The cellulose is enormous source of organic carbon on the earth. It has varied industrial applications; the most important of these in 21st century is bio-ethanol production. The cellulose degradation requires extremes of pH and temperature, and thereby it is expensive and hazardous to the environment. This signifies usage of enzymes for cellulose hydrolysis for its conversion to ethanol. A large number of cellulases have been identified from bacteria and fungi, but there is need of more efficient cellulases. It is observed that majority of microbes defy cultivation under laboratory conditions so the metagenomics offer new avenues in search of novel cellulases, capable of efficient bioconversion of cellulosic materials. Metagenomics is complementary method to traditional culture-based method, as it allows exhaustive mining of microbial genomes from their habitats. This review covers the current status of cellulase gene retrieved from metagenomes of various environments.
Cellulase, Biofuel, Metagenomic library, Function-based screening, Bio-ethanol.
The combustion of petroleum-based fossil fuels has become a concern due to accelerated carbon emissions, unstable and uncertain petroleum sources, as well as, the fluctuations in cost of fuels. These concerns led the global efforts towards utilization of renewable resources and incessant production of green fuels. Plant biomass is the most viable renewable resource for production of biofuel because it is largely abundant and inexpensive resources. The primary obstacle impeding the commercial production of energy from cellulose rich biomass is the absence of low-cost technology for conversion of cellulose to ethanol. Cellulose is mainly crystalline, but varies with plant species, having tightly packed bundles of microfibrils, preventing penetration of hydrolytic enzymes and in turn its conversion to glucose. Therefore conversion of cellulose into glucose is slow and economically unviable. Huge amounts of agricultural, industrial and municipal cellulosic waste is accumulating or used inefficiently due to the high cost of their management (Edwards et al. 2006). Therefore, it has become an environmental and economic interest to develop processes for the effective treatment and utilization of cellulosic wastes as inexpensive carbon sources. In this scenario, cellulases are the enzymes, providing an opportunity to achieve advantage of biomass utilization (Wen, Liao, and Chen 2005). Cellulases are inducible enzymes which are synthesized and produced by microorganisms during their growth on cellulosic substrates (Kuhad, Gupta, and Singh 2011). The complete enzymatic hydrolysis of cellulosic materials needs different types of cellulase; endoglucanase (1,4-b-D-glucan-4-glucanohydrolase; EC 3.2.1.4), exocellobiohydrolase (1,4-b-D-glucan glucohydrolase; EC 3.2.1.74) and b-glucosidase (b-D-glucoside glucohydrolase; EC 3.2.1.21) (Pandey et al. 2013). The endoglucanases randomly hydrolyzes the b-1, 4 bonds in the cellulose molecule, and the exocellobiohydrolases in most cases release a cellobiose unit showing a recurrent reaction from chain extremity. Lastly, the cellobiose is converted to glucose by b-glucosidase (Pandey et al. 2013). Many organisms, including animals, plants, and microorganisms, produce cellulases; however, the majority of cellulases are of microbial origin. Aerobic microorganisms usually use the free cellulase mechanism (non-complexed cellulase systems) to digest cellulose; for example the fungi, Trichoderma reesei, Humicola insolens and Phanerochaete chrysosporium, and bacteria belonging to the genera Cellulomonas, Thermobifida, and Bacillus, always produce individual cellulases, including endoglucanase, exoglucanase, and b-glucosidase (Lynd et al. 2002). Most anaerobic microorganisms digest cellulose using their cellulosome, which is a large cellulase complex present on the outer surface of the host cell. Cellulosomes from different clostridia (Clostridium thermocellum, Clost. cellulolyticum, Clost. cellulovorans, and Clost. josui) and Ruminococcus species in the rumen have been studied in detail (Bayer et al. 2008).
Although a large number of cellulases are isolated from different hosts, but still there is demand for new cellulases having better properties for e.g higher catalytic activity on insoluble substrates, increased stability at higher temperature and increased resistance to end product inhibition. The two common strategies to get ideal cellulases are; (1) molecular evolution of cellulase through DNA shuffling. (2) Cloning and identification of novel cellulase from cultured microorganisms (Rappé and Giovannoni 2003). Apart from this, there is huge amount of genetic resources locked in the uncultured microbes. Metagenomics is one of the key technology can be used to access and investigate this potential. This review describes the mining of novel cellulase genes from various environments using metagenomics and assessment of their applications in production of biofuel.
Strategies for prospecting cellulases from metagenomes
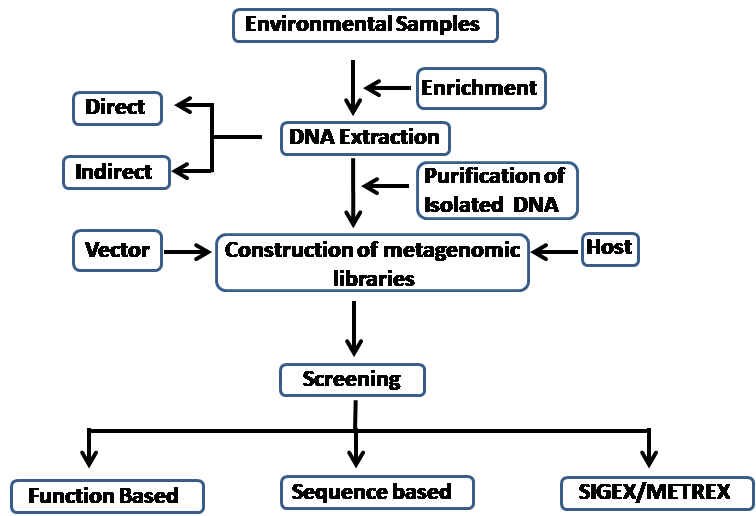
The function-based screening of metagenomic libraries and sequence-based search of novel genes are the two common approach, used for prospecting of novel enzymes or genes (Fig.1). In the function-based screening, metagenomic expression libraries are constructed and screened for the enzyme of interest. Whereas, in the case of sequence-based search the gene of interest is amplified by polymerase chain reaction from metagenomic DNA and cloned. Another alternate method is that gene can be discovered from metegenome sequence database and then can be amplified and cloned in the suitable expression vector.
