ISSN: 0973-7510
E-ISSN: 2581-690X
Methicillin-resistant Staphylococcus aureus (MRSA) is a prominent pathogenic, antibiotic-resistant microorganism that contains a variety of virulent characteristics having the capacity to develop tolerance to several major classes of antibiotics. The ongoing creation of clones enhances this potential, transforming S. aureus into an “Anti-Infective.” MRSA has started to rise as a Hospital-Acquired MRSA, but due to evolution, new strains of MRSA have been discovered throughout the past several years. The new strains of MRSA as Community-Acquired MRSA, and Livestock-Associated MRSA are infecting the patients despite preexisting medical conditions, being as susceptible to any treatment. The continuous expansion of MRSA is still ongoing. The main goal of this article is to improve reading comprehension of MRSA by studying the prominent classes of antibiotics and their mechanism of resistance which are now susceptible or getting susceptible to the MRSA.
Antibiotic-resistant, Anti-effective, MRSA, Pathogenic, Staphylococcus aureus
Methicillin-resistant Staphylococcus aureus is a gram-positive bacterium (0.8µm) whose colony seems like a cluster of grapes under a microscope. It belongs to the family Staphylococci and the genus Staphylococcus aureus, which is frequently linked to the rise in bacterial resistance against antibiotics. It is now a part of the ESKAPE group, a subset of the most significant bacteria that cause illness and are known for their resistance treatment.1 In 1881, Sir Alexander Ogston discovered that Staphylococcus has the potential to lead to wound infections in living organisms.2 In 1882, Staphylococcus was named to the genus, which was later parted by Rosenbach (1884) to S. aureus and S. albus.3 Around 81 species of this genus are yet known.4 Most of this genus (S. aureus) causes opportunistic infections showing significance in both veterinary and medical studies. It is one of the most prevalent and prominent species for human pathogenicity.5 S. aureus lives asymptomatically or primarily doesn’t cause any severe infection but causes serious infections by entering the internal tissues or bloodstream.6 Minor skin infections caused by S. aureus include impetigo, scalded skin, pimples, boils, abscesses, etc. Moreover, it also causes fatal illnesses like sepsis, bacteremia, meningitis, pneumonia, and endocardi.6-9
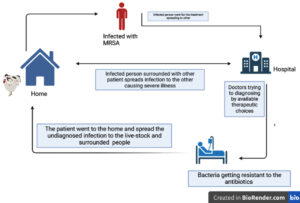
The increasing rate of MRSA in both hospitals and communities is frightening. The spread of MRSA is considerably linked to high morbidity, rise in mortality, poor practices, and expensive treatment.10 MRSA was first identified in 1960, showing resistance to penicillin and many lactam-like drugs. The extensive use of antibiotics and the spread of MRSA was seen from 1970-1980. Till 2010 cephalosporins were active against MRSA but in a very short period, MRSA has shown resistance against these antibiotics. It has been acknowledged that this bacteria is naturally gaining or resisting several antibiotic classes and severely curbing the current treatment options.11 Resistance against all therapeutics, β-lactams, MRS-CN in MRSA strains was linked to transferable genomic material in the bacterial genome called SCCmec (Staphylococcal Chromosomal Cassette mec). Here, methicillin resistance is controlled by the mec gene which further involves a high rate of genetic mobility and fast evolution. In different types of SCCmec, the mecA and mecC with other resistant genes render resistance against other classes of antibiotics such as aminoglycosides, macrolides, lincosamides, streptogramins B, and tetracycline.12 Previously MRSA was related to hospital as Hospital-Acquired MRSA but in 1990 new strains of MRSA were found to be linked with infection among patients who were not hospitalized i,e. community-associated and at the start of 21 century LA-MRSA (Livestock-associated) was also identified13 (as shown in Figure 1).
Literature Search
The information was gathered through PubMed, Google Scholar, and Scopus articles. The keywords used as search terms were “Staphylococcus aureus,” “Methicillin,” “Resistance,” “MRSA,” “Antibiotic,’’ “Anti-effective,” “β-Lactams,” “Daptomycin”, “Glycopeptides & lipoglycopeptides”, “vancomycin”, “Teicoplanin”, “Telavancin”, “Oxazolidinone”, “Linezolid”, “Tedizolid”, “Tetracyclines”, “Doxycycline”, “Minocycline”, “clindamycin”, “Dalbavancin” and “Heavy metal”. After searching the electronic databases, 450 articles were retrieved. There were 350 articles left after duplicate entries were removed. After applying the inclusion (inclusion of specifically matched articles from our study related to plant growth regulators and stress) and exclusion criteria (exclusion of duplicate articles, personal opinions, book chapters, conference abstracts, full copies not available and low-quality paper), a total of 222 studies were chosen for the investigation.
