosols increases, the request for advanced and predominantly appropriate techniques for the recognition of bacterial portions in such releases is increasing congruently32,33,34. In the present investigation, it was aimed to establish MBT as an effective mass spectrometry technology for identification of various airborne bacteria with slight workload.
In the current study, a huge number of isolates recovered from air samples examined by MBT Compass software, and it was stated that MBT is considered as a suitable technique for the precise recognition of airborne pathogens at the genus and species levels. Throughout Al-Qassim region, samples of air were collected from different localities and showed a huge contamination by numerous environmental non-pathogenic and pathogenic bacterial species. One major restriction of the current research was that the samples were only examined for airborne bacteria, whereas, other types of microorganisms and their toxins were excluded from the study. Furthermore, the isolation was depended mainly on the culture technique, a restriction that perhaps reduced variety of the strains. Lastly, the amount of strains initially observed in the agar media didn’t calculated to define which location was the greatest tainted with microorganisms.
MBT is used routinely in our lab. to recognize various types of bacteria at species and subspecies levels15,35. In our microbiology laboratory, we identify annually about 9,500 isolates by MBT. Since we examined the isolates recovered from air samples, the MBT database didn’t comprise their spectra, and only 119 (92.25%) of the isolates were properly recognized with score values ³2.000. Due to the existence of a large stored spectra in the Bruker Daltonics databank of certain types permitted the MBT to recognize a large number of bacteria within the species levels. Similarly, in a former study conducted Seng et al.36 on samples isolated from patients, who found that MBT was able to identify approximately 95% of the bacterial isolates.
Proteomic identification of airborne bacteria by MBT confirmed by RT-PCR provided us with data regarding the bacterial pathogenicity in different localities at Al-Qassim region. Throughout our study, the identified bacteria seemed to be similar to the former studies reported by Papadopoulou et al.37 and Martin et al.38, who found that the most commonly pathogens were belonged to Staphylococcus, Microccocus, Kocuria, Arthrobacter and Bacillus genera. Furthermore, Hernבndez et al.39 identified 19 bacterial genera recovered from Biosafety level 2 laboratory by MBT and they found that the most predominant bacteria were Staphylococcus aureus and Micrococcus luteus. Nevertheless, in the present study, the predominant genera recognized from air samples of three different localities in Al-Qassim region were Staphylococcus aureus, Escherichia coli, Staphylococcus epidermidis, Enterococcus faecalis and Acinetobacter baumanni.
In our study, among the three localities Al-Qassim Hospitals were the most commonly contaminated site with a huge number of bacteria (41.10%) and the genus Staphylococcus was the predominant isolated bacteria. Similar results were obtained by Kausar et al.40 who indicated that a large number of Staphylococcus sp. were isolated from the air of Hail hospitals, Saudi Arabia. Our results, propose that hospitals have more microbial contamination than other sites, this is due to the presence of several kinds of patients. The presence of Staphylococcus, may has a possible hazard to the health of patients of these rooms40. This type of bacteria may effect on humans by different ways and are able to cause various illnesses such as shocking infections in these high-risk patients41. Moreover, surface contamination with settled microbes could also present a source of potential health risk. In the current study, we found a high incidence of airborne bacteria in Al-Qassim Hospitals, Qassim University and poultry slaughter house, Al-Qassim region, Saudi Arabia. As well, the RT PCR proven here was successfully applied to settle the recognition of nuc, gelE, bsub, recA and iss genes specific for Staphylococcus aureus, Enterococcus faecalis, Bacillus subtilis, Acinetobacter baumanni and Escherichia coli, respectively. A respectable link between MBT and PCR investigation was found.
The current study demonstrates that MBT is a powerful and rapid alternative technique to the conservative methods for airborne bacterial identification. Accurate and fast identification of bacterial contamination in bioaerosols is compulsory to avoid potential eruptions. Various types of bacteria (pathogenic and nonpathogenic) were identified in the indoor and outdoor environment of Qassim University, hospitals and poultry slaughterhouse, Al-Qassim region. The most commonly isolated bacteria were Staphylococcus aureus, Enterococcus faecalis, Bacillus subtilis, Acinetobacter baumanni. Therefore, students, employees, staff members, patients and doctors are potentially exposed to diverse airborne microorganisms, which may perhaps result in diversity of illnesses comprising respiratory distresses. Governmental organizations must increase the research activities to collect information concerning health influences that effect on both human and animal populations. We suggest that increasing the awareness regarding the possible health threats of indoor and outdoor bio-contaminants are considered as the first step in handling and eventually decreasing the diseases they persuade.