The metagenomic expression libraries are created by insertion of fragmented metagenomic DNA into expression vectors based on plasmids, cosmids, fosmids, or phages. The expression of gene is then checked in suitable host. This method of direct screening facilitate discovery of completely unknown genes and their enzymes. After discovery of unknown gene or enzymes, it can be functionally characterized. However, for expression of an active enzyme, the clone must contain the complete gene sequence. The selection of suitable vector for e.g Libraries containing 2-10 Kilobase (Kb) can be constructed in plasmids or lambda expression vectors. Larger gene fragments having 20-40 Kb size can be cloned in cosmids or fosmids. The 100 -200 Kb fragment are suitable in bacterial artificial chromosome (BAC) vectors. Another problem faced during expression in heterologous host is difference in use of codon, transcription/translation initiation signals, protein folding or certain post-translational modifications. The way of sorting out these problems are selection of vector having appropriate transcription and translation – initiation sequences and use of appropriate hosts for e.g E.coli Rosetta strains ( Novagen, Madison, Wisconsin, USA), containing the tRNA genes for rare amino acid codons (Duan et al. 2009), or simultaneous expression of chaperone proteins such as GroES, GroEL and heat shock proteins.(Nishihara et al. 1998). Several efforts were made to improve the host such as Pichia pastoris, Pseudomonas putida, Streptomyces lividans and Bacillus subtilis for improved heterologous expression (Duan et al. 2009). Apart from these examples several modified function-based methods are designed specifically for exploration of metagenomic libraries. A substrate induced gene-expression system was developed by Uchiyama and colleagues (Uchiyama, Miyazaki, and Yaoi 2013) to identify the clones rapidly that can be induced by a target substrate and exhibit catabolic gene expression, while clones generating quorum sensing gene inducing compounds can be regulated by metabolite (Williamson et al. 2005). Function-based metagenome library screening has revealed a wide range of biocatalysts. In this manuscript, we report several published results that screened for cellulase enzyme involved in degradation of lignocellulosic biomass. The known conserved sequences are searched for in sequence-based approach so it cannot unearth the non-homologous enzymes. Therefore, this method is not suitable for detection of novel genes. However, this is better than function-based approach because it can uncover the gene of interest, regardless of gene expression, protein folding and completeness of gene of interest. However, success rate of this method depends on various factors (1) Sequencing at larger scale for very complex communities. However, the development of new sequencing technology like next-generation 454-pyrosequencing has made the process very easy. There are so many examples, where it is exploited. For example explorations of microbial communities in the drainage of acid mines.(Tyson et al. 2004). The major advantage of metagenomic projects using new sequencing technologies generate huge base pair reads and cover species evenly within the community (Dalevi et al. 2008). (2) Although metagenomic DNA represents DNA samples from diverse organisms, but sizes of environmental genomes and their presence are not uniform so many sequence reads remain unassembled. Therefore, bulk sequencing of maximum possible genes took over the complete metagenomic sequencing. In case of bulk sequencing, assembling of sequences into contigs is required so length of the fragments obtained for high throughput screening and cloning becomes a limiting factor. The gene fragment should be long enough to contain complete open reading frame for the functions of interest. Hence, optimized 454 sequencing ( about 450 nucleotide) seems to be more favorable than extremely high-volume short-run (Edwards et al. 2006; Dalevi et al. 2008), but downstream cloning and expression of genes like GHase, varying in length from less than 1 Kb to more than 20 Kb becomes major limitation. It has been reported that Meta Gene, one of the Gene-finding tools can predict 90% of shortgun sequences (Noguchi, Park, and Takagi 2006). (3) There is a need for more data mining tools, that can predict protein structures, putative catalytic sites and functions in addition to prediction based on primary sequence homology. With the advancement of protein classification tools, models can be designed to correlate protein folding and mechanism of enzyme function (Claudel-Renard et al. 2003; Selengut et al. 2007). We anticipate that in future, sequence based metagenome database search with bioinformatic tools will have a greater influence on mining of novel biocatalytic genes than function-based methods. There are many reports in literature, describing prospect of genes and enzymes involved in biofuel production in metagenome sequence databases. For example, metagenomic library of hindgut microbiota of wood-feeding termites was sequenced, From this, Warnecke and colleagues (Warnecke et al. 2007) generated 71 million base pairs of sequence data. Using global al;ignment, they identified more than 700 domains homologous to glycoside-hydrolase catalytic to 45 different homologous carbohydrate active enzymes (CAZy) families (Henrissat 1991), containing diverse range of putative cellulases and hemicellulsaes. A metagenomic library of the microbial community from the biogas fermentor was sequenced by Schlüter and colleagues (Schluter et al. 2008; Krause et al. 2008). Bacteria playing dominant roles in methanogenesis and genes coding for cellulolytic activity were identified from the Clostridia spp. out of 141 million base pair sequences generated (Schluter et al. 2008; Krause et al. 2008).