Expanded Information LACTAMS
β-Lactams
β-Lactams as microbiological arsenal comprised more than 65% of the global market of antibiotics consisting of leading resistance mechanisms majorly in gram-negative. It was first reported in Escherichia coli introducing the first β-lactam drug penicillin.14 Later on, various semi-synthetic and natural classes of antibiotics were derived from the β-lactams like methicillin, and cephalosporin C which raised the new family of β-Lactams.15,16 According to the scientific literature β-Lactam antibiotic affects the host by PBP (Penicillin-Binding-Protein) which inhibits the production of mucin secreted by the cell wall causes cell lysis and stimulates microbial activity or by activating the cell lysis activity which further promotes cell death.17 They acquire resistance by introducing the peptidoglycan progenitors of peptidoglycan in the developing wall by penultimate the cell wall and binding a group of active membrane-bound catalysts.18,19
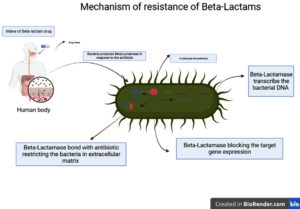
Exposure to MRSA β-Lactams was the most effective antibiotic, but the production of β-Lactamase in contrast with MRSA strains becomes ineffective.20 β-Lactamase, hydrolyze β-lactam antibiotics by transferrable transcribed bacterial chromosomal DNA.21 The massive quantity of β-lactamase strongly forms a bond with the antibiotic in the extracellular matrix, restricting the drug from entering the intracellular matrix. Hence, block the target gene expression and acquired resistance (Figure 2).22,23
Daptomycin
Daptomycin is a lipopeptide medication obtained by fermentation from the bacterial species Streptomyces roseporus.24 It produces oligosaccharides by binding Ca2+ to the phosphatidylglycerol H-bond by inserting Ca2+ into the biomembranes.25,26 Daptomycin doesn’t block lipoteichoic acid due to its effect, which evolves to destroy the electrostatic attraction of cytoplasmic membrane in the calcium-containing environment. Daptomycin’s distinct method of operation mechanisms prevents it from developing resistance as an alveolar suppressor activator to certain MRSA-causing nosocomial infections, skin infection and surrounding tissues damage.27 It is an essential medication or instrument for treating infections that pose a severe risk of death.28 As per the literature the bacteriocidic efficacy of daptomycin is strengthened by β-lactam and the synergid combination is effective in improvising the endovascular disease.29 The synergid combination of β-lactams and daptomycin causes inhibition of b-lactams against PBP1.30 As PBP1 principally initiates cell proliferation whereas β-lactam primarily synthesizes peptidoglycan.31 Daptomycin induces cell death by the pbpA gene encoding for PBP1 which makes cellular clustering after exposure to certain stimuli, the cell becomes vulnerable, especially to PBP1 targeting Carabapenes which further stimulates stress.32,30
Moreover, several life-threatening and gram-positive bacteria like penicillin-resistant Streptococcus pneumonia, vancomycin-resistant enterococci, coagulase-negative staphylococci, and glycopeptide-sensitive Staphylococcus aureus are resistant to daptomycin.33 It is the only accessible drug as an injectable while research is underway for an oral dose formulation. The government of the United States authorizes Daptomycin intravenous infusion to cure severely complicated skin and soft tissue infections.24,34
Mechanism of Daptomycin
Daptomycin’s mode of operation can be as special; it is not completely recognized and likely considerably more complicated and multifunctional than all therapeutics. The antimicrobial agent connects to the cellular membrane more than the therapeutic quantities of calcium ions causing (50 g/ml) dysregulation which results in the efflux of potassium. This results in numerous microbial cell wall components being disrupted without entering the cytoplasmic phase. When the cellular equilibrium of cells is altered critical microbial processes are inhibited, which results in apoptosis in the bacteria 9-12 RES paper. Transformation of the phosphatidylglycerol to lysyl-phosphatidylglycerol frequently causes daptomycin failure, which is altered by the gain of function in the MPrF protein.25,26 Ca2+ daptomycin was rejected by lysine residues when added to the extracellular side barrier and stopped penetration. The absence of phospholipids affects the deposition by LGT acetylase.35 The PsrA, which is necessary for PBP2a’s proper folding, due to loss of function daptomycin – insensitive variants of MRSA unexpectedly acquire vulnerability to β-lactams which inhibit PBP2 causes resistance.36,37
Expanded information about glycopeptides & lipoglycopeptides