Acknowledgments
We would like to thank the Deanship of Scientific Research, Qassim University, Buraidah,
Saudi Arabia for the financial support of this research.
Conflict Of Interest
The authors declare that there is no conflict of interest.
Authors’ Contributions
AE, MA, EM, FM, MR, FA and AA designed the experiments. AE, MA, EM and MR performed the experiments. AE, FM, FA and AA analyzed the data. AE, MA and EM wrote the manuscript. AE, MA, EM, FM, MR, FA and AA reviewed the manuscript.
Funding
This work was supported by the Deanship of Scientific Research, Qassim University, Buraidah, Saudi Arabia (Grant number bhsc-2016-1-12-I-1813).
Data Availability
All datasets generated or analyzed during this study are included in the manuscript.
Ethics Statement
This article does not contain any studies with human participants or animals performed by any of the authors.
- Gorny R., Reponen T., Willeke K., Schmechel D., Robine E., Boissier M., Grinshpun S.A. Fungal fragments as indoor air biocontaminants. Appl. Environ. Microbiol., 2002; 68: 3522-3531.
- Fracchia L., Pietronave S., Rinaldi M., Martinotti M.G. The assessment of airborne bacterial contamination in three composting plants revealed site related biological hazard and seasonal variations. J. Appl. Microbiol., 2006; 100: 973-984.
- Yassin M.F., Almouqatea S. Assessment of airborne bacteria and fungi in an indoor and outdoor environment. Int. J. Environ Sci. Tech., 2010; 7: 535-544.
- Bomala K., Saramanda G., Reddy B., Kaparapu J. Microbiological indoor and outdoor air quality of selected places in Visakhapatnam city, India. International Journal of Current Research, 2016; 8(4): 29059-29062.
- Jones A.M., Harrison R.M. The effects of meteorological factors on atmospheric bio-aerosol concentrations–a review. Science of the total environment, 2003; 326 (1-3): 151-180.
- Douwes J., Thorne P., Pearce N., Heederik D. Bioaerosol health effects and exposure assessment: progress and prospects. Ann. Occup. Hyg., 2003; 47: 187-200.
- Wojtatowicz M., Stempniewicz R., Zarowska B., Rymowicz W., Robak M. Mikrobiologia ogolna. Wydawnictwo Uniwersytetu Przyrodniczego we Wroclawiu, Wroclaw 2008.
- Valen A. Characterization of Airborne Micro-organisms at Nationaltheatret. Subway Station. Master thesis, Norwegian University of Science and Technology, Faculty of Natural Sciences and Technology, Department of Biotechnology 2011.
- Gilbert Y., Duchaine C. Bioaerosols in industrial environments: A review. Canadian Journal of Civil Engineering, 2009; 36: 1873–1886.
- Mandal J., Brandl H. Bioaerosols in indoor environment—A review with special reference to residential and occupational locations. Open Environmental and Biological Monitoring Journal., 2011; 4: 83–96.
- Despres V.R., Huffman J.A., Burrows S.M., Hoose C., Safatov A.S., Buryak G., Frohlich-Nowoisky J., Elbert W., Andreae M., Poschl U., Jaenicke R. Primary biological aerosol particles in the atmosphere: A review. Tellus. B., 2012; 64: 1-58.
- Fykse E.M., Tjaernhage T., Humppi T., Eggen V.S., Ingebretsen A., Skogan G., Olofsson G., Waesterby P., Gradmark P., Larsson A., Dybwad M., Blatny J.M. Identification of airborne bacteria by 16S rDNA sequencing, MALDI-TOF MS and the MIDI microbial identification system. Aerobiologia, 2015; 31:271-281.