Cellulases from metagenome expression libraries (function-based screening)
The first report of isolation of a cellulase gene from metagenomic library was from microbial consortia in a thermophillic, anaerobic digestor maintained on lignocellulose (Healy et al. 1995). In that report, 12 clones exhibiting CMCase activity and 11 clones revealing 4-methylumbelliferyl-â-D-cellobioside (MUC) hydrolase activity were detected. Out of these clones, four were further characterized and they revealed optimum temperature at 60-65ºC and optimum pH at 6-7. One clone SE1402 (pFGH1) showing CMCase activity was sequenced, which exhibited less than 50% similarity with known cellulases. Afterwards, metagenomic approaches have been applied extensively in various environments where plant materials are decomposed intensively, including soil (Jiang et al. 2011; Kim et al. 2008), hindgut of termites (Warnecke et al. 2007)23, compost (Pang et al. 2009), cecum of rabbit (Feng et al. 2007), sludge of biogas reactor (Jiang et al. 2010) and enrichment cultures (Grant et al. 2004; Rees et al. 2003; Voget, Steele, and Streit 2006) to isolate cellulases. Rumen is one of the important fiber degradation system. Microbes of rumen play an essential role in degradation of cellulose of plants, which could be a very good source of cellulases so metagenomic studies are focussed on this environment (Duan et al. 2009; Ferrer, Golyshina, Chernikova, Khachane, Martins dos Santos, et al. 2005; Wang et al. 2009). Ferrer et al (2005) constucted a metagenomic expression library from rumen of cow and screened for cellulase positive clone, 7 clones showing b-1,4-endoglucanase activity was found. Sequence analysis of the retrieved cellulases were completely new and distantly related to other reported cellulases. Fang et al (2009) reported six positive clones showing b-glucosidase activity, through functional screening of a metagenomic library of the microbes from the surface water of South China Sea. The sequence analysis of one of these clones, pSB47B2 was done and it was found that it contain an open reading frame for a novel b-glucosidase (bgl1B). Bgl1B was overexpressed with high yield and considerable enzymatic activity using pET22b (+) as vector and Escherichia coli BL21(DE3) as host. The biochemical characterization of purified recombinant protein (rBgl1B) indicated that with pNPG as substrate the hydrolytic activity of rBgl1B was optimum at pH and temperature 6.5 and 40ºC respectively. The Km and Vmax of rBgl1B was 0.288×10-3 mol/L and 36.9×10-6 mol/L respectively. It hydrolyzed the pNPG with an activity upto 39.7U/mg. It was also observed that rBgL1B could hydrolyze cellobiose with a Km of 0.173×10-3mol/L and Vmax of 35×10-6 mol/L. There was no significant activity of rBgl1B was observed against lactose, maltose, sucrose and CMC. A small concentration of Ca2+ or Mn 2+ stimulated the enzymatic activity of rBgl1B to pNPG. A novel b-glucosidase gene (bgl1A) encoding a 442-amino-acid protein was isolated from a marine microbial metagenomic library through functional screening by Fang and colleagues (Fang et al. 2010) reported a new gene, RuCelA, coding for a bifunctional xylanase / endoglucanase from a metagenomic library of yak rumen microorganisms. It had both activity against xylan and carboxymethylcellulose (CMC), indicating the bifunctional xylanase/endoglucanase activity. The optimum conditions for xylanase and endoglucanase activities were 65ºC, pH 7.0 and 50ºC, pH 5.0, respectively. Above all this, presence of Co (+) and Co (2+) significantly improved the endoglucanase activity, while it inhibited the xylanase activity. The substrate preference was tested and higher activity against barley glucan and lichenin was observed than against xylan and CMC. The various identified cellulase genes from different environments displayed certain features as listed in Table 1. The first common observed feature, based on module analysis is that most of the encoded products of cloned cellulase genes belonged to GH5 family, followed by GH9. The reason of abundance of GH5 and GH9 cellulases is due to their expression characteristics. The GH5 and GH9 cellulase genes might be easily expressed in E. coli. Another potential reason could be that in bacterial genomes, there are many genes for GH5 and GH9 cellulases. This hypothesis is supported by Genome sequencing of cellulase producing bacteria such as F. succinogenes, Saccharophagus degradans, and C. hutchinsonii, revealing that there are more cellulases belonging to family GH5 and GH7 than other families (Duan and Feng 2010; Xie et al. 2007). The second reason for this could be that, genes encoding exoglucanases (cellobiohydrolases), belonging to family GH6, GH7 and GH48 could not be isolated from any metagenomic library, even if MUC was used as substrate to screen clones expressing cellobiohydrolase activities. The cellulase genes identified by sequencing of MUC positive clones were either endoglucanase (Healy et al. 1995) or cellodextrinases (Duan et al. 2009). Among cellobiohydrolases, enzymes of family GH7 are only found in fungi, GH6 enzymes are found both in fungi and bacteria (Edwards, Upchurch, and Zak 2008) and cellulases of family GH48 are common in cellulase producing bacteria (Berger et al. 2007). As most of the expression vectors are based on E. coli hosts so fungal genes are not expressed because promoter and intron sequences of gene of interest are not recognized. Apart from this, GH48 cellobiohydrolases are very large proteins so genes may be not expressed properly in E. coli system. The third reason for abundance of GH5 and GH7 may be due to the fact that cloned cellulase genes shared less than 70% similarity with already reported cellulases (Duan and Feng 2010).