The glycopeptides antibacterial drugs belong to the free-liberated mRNA or pseudo-ribosome peptides group. which impede the formation of Gram-positive bacterial cell membranes. these chemical compounds behave like binding agents compared to effective site enzyme blockers, which differs from other anti-bacterial drugs.38,39 The first glycopeptide antimicrobial drug remaining in medical use includes vancomycin, derived from the Latin word “Vanquish” and added to the WHO’s standard classification of essential drugs.39 The glycopeptide class was raised by “ Vancomycin’s carbohydrates-decorated polypeptide organization, characterized by an essential heavily interconnected, multi-annuli, polar substance, free of charge amino acid,40 Several plant-based glycopeptides (therapeutics) differ from one another due to differences in the amino acid sequences between the genera,39 The difference between glycopeptides and lipoglycopeptides is that polypeptides consist of a polar moiety with a noticeable presence of partially synthetic glycopeptides. In particular, N-alkyl alterations to the amino acids and amide bonding change in the terminal carboxylic boost of these drugs, antibacterial effectiveness despite affecting the second lipid interaction,41-43 Teicoplanin is the molecular ancestor of dalbavancin, a longer hydrophilic terminal chain that increases strength that prolongs its life span, while an animated carboxylic terminal unit improves antibacterial function.44 From the reductive alkylating regiochemistry of nitrogen in vancomycin (N-decyl aminoethyl). Tealvancin was produced in contrast to the primary variants. Telavancin has an enormous rise in discharge and a significant decrease in real and hepatic transportation after introducing the polar supplementary group (methylamino phosphonate) acid to exhibit decreased antibacterial properties compared to the initial prelude.45
Significance of vancomycin
Vancomycin was a preferred medication for treating severe infections caused by methicillin-resistant Staphylococcus aureus. It is a complicated tricyclic glycopeptide as an “Ultimate choice “for treating Gram-positive bacterial infection.46 It was first successfully used to treat methicillin-resistant S.aureus in 1952 by Kornfield’s discovery and continued to be actively used till 2012, when it showed resistance towards staphylococcus aureus.33,47 The first strain of S. aureus with diminished susceptibility towards vancomycin was reported in Japan in 1997 and shortly after many reports confirmed the same, including India.48-50 Biofilm production in species that usually didn’t produce biofilm promoted vancomycin resistance by altering the breakdown of cells using their enzymes, particularly those secreted by lysosomes.51,52 It was contributed by the overuse and/or misuse of vancomycin, which consequently encouraged the development of resistance mechanisms that eventually resulted in the rise of multidrug-resistant S. aureus.53,54 As the genetic component driving resistance could be passed from species to species.55 These findings expand the list of targets of RecA to recruit the repressor of another DNA element following a mechanism by which SOS promotes genetic alteration and can increase antibiotic resistance by facilitating the transition of a recombinant resistance element, antibiotics that have not induced the SOS mechanism would not be susceptible to such resistance.56 The criteria are given by NCCLS (National Committee for Clinical Laboratory Standards) for measuring the Vancomycin concentration required by the Staphylococci. The concentration of <4µg/ml for growth inhibition is considered susceptible, 8-16 µg/ml is considered as VISA (Vancomycin-intermediate S. aureus), and those whose required concentration is >8 µg/ml are considered as VRSA (Vancomycin-resistant S. aureus), Hetero-VRSA (Heteroresistant VRSA) required concentration 1-4µg/ml for MICs constitute of different sub-population of vancomycin, these rPAP determines these rations-AUC ration.49,57-59
Mechanism of Resistance of Vancomycin
The resistance mechanism shown by vancomycin towards S. aureus was seen in vanA determinant from Enterococci which has been transferred in vitro to S. aureus.60 The VanA operon encodes on transposon Tn1546 component of a vancomycin-resistant enterococci (VRE) plasmid, which confers total vancomycin resistance in S. aureus (Vancomycin-resistant S. aureus) VRSA (MIC 16>µg/ml).61
The mechanism was due to a rise in cell wall disintegration, which increases non-cross-linked d-alanyl-d-alanine side chains, which can bind vancomycin outside the cell wall, reducing the vancomycin accessible for the intracellular target molecule.62 Understanding the essential elements of the S. aureus cell wall and vancomycin mechanism of action is crucial to comprehend how the VanA operon imparts resistance at a molecular level. The Gram-positive bacteria S. aureus cell wall maintains cellular integrity and promotes interactions between hosts and pathogens just below the topmost polysaccharide capsule layer.63