- Killingley B., Greatorex J., Cauchemez S., Enstone J.E., Curran M., Read R.C., Lim W.S., Hayward A., Nicholson K.G., Nguyen-Van-Tam J.S. Virus shedding and environmental deposition of novel A (H1N1) pandemic influenza virus: interim findings. Health Technol. Assess., 2010; 14: 237-354.
- National Research Council. Sensor Systems for Biological Agent Attacks: Protecting Buildings and Military Bases. Washington DC, The National Academies Press, 2005.
- Angelakis E., Yasir M., Azhar E.I., Papadioti A., Bibi F., Aburizaiza A.S., Metidji S., Memish Z.A., Ashshi A.M., Hassan A.M., Harakeh S., Gautret P., Raoult D. MALDI-TOF mass spectrometry and identification of new bacteria species in air samples from Makkah, Saudi Arabia. BMC Res. Notes, 2014; 7: 892.
- Hope A.P., Simon R.A. Excess dampness and mold growth in homes: an evidence-based review of the aeroirritant effect and its potential causes. Allergy Asthma Proc., 2007; 28: 262-270.
- Rotz L.D., Khan A.S., Lillibridge S.R., Ostroff S.M., Hughes J.M. Public health assessment of potential biological terrorism agents. Emerg. Infect. Dis., 2002; 8: 225-230.
- Mellmann A., Bimet F., Bizet C., Borovskaya A.D., Drake R.R., Eigner U., Fahr A.M., He Y., Ilina E.N., Kostrzewa M., Maier T., Mancinelli L., Moussaoui W, Prיvost G., Putignani L., Seachord C.L., Tang Y.W., Harmsen D. High inter-laboratory reproducibility of matrix-assisted laser desorption ionization-time of flight mass spectrometry-based species identification of non-fermenting bacteria. J. Clin. Microbiol., 2009; 47: 3732-3734.
- Cherkaoui A., Hibbs J., Emonet S., Tangomo M., Girard M., Francois P., Schrenzel J. Comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J. Clin. Microbiol., 2010; 48: 1169-1175.
- Elbehiry A, Marzouk E, Abdeen E, Al-Dubaib M, Alsayeqh A., Ibrahem M., Hamada M., Alenzi A., Moussa I., Hemeg H.A. Proteomic characterization and discrimination of Aeromonas species recovered from meat and water samples with a spotlight on the anti-microbial resistance of Aeromonas hydrophila. Microbiologyopen, 2019; e782.
- Chalupova J., Raus M., Sedlářová M., Sebela M. Identification of fungal microorganisms by MALDI-TOF mass spectrometry. Biotechnol. Adv., 2014; 32: 230-41.
- Tomazia T., Gonחalvesa J.L., Barreiroa J.R., de Campos Bragab P.A., Prada e Silva L.F., Eberlin M.N., dos Santos M.V. Identification of coagulase-negative staphylococci from bovine intramammary infection by matrix assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol., 2014; 52: 1658-1663.
- Elbehiry A., Marzouk E., Hamada M., Al-Dubaib M., Alyamani E., Moussa I.M., Al-Rowaidhan A., Hemeg H.A. Application of MALDI-TOF MS fingerprinting as a quick tool for identification and clustering of foodborne pathogens isolated from food products. New Microbiol., 2017; 40(4): 269-278.
- Dybwad M., Skogan G., Blatny J.M. Temporal variability of the bioaerosol background at a subway station: Concentration level, size distribution and diversity of airborne bacteria. Applied and Environmental Microbiology, 2014; 80: 257–270.
- Chen Y.S., Liu Y.H., Teng S.H., Liao C.H., Hung C.C., Sheng W.H., Teng L.J., Hsueh P.R. Evaluation of the matrix-assisted laser desorption/ionization time-of-flight mass spectrometry Bruker Biotyper for identification of Penicillium marneffei, Paecilomyces species, Fusarium solani, Rhizopus species, and Pseudallescheria boydii. Front Microbiol., 2015; 6: 679.
- Deng J., Fu L., Wang R., Yu N., Ding X., Jiang L., Fang Y., Jiang C., Lin L., Wang Y., Che X. Comparison of MALDI-TOF MS, gene sequencing and the Vitek 2 for identification of seventy-three clinical isolates of enteropathogens. J. Thorac. Dis., 2014; 6: 539-544.