Sequence based approach
The sequence-based approach was also used to mine cellulase genes from uncultured microbes. Ohotoko et al (2000) reported GH45 cellulase homologs from the symbiotic protists in the hindgut of termite Reticulitermes speratus using consensus PCR and cDNA library screening. Edwards and others (2008) developed new oligonucleotide primers for fungal cellobiohydrolase I (CBHI) genes and used this to isolate and clone CBHI homologs from forest soil by PCR. The diversity of GH48 cellulases in cellulolytic consortia enriched with thermophillic compost was analyzed by Izquierdo and colleagues (2010). The major problem with sequence-based method is that identified cellulases exhibit high percentage of similarity with reported genes and with each other. The sequence based approach should be used along with function-based method to overcome the problem of biased and insufficient expression of the target gene in E. coli. The several important ecosystems, where cellulose is degrading including rumen (Brulc et al. 2009) and hindgut of higher termites (Warnecke et al. 2007) metagenomic sequencing projects are carried out. The analysis of these metagenomic sequence data of those environments revealed that there is abundance of glycoside hydrolases involved in degradation of cellulose and xylan. A metagenomic fosmid library was created from contents of biogas digestor and after screening 341, 246 and 386 positive clones with b-1, 4-endoglucanase, b- glucosidase and b-1,4-xylanase activities respectively was observed (Yan et al. 2013). After that 4, 10 and 16 positive clones were pooled together and subjected to 454 pyrosequencing. From this, 21 unique glycosyl hydrolase (GH) genes were anticipated by bioinformatics analysis, indicating similarities to their nearest neighbors from 39% to 72%. Nine GH genes were expressed and purified to find their activity on four kinds of substrates besides bioinformatics analysis. The activities of the most expressed proteins were in agreement with their annotation based on their bioinformatics analysis, however only three genes of family GH5 revealed different activities from their annotation. A new method called metagenomic gene specific multi-primer PCR (MGSM-PCR) was introduced that uses multiple gene-specific primers based on isolated gene from metagenomic library rather than degenerate primers (Xiong et al. 2012). The major application of MGSM-PCR was displayed by applying it to search for homologues of cellulase belonging to GH9 family in metagenomic DNA. In metagenomic data of the contents termite hindgut, more than 100 gene modules involved in cellulose hydrolysis were identified, corresponding to catalytic domains of GH5, GH94 and GH51. The rate of finding cellulase gene was 1/0.4Mb metagenomic DNA. Whereas, gene sequences coding for the catalytic domains of endoglucanases and cellobiohydrolases of family GH6, GH7, GH48 and cellulase systems of well described fungi Trichoderma reesei and bacterial genera Cellulomonas were absent (Warnecke et al. 2007). The characterization of the microbial community by metagenomics, helped in discovering carbohydrate-active genes of an enriched thermophillic cellulose-degrading sludge. It was found by 16S analysis that sludge microbiome was dominated by cellulolytic Clostridium and methanogen Methanothermobacter. The de novo assembly of the 11,930,760 Illumina 100 base paired-ends was performed to retrieve gene of interest from metagenome. Out of this 75% of all reads was utilized in the denovo assembly, and 64% of these open reading frame having average length of 852 bp were projected from the assembly, and 64% of these open reading frames were told to contain full length genes. The Hidden Markol Model 253 was used to predict number of genes, which indicated 253 genes were thermostable and putatively carbohydrate-active. The GH9 and corresponding CBM3 was dominant and revealed a cellulosome-based attached metabolism of polysaccharide in the thermophillic sludge. The putative carbohydrate acting genes exhibited sequence similarity ranging from 20-100% amino acid sequence in proteins in NCBI database (Xia et al. 2013).
Potential applications of metagenomic cellulases
Although a number of cellulase genes and enzymes have been obtained from metagenomic library, but only few of them could fulfill the bioprocessing conditions prevalent during bioethanol production. An endoglucanase Cel5A, obtained from soil was found very suitable for industrial applications (Voget, Steele, and Streit 2006). It was stable over a wide range of pH and temperature, presence of high concentration of salt, presence of divalent cations, detergents and chelating agents, common in detergents (Voget, Steele, and Streit 2006). A hybrid glycosyl hydrolase, GH6248, having two independent catalytic modules of GH5 and GH26, showing glucanase and mannase activity respectively was reported. The cellulases obtained from rumen by metagenomic approach were mostly acidic and mesophillic, which is similar to the fermentation condition of ethanol by yeast. Acidic and mesophillic enzymes are very useful as it helps in simultaneous sacchrification and fermentation of lignocellulose (Duan et al. 2009; Liu et al. 2009). Pottka ¨mper et al. (2009) reported three novel cellulases from metagenome of soil, suitable in degradation of cellulose under high concentration of various ionic liquids. BglA, derived from soil has ability to convert the major ginsenoside Rb1 into pharmaceutically active minor ginsenoside Rd (Kim et al. 2011). Two promising alkaline b-glucosidase was reported by Biver and colleagues (2014) derived from metagenome of agricultural soil, including one AS-Esc10 showing high tolerance towards harsh detergents, oxidants and glucose. Another b-glucosidase unbg1A, tolerant to glucose concentration as high as 2M with Ki value 1.5M and NaCl concentration 0.6M. Transglucosylation activities was also observed in this enzyme, leading to formation of cellotriose from cellobiose (Lu et al. 2013).
One more b-glucosidase coded by the gene td2f2, obtained from metagenome of compost, the hydrolysis activity of p-nitrophenyl-b-D-glucopyranoside was stimulated by various monosacchrides and sugar alcohols demonstrating its transglucosylation activity. A novel b-glucosidase encoding gene Bgl-gsl, derived from a metagenomic library of contents of the gut of Globitermes brachycerastes was reported by Wang and colleagues (Wang et al. 2012). It was observed that the residual activity of Bgl-gsl was retained above 70% after the recombinant enzyme was incubated at 75ºC and at pH 6.0 for 2 hour and its half- life was 1hour at 90ºC in the presence of 4×10-3 M pNPG. A synergistic effect between Bgl-gsl and with crude enzyme of either fungus Trichoderma reesei Rut-C30 or with a fusion protein (TcE1) made from cellobiohydrolase cbh1 gene of T. reesei and endoglucanase of Acidothermus cellulolyticus was also observed. The above results indicate that the b-glucosidase Bgl-gsl is the possible contender for its application in biofuel production. A â-glucosidase (Bgl1269) having high hydrolyzing capacity for soyabean isoflavone glycosides and tolerant to glucose was reported from metagenomic library of soil (Li et al. 2012). After further investigation, these properties of enzymes can be exploited for production of soyabean isoflavone aglycones.
Challenges in digging out cellulases from metagenome
The first challenge is the extraction of pure metagenomic DNA from various environmental samples for isolation and identification of cellulases. It is observed that the DNA extraction process lead to co-extraction of humic acid and other inhibitory substances that interfere with different cloning steps e.g restriction enzyme digestion, PCR amplification, transformation efficiency and specificity of DNA hybridization (Alawi, Schneider, and Kallmeyer 2014; Tsai and Olson 1992). The other challenges includes: Microorganisms in different environment have different susceptibilities to cell lysis methods so biased extraction of DNA occur, so the sequences present in the isolated DNA and metagenomic libraries is dependent on DNA extraction method. The degree of biasness due to DNA extraction method needs to be studied intensively in different metagenomic DNA. It is supposed that DNA isolated through direct lysis method have better representation of microbial diversity in a soil sample because cell separation step is not there, so microorganisms sticking to soil particles are also lysed (Alawi, Schneider, and Kallmeyer 2014; Leff et al. 1995). The common challenge faced during functional screening of metagenomic DNA is insufficient or biased expression of foreign genes in E. coli. It is important to explore the possibilities to overcome these limitations in order to find out novel cellulase from metagenomic DNA. There are several literature suggesting solution to these problems (Uchiyama, Miyazaki, and Yaoi 2013; Ferrer, Golyshina, Chernikova, Khachane, Martins dos Santos, et al. 2005). Only one cellobiohydrolase gene have been detected so far from any metagenomic library (Table 1). The cellobiohydrolase of family GH48, but none of family GH6 and GH7 were detected by metagenomic sequencing in the contents of bovine rumen (Brulc et al. 2009) and in higher termite hindgut (Warnecke et al. 2007). These findings suggest that fewer cellobiohydrolases exist in natural microbes or there is a novel family of cellobiohydrolase genes in metagenomes that could not be detected by homology searching. Therefore, one of the main challenges for mining cellulases from metagenome of various environment is to develop a robust screening or selection system to select cellobiohydrolases, playing significant role in degrading crystalline cellulose. One of the possible solution to this problem could be construction of metagenomic libraries having larger capacity. The MUC have been used for screening of exoglucanase activity, but this substrate is reported to exhibit activity towards â-glucosidases, cellodextrinases, endoglucanases and some xylanases. In wild strains Avicel is also used as substrate to screen for cellobiohydrolase activity, but in case of clone hydrolyzing activity is not shown towards Avicel and Congo red on agar plate due to limited presence of the endoglucanase and cellobiohydrolase (Duan et al. 2009). These limitations could be overcome by use of alternative host for library construction. One example for this could be construction of recombinant E.coli host constituitively expressing endoglucanase, the synergistic action of endoglucanase and exoglucanase could hydrolyze Avicel easily and will help in detection of cellobiohydrolase activity on Avicel/Congo red plates (Duan and Feng 2010). Metaproteomics is another alternative way of obtaining novel cellobiohydrolases (Warnecke et al. 2007). These strategies were used to isolate cellobiohydrolases from moldy silage and sheep rumen (Toyoda et al. 2009; Yu et al. 2007). The metaproteomics have been used to identify cellulases of family GH1, 3 and 5 (Warnecke et al. 2007). Above all this, sequencing of whole metagenome derived from niche undergoing cellulose decomposition at larger scale may mitigate the problem of biased gene expression. (Warnecke et al. 2007; Brulc et al. 2009). The first metagenomic cDNA library was constructed by Grant et al., (Grant et al. 2004) using RNA derived from water of hot springs and activated sludge. After that, very few metagenomic cDNA library was constructed from mRNA of different environments (Bailly et al. 2007; Frias-Lopez et al. 2008). The construction of less number of cDNA metagenomic libraries than metagenomic DNA libraries may be due to difficulties in RNA isolation, separation of mRNA from other RNAs and less stability of mRNA. It is reported that fungi are important source of cellobiohydrolases, larger number of cellobiohydrolases have been obtained from fungi than from bacteria (Wen, Liao, and Chen 2005). Therefore cDNA metagenomic library will help in extracting cellobiohydrolases of eukaryotes including fungus The metagenomic cDNA of the termite gut have also been reported to contain cellulases (Todaka et al. 2011). This library possessed many cellulase genes involved in protistan cellulose degradation, containing glycosyl hydrolase family 7. The gene of GH7 is the most commonly expressed cellobiohydrolase, accounting for 4% of 910 sequences retrieved. All above examples indicate that the environmental cDNA library approach might be better than metagenomic DNA library method for isolation of cellobiohydrolases. Despite this fact, there is no report of function-based screening of cellulases from any metagenomic cDNA library. The major challenge in case of metagenome is how to get ideal cellulases with desired characteristics fulfilling the requirements of biorefineries. This problem can be resolved to some extent by retrieving enzymes from certain environments having conditions similar to bio-refineries. It was reported that 10 of the 11 cloned cellulases from rabbit cecum exhibited their maximum activities at pH 5.5-7.0 and at temperature 40-55ºC, conditions similar to those prevailing in rabbit cecum (Feng et al. 2007). Similarly an alkaline b-glucosidase was identified from the alkaline soil (Jiang et al. 2010) But there are reports suggesting properties of enzyme being different from the source environments, such as from a high-temperature compost a low temperature tolerant cellulase was isolated (Pang et al. 2009), and from non-saline soil a halotolerant cellulase was isolated (Voget, Steele, and Streit 2006). Therefore, the clones expressing enzymes can be characterized for their enzyme activities as reported previously in several studies (Duan et al. 2009). After obtaining the primary characteristics of crude enzymes, the clone exhibiting remarkable properties could be selected for detailed study. Another alternative process is to find out the activity of clone expressing enzyme in specific conditions mimicking the condition under which enzyme will be used. For example, three cellulase active clones stable in ionic liquids were selected from 24 metagenomic cellulase-active clones by testing their performance in the presence of ionic liquids (Pottkämper et al. 2009).
Concluding remarks
The optimization of DNA extraction process, choice of suitable host-vector for unbiased expression, efficient and robust screening strategies will help in identification of new cellulases from metagenome of various habitats. Although variety of novel genes encoding cellulases have been discovered from metagenomic library, but only few of them possess novel properties in comparison to previously described ones. Therefore one of the major challenge before metagenome derived cellulases is characterization of their properties and finding ways of using them. At this point culture-based method has advantages over culture-independent approach because cellulases possessing desirable properties and fulfilling the criteria of biorefineries can be easily obtained by culture-based methods. After obtaining the suitable cellulases, the shortcomings in properties could be further improved by molecular techniques. Therefore, both culture based approach and culture independent approach are complementary to each other and both could be used together for getting ideal cellulases.