- Zhang K., Sparling J., Chow B.L., Elsayed S., Hussain Z., Church D.L., Gregson D.B., Louie T., Conly J.M. New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J. Clin. Microb., 2014; 42(11): 4947-4955.
- Asmat A., Dada A.C., Gires U. Biofilm Formation, gel and esp Gene Carriage among Recreational Beach Enterococci. Global Journal of Health Science, 2014; 6(5): 241-253.
- Wattiau P., Renard M.E., Ledent P., Debois V., Blackman G., Agathos S.N. A PCR test to identify Bacillus subtilis and closely related species and its application to the monitoring of wastewater biotreatment. Applied Microbiology and Bio-technology, 2001; 56: 816-819.
- AL-Jubori S.S., Al-kadmy I.M.S., Al-Ani Z.J. Emergence of multidrug resistance (MDR) Acinetobacter baumannii isolated from Iraqi hospitals, AENSI, 2016; 10(5): 265e275.
- Yaguchi K., Ogitani T., Osawa R., Kawano M., Kokumai N., Kaneshige T., Noro T., Masubuchi K., Shimizu Y. Virulence Factors of Avian Pathogenic Escherichia coli Strains Isolated from Chickens with Colisepticemia in Japan. Avian. Dis., 2007; 51: 656-662.
- Jahne M.A., Rogers S.W., Holsen T.M., Grimberg S.J., Ramler I.P. Emission and dispersion of bioaerosols from dairy manure application sites: human health risk assessment. Environ. Sci. Technol., 2015; 49: 9842–9849.
- Jahne M.A., Rogers S.W., Holsen T.M., Grimberg S.J., Ramler I.P., Kim S. Bioaerosol deposition to food crops near manure application: quantitative microbial risk assessment. J. Environ. Qual. 2016; 45: 666–674.
- Druckenm ller K., Gartner A., Jeckel U., Klug K., Schiffels J., G nther K., Elbers G. Development of a methodological approach for the characterization of bioaerosols in exhaust air from pig fattening farms with MALDI-TOF mass spectrometry. Int. J. Hyg. Environ. Health 2017; 220(6): 974-983.
- Seng P., Abat C., Rolain J.M., Colson P., Lagier J.C., Gouriet F., Fournier P.E., Drancourt M., La S.B., Raoult D. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol., 51(7): 2182-2194.
- Seng P., Drancourt M., Gouriet F., La S.B., Fournier P.E., Rolain J.M., Raoult D. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis., 2009; 49(4): 543-551.
- Papadopoulou C., Economou V., Sakkas H., Gousia P., Giannakopoulos X., Dontorou C., Filioussis G., Gessouli H., Karanis P., Leveidiotou S. Micro-biological quality of indoor and outdoor swimming pools in Greece: Investigation of the antibiotic resistance of the bacterial isolates. Int. J. Hyg. Environ. Health, 2008; 211: 385-397.
- Martin P.G., Gonzalez M.B., Martinez A.R., Lara V.G., Naveros B.C. Isolation and characterization of the environmental bacterial and fungi contamination in a pharmaceutical unit of mesenchymal stem cell for clinical use. Biologicals, 2012; 40: 330-337.
- Hernandez I.A., Flores J.L.H., Gomez S.R., Hernandez A.C., Gutierrez C.S., Xechit I., Martinez P., Jone G.H., Campos-Guillen J. Identification by MALDI-TOF Mass Spectrometry of Bacteria in Air Samples in a Biosafety Level 2 Laboratory. Modern Environmental Science and Engineering, 2016; 2(4): 238-245.
- Kausar M.A., Arif J.M., Alanazi S.M.M., Alshmmry A.M.A., Alzapni Y.A.A., Alanazy F.K.B., Shahid S.M.A., Hossain A. Assessment of microbial load in indoor environment of university and hospitals of hail, ksas. Biochem. Cell Arch., 2016; 16(1): 177-183
- Alhambra A., Catalin M., Moragues M.D., Brena S., Ponton J., Montejo J.C., del Palacio A. Isolation of Aspergillus lentulusin Spain from a critically ill patient with chronic obstructive pulmonary disease. Rev. Iberoam. Micol., 2008; 25(4): 246-249.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.