ACKNOWLEDGMENTS
The author is thankful to the Indian Agricultural Research Institute, New Delhi and Amity Institute of Organic Agriculture, Noida. The author declares that there is no conflict of interest regarding the publication of this paper.
- Alawi, M., B. Schneider, and J. Kallmeyer. ‘A procedure for separate recovery of extra- and intracellular DNA from a single marine sediment sample’, J Microbiol Methods, 2014; 21: 36-42.
- Bailly, Julie, Laurence Fraissinet-Tachet, Marie-Christine Verner, Jean-Claude Debaud, Marc Lemaire, Micheline Wésolowski-Louvel, and Roland Marmeisse. ‘Soil eukaryotic functional diversity, a metatranscriptomic approach’, The ISME journal, 2007; 1: 632-42.
- Bayer, Edward A, Raphael Lamed, Bryan A White, and Harry J Flint. ‘From cellulosomes to cellulosomics’, The Chemical Record, 2008; 8: 364-77.
- Berger, Emanuel, Dong Zhang, Vladimir V Zverlov, and Wolfgang H Schwarz. ‘Two noncellulosomal cellulases of Clostridium thermocellum, Cel9I and Cel48Y, hydrolyse crystalline cellulose synergistically’, FEMS microbiology letters, 2007; 268: 194-201.
- Biver, S., A. Stroobants, D. Portetelle, and M. Vandenbol. ‘Two promising alkaline beta-glucosidases isolated by functional metagenomics from agricultural soil, including one showing high tolerance towards harsh detergents, oxidants and glucose’, J Ind Microbiol Biotechnol, 2014; 41: 479-88.
- Brulc, Jennifer M, Dionysios A Antonopoulos, Margret E Berg Miller, Melissa K Wilson, Anthony C Yannarell, Elizabeth A Dinsdale, Robert E Edwards, Edward D Frank, Joanne B Emerson, and Pirjo Wacklin. ‘Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases’, Proceedings of the National Academy of Sciences, 2009; 106: 1948-53.
- Claudel-Renard, C., C. Chevalet, T. Faraut, and D. Kahn. ‘Enzyme-specific profiles for genome annotation: PRIAM’, Nucleic Acids Res, 2003; 31: 6633-9.
- Dalevi, D., N. N. Ivanova, K. Mavromatis, S. D. Hooper, E. Szeto, P. Hugenholtz, N. C. Kyrpides, and V. M. Markowitz. Annotation of metagenome short reads using proxygenes’, Bioinformatics, 2008; 24.
- Duan, Cheng-Jie, and Jia-Xun Feng. ‘Mining metagenomes for novel cellulase genes’, Biotechnology letters, 2010; 32: 1765-75.
- Duan, CJ, L Xian, GC Zhao, Y Feng, H Pang, XL Bai, JL Tang, QS Ma, and JX Feng. ‘Isolation and partial characterization of novel genes encoding acidic cellulases from metagenomes of buffalo rumens’, Journal of applied microbiology, 2009; 107: 245-56.
- Edwards, Ivan P, Rima A Upchurch, and Donald R Zak. ‘Isolation of fungal cellobiohydrolase I genes from sporocarps and forest soils by PCR’, Applied and environmental microbiology, 2008; 74: 3481-89.
- Edwards, R. A., B. Rodriguez-Brito, L. Wegley, M. Haynes, M. Breitbart, D. M. Peterson, M. O. Saar, S. Alexander, E. C. Alexander, Jr., and F. Rohwer. ‘Using pyrosequencing to shed light on deep mine microbial ecology’, BMC Genomics, 2006; 7: 57.
- Fang, W., Z. Fang, J. Liu, Y. Hong, H. Peng, X. Zhang, B. Sun, and Y. Xiao. ‘[Cloning and characterization of a beta-glucosidase from marine metagenome]’, Sheng Wu Gong Cheng Xue Bao, 2009; 25: 1914-20.
- Fang, Z., W. Fang, J. Liu, Y. Hong, H. Peng, X. Zhang, B. Sun, and Y. Xiao. ‘Cloning and characterization of a beta-glucosidase from marine microbial metagenome with excellent glucose tolerance’, J Microbiol Biotechnol, 2010; 20: 1351-8.
- Feng, Yi, Cheng-Jie Duan, Hao Pang, Xin-Chun Mo, Chun-Feng Wu, Yuan Yu, Ya-Lin Hu, Jie Wei, Ji-Liang Tang, and Jia-Xun Feng. ‘Cloning and identification of novel cellulase genes from uncultured microorganisms in rabbit cecum and characterization of the expressed cellulases’, Applied microbiology and biotechnology, 2007; 75: 319-28.
- Ferrer, Manuel, Olga V Golyshina, Tatyana N Chernikova, Amit N Khachane, Vitor AP Martins dos Santos, Michail M Yakimov, Kenneth N Timmis, and Peter N Golyshin. ‘Microbial enzymes mined from the Urania deep-sea hypersaline anoxic basin’, Chemistry & biology, 2005; 12: 895-904.
- Ferrer, Manuel, Olga V Golyshina, Tatyana N Chernikova, Amit N Khachane, Dolores Reyes Duarte, Vitor AP Santos, Carsten Strompl, Kieran Elborough, Graeme Jarvis, and Alexander Neef. ‘Novel hydrolase diversity retrieved from a metagenome library of bovine rumen microflora’, Environmental Microbiology, 2005; 7: 1996-2010.
- Frias-Lopez, Jorge, Yanmei Shi, Gene W Tyson, Maureen L Coleman, Stephan C Schuster, Sallie W Chisholm, and Edward F DeLong. ‘Microbial community gene expression in ocean surface waters’, Proceedings of the National Academy of Sciences, 2008; 105: 3805-10.
- Grant, Susan, Dimitry Y Sorokin, William D Grant, Brian E Jones, and Shaun Heaphy. ‘A phylogenetic analysis of Wadi el Natrun soda lake cellulase enrichment cultures and identification of cellulase genes from these cultures’, Extremophiles, 2004; 8: 421-29.
- Healy, FG, RM Ray, HC Aldrich, AC Wilkie, LO Ingram, and KT Shanmugam. ‘Direct isolation of functional genes encoding cellulases from the microbial consortia in a thermophilic, anaerobic digester maintained on lignocellulose’, Applied microbiology and biotechnology, 1995; 43: 667-74.
- Henrissat, Bernard. ‘A classification of glycosyl hydrolases based on amino acid sequence similarities’, Biochem. J, 1991; 280: 309-16.
- Izquierdo, Javier A, Maria V Sizova, and Lee R Lynd. ‘Diversity of bacteria and glycosyl hydrolase family 48 genes in cellulolytic consortia enriched from thermophilic biocompost’, Applied and environmental microbiology, 2010; 76: 3545-53.
- Jiang, C., S. X. Li, F. F. Luo, K. Jin, Q. Wang, Z. Y. Hao, L. L. Wu, G. C. Zhao, G. F. Ma, P. H. Shen, X. L. Tang, and B. Wu. ‘Biochemical characterization of two novel beta-glucosidase genes by metagenome expression cloning’, Bioresour Technol, 2011; 102: 3272-8.
- Jiang, Chengjian, Zhen-Yu Hao, KE Jin, Shuang-Xi Li, Zhi-Qun Che, Ge-Fei Ma, and Bo Wu. ‘Identification of a metagenome-derived â-glucosidase from bioreactor contents’, Journal of Molecular Catalysis B: Enzymatic, 2010; 63: 11-16.
- Kim, D., S. N. Kim, K. S. Baik, S. C. Park, C. H. Lim, J. O. Kim, T. S. Shin, M. J. Oh, and C. N. Seong. ‘Screening and characterization of a cellulase gene from the gut microflora of abalone using metagenomic library’, J Microbiol, 2011; 49: 141-5.
- Kim, SJ, CM Lee, BR Han, MY Kim, YS Yeo, SH Yoon, BS Koo, and HK Jun. ‘Characterization of a gene encoding cellulase from uncultured soil bacteria’, FEMS microbiology letters, 2008; 282: 44.
- Krause, L., N. N. Diaz, R. A. Edwards, K. H. Gartemann, H. Kromeke, H. Neuweger, A. Puhler, K. J. Runte, A. Schluter, J. Stoye, R. Szczepanowski, A. Tauch, and A. Goesmann. ‘Taxonomic composition and gene content of a methane-producing microbial community isolated from a biogas reactor’, J Biotechnol, 2008; 136: 91-101.
- Kuhad, Ramesh Chander, Rishi Gupta, and Ajay Singh. ‘Microbial cellulases and their industrial applications’, Enzyme research, 2011.
- Leff, L. G., J. R. Dana, J. V. McArthur, and L. J. Shimkets. ‘Comparison of methods of DNA extraction from stream sediments’, Appl Environ Microbiol, 1995; 61: 1141-3.
- Li, G., Y. Jiang, X. J. Fan, and Y. H. Liu. ‘Molecular cloning and characterization of a novel beta-glucosidase with high hydrolyzing ability for soybean isoflavone glycosides and glucose-tolerance from soil metagenomic library’, Bioresour Technol, 2012; 123: 15-22.
- Liu, Li, Yi Feng, Cheng-Jie Duan, Hao Pang, Ji-Liang Tang, and Jia-Xun Feng. ‘Isolation of a gene encoding endoglucanase activity from uncultured microorganisms in buffalo rumen’, World Journal of Microbiology and Biotechnology, 2009; 25: 1035-42.
- Lu, J., L. Du, Y. Wei, Y. Hu, and R. Huang. ‘Expression and characterization of a novel highly glucose-tolerant beta-glucosidase from a soil metagenome’, Acta Biochim Biophys Sin, 2013; 45: 664-73.
- Lynd, Lee R, Paul J Weimer, Willem H Van Zyl, and Isak S Pretorius. ‘Microbial cellulose utilization: fundamentals and biotechnology’, Microbiology and molecular biology reviews, 2002; 66: 506-77.
- Nishihara, Kazuyo, Masaaki Kanemori, Masanari Kitagawa, Hideki Yanagi, and Takashi Yura. ‘Chaperone Coexpression Plasmids: Differential and Synergistic Roles of DnaK-DnaJ-GrpE and GroEL-GroES in Assisting Folding of an Allergen of Japanese Cedar Pollen, Cryj2, inEscherichia coli’, Applied and environmental microbiology, 1998; 64: 1694-99.
- Noguchi, H., J. Park, and T. Takagi. ‘MetaGene: prokaryotic gene finding from environmental genome shotgun sequences’, Nucleic Acids Res, 2006; 34: 5623-30.
- Ohtoko, Kuniyo, Moriya Ohkuma, Shigeharu Moriya, Tetsushi Inoue, Ron Usami, and Toshiaki Kudo. ‘Diverse genes of cellulase homologues of glycosyl hydrolase family 45 from the symbiotic protists in the hindgut of the termite Reticulitermes speratus’, Extremophiles, 2000; 4: 343-49.
- Pandey, S., S. Singh, A. N. Yadav, L. Nain, and A. K. Saxena. ‘Phylogenetic diversity and characterization of novel and efficient cellulase producing bacterial isolates from various extreme environments’, Biosci Biotechnol Biochem, 2013; 77: 1474-80.
- Pang, Hao, Peng Zhang, Cheng-Jie Duan, Xin-Chun Mo, Ji-Liang Tang, and Jia-Xun Feng. ‘Identification of cellulase genes from the metagenomes of compost soils and functional characterization of one novel endoglucanase’, Current microbiology, 2009; 58: 404-08.
- Pottkämper, Julia, Peter Barthen, Nele Ilmberger, Ulrich Schwaneberg, Alexander Schenk, Michael Schulte, Nikolai Ignatiev, and Wolfgang R Streit. ‘Applying metagenomics for the identification of bacterial cellulases that are stable in ionic liquids’, Green chemistry, 2009; 11: 957-65.
- Rappé, Michael S, and Stephen J Giovannoni. ‘The uncultured microbial majority’, Annual Reviews in Microbiology, 2003; 57: 369-94.
- Rees, Helen C, Susan Grant, Brian Jones, William D Grant, and Shaun Heaphy. ‘Detecting cellulase and esterase enzyme activities encoded by novel genes present in environmental DNA libraries’, Extremophiles 2003; 7: 415-21.
- Schluter, A., T. Bekel, N. N. Diaz, M. Dondrup, R. Eichenlaub, K. H. Gartemann, I. Krahn, L. Krause, H. Kromeke, O. Kruse, J. H. Mussgnug, H. Neuweger, K. Niehaus, A. Puhler, K. J. Runte, R. Szczepanowski, A. Tauch, A. Tilker, P. Viehover, and A. Goesmann. ‘The metagenome of a biogas-producing microbial community of a production-scale biogas plant fermenter analysed by the 454-pyrosequencing technology’, J Biotechnol, 2008; 136: 77-90.
- Selengut, J. D., D. H. Haft, T. Davidsen, A. Ganapathy, M. Gwinn-Giglio, W. C. Nelson, A. R. Richter, and O. White. ‘TIGRFAMs and Genome Properties: tools for the assignment of molecular function and biological process in prokaryotic genomes’, Nucleic Acids Res, 2007; 35: 6.
- Todaka, Nemuri, Risa Nakamura, Sigeharu Moriya, Moriya Ohkuma, Toshiaki Kudo, Haruo Takahashi, and Nobuhiro Ishida. ‘Screening of optimal cellulases from symbiotic protists of termites through expression in the secretory pathway of Saccharomyces cerevisiae’, Bioscience, biotechnology, and biochemistry, 2011; 75: 2260-63.
- Toyoda, Atsushi, Wataru Iio, Makoto Mitsumori, and Hajime Minato. ‘Isolation and identification of cellulose-binding proteins from sheep rumen contents’, Applied and environmental microbiology, 2009; 75: 1667-73.
- Tsai, Y. L., and B. H. Olson. ‘Detection of low numbers of bacterial cells in soils and sediments by polymerase chain reaction’, Appl Environ Microbiol, 1992; 58: 754-7.
- Tyson, G. W., J. Chapman, P. Hugenholtz, E. E. Allen, R. J. Ram, P. M. Richardson, V. V. Solovyev, E. M. Rubin, D. S. Rokhsar, and J. F. Banfield. ‘Community structure and metabolism through reconstruction of microbial genomes from the environment’, nature, 2004; 428: 37-43.
- Uchiyama, T., K. Miyazaki, and K. Yaoi. ‘Characterization of a novel beta-glucosidase from a compost microbial metagenome with strong transglycosylation activity’, J Biol Chem, 2013; 288: 18325-34.
- Voget, S, HL Steele, and WR Streit. ‘Characterization of a metagenome-derived halotolerant cellulase’, Journal of biotechnology, 2006; 126: 26-36.
- Wang, Fengchao, Fan Li, Guanjun Chen, and Weifeng Liu. ‘Isolation and characterization of novel cellulase genes from uncultured microorganisms in different environmental niches’, Microbiological research, 2009; 164: 650-57.
- Wang, W., T. Archbold, M. S. Kimber, J. Li, J. S. Lam, and M. Z. Fan. ‘The porcine gut microbial metagenomic library for mining novel cellulases established from growing pigs fed cellulose-supplemented high-fat diets’, J Anim Sci, 2012; 4: 400-2.
- Warnecke, Falk, Peter Luginbühl, Natalia Ivanova, Majid Ghassemian, Toby H Richardson, Justin T Stege, Michelle Cayouette, Alice C McHardy, Gordana Djordjevic, and Nahla Aboushadi. ‘Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite’, Nature, 2007; 450: 560-65.
- Wen, Z., W. Liao, and S. Chen. ‘Production of cellulase by Trichoderma reesei from dairy manure’, Bioresour Technol, 2005; 96: 491-9.
- Williamson, L. L., B. R. Borlee, P. D. Schloss, C. Guan, H. K. Allen, and J. Handelsman. ‘Intracellular screen to identify metagenomic clones that induce or inhibit a quorum-sensing biosensor’, Appl Environ Microbiol, 2005; 71: 6335-44.
- Xia, Yu, Feng Ju, Herbert HP Fang, and Tong Zhang. ‘Mining of novel thermo-stable cellulolytic genes from a thermophilic cellulose-degrading consortium by metagenomics’, PloS one, 2013; 8: e53779.
- Xie, Gary, David C Bruce, Jean F Challacombe, Olga Chertkov, John C Detter, Paul Gilna, Cliff S Han, Susan Lucas, Monica Misra, and Gerald L Myers. ‘Genome sequence of the cellulolytic gliding bacterium Cytophaga hutchinsonii’, Applied and environmental microbiology, 2007; 73: 3536-46.
- Xiong, X., X. Yin, X. Pei, P. Jin, A. Zhang, Y. Li, W. Gong, and Q. Wang. ‘Retrieval of glycoside hydrolase family 9 cellulase genes from environmental DNA by metagenomic gene specific multi-primer PCR’, Biotechnol Lett, 2012; 34: 875-82.
- Yan, X., A. Geng, J. Zhang, Y. Wei, L. Zhang, C. Qian, Q. Wang, S. Wang, and Z. Zhou. ‘Discovery of (hemi-) cellulase genes in a metagenomic library from a biogas digester using 454 pyrosequencing’, Appl Microbiol Biotechnol, 2013; 97: 8173-82.
- Yu, Rentao, Lushan Wang, Xinyuan Duan, and Peiji Gao. ‘Isolation of cellulolytic enzymes from moldy silage by new culture-independent strategy’, Biotechnology letters, 2007; 29: 1037-43.
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.