Assembly of a new cell is generated by the precursor element produced inside the cytoplasm and further transferred to the expanding cell wall division building septum.64 Vancomycin disrupts the peptidoglycan assembly by terminating the D-Ala-D-Ala with newly synthesized UDP-MurNAc-pentapeptides by interfering with the peptidoglycan production in the last stage, it forms vancomycin-pentapeptide complexes build up inside hindering cell wall formation.38 Vancomycin antibiotic resistance depends on crucial events mediated by the Van A operon binds with the dipeptide D-Ala-D-Ala peptidoglycan by hydrolyzing the precursor.65 The molecular mechanism evolved by the VanA operon is, it is comprised of seven coding genes VanA, VanH, Vanx, VanS, VanR, VanY, and VanZ, in which VanH and Vanx are sense regulators which regulate the transcription of vancomycin operon by sensors, VanA and VanX produced D-Ala-D-Ala depsipeptide in which VanA is catalysis which catalysis the ester bonds forms the D-Ala-D-Lac and VanH act as a dehydrogenase forms D-Ala-D-Lac by using pyruvate. VanA, VanH, and VanX act as a resistance phenotype. VanX is a D, D-peptidase that hydrolyzes the ester bond of D-Ala-D-Ala and ensures the D-Ala-D-Lac by UDP-linked tripeptide precursor. In contrast, D, D Carbopeptidase VanY Cleave the D-Ala-D-Ala at the C terminal pentapeptideVanZ function is unclear. However, it may cause teicoplanin resistance.65 Vancomycin-resistant cell walls are produced when changed D-Ala-D-Lac is incorporated into the peptidoglycan.20 Regardless of how the VanA gene expressed resistance against S. aureus strains, many others like Tetracyclines, MLS-B, aminoglycosides, streptomycin, streptogramins B, efflux macrolides, aminoglycosides expressing genes (tet(S) and tet(U), ermB, aac(6) – sph(2) – la, aadE(ant(6)-la, msrA, aphA-3, msrA may be conferred from E. faecium.66
Dalbavancin
Dalbavancin is a newly developed second-generation drug belonging to the lipoglycopeptides, half-line drugs that were authorized to cure serious skin infections. It is effectively active in gram-positive bacteria, especially MRSA.67 It is regarded as a revolutionary therapy due to its efficacy, long dose range, and unique application schedule. It became the initial antimicrobial drug recognized by the FDA as a qualified infectious product by the results. Marion Merrell Dow found Dalbavancin from a chemical compound BI397. Vicuron, Pfizer, and Durata all proceeded with the research and came up with the drugs in (2003, 2005, 2009). Dalbavancin has advanced pharmacological features which must be administered intravenously as substantially associated with the protein.68,69 It can be appropriately characterized based on a 3-compartment system having a long (187 hours) ultimate degradation period. 20% of excretion occurs via feces and 35% o it is unaltered by the urinary tract in healthy human beings. Individuals with severe kidney damage (CrCl, 30ml/min) received dalbavancin and experienced a 50% discharge and an exponential increase in the urine.70 Dalbavancin exhibits more than 4-8 % antibacterial properties than vancomycin against MRSA, according to an examination of time-kill graph experiments searching for effective antibiotics against MRSA dalbavancin is formed.71,72 Dalbavancin is comprised of various antimicrobial resistance for e.g.- (VISA, hVISA, VSSA, MSSA, MRSA).73,74
Mechanism of Dalbavancin
Dalbavancin is a naturally occurring antimicrobial drug derived (A-40926) derived from teicoplanin.75 In practice to create antibiotics against MRSA, many experiments were performed which has further given rise to the continuance of Dalbavancin use.75 Introduction of the polar chain to the (A-40926) teicoplanin resulted in enhanced binding of Dalbavancin to the bacterial cell wall thereby increasing its efficacy as well as its systemic circulation.68 The presence of aminated carboxylic side chains poses anti-MRSA significance, D-alanyl-D-alanine allows Dalbavancin to form bond-terminating sequences which inhibit the ability of GH enzyme and proteolytic enzyme to catalyze amino acids cross-linkage, preventing the structural stability of the cytoplasmic wall from being destroyed and eventually leading to the death of cells.68 Its minimum inhibiting concentration approved by FDA against MRSA is ≤ 0.125µg/ml.76
Teicoplanin
Teicoplanin was previously known as teichomycin, which was obtained by the collagenolytic activities against moenomycin from a separate forming gram-positive bacteria actinomycetes known as ATCC31121 (Actinoplanes teichomyceticus) was obtained from the soil in India.77 For the diagnosis of mild to life-threatening MRSA in people of all generations, teicoplanin was approved as therapeutic by the “Union of Europe”.78 It belongs to the glycopeptide class with antimicrobial properties similar to vancomycin.78,79 Teicoplanin has been created besides modern pharmacological approaches. Thus, therefore, aren’t many consent documents to support the most beneficial usage of this medication.80 The most recent healthcare regulations released on behalf of IDSA emphasized the significance of practical application along with surveillance of bloodstream levels of drugs for maintaining human serum vancomycin quantities sufficient against life-threatening disease caused by MRSA, surveillance of forbearing is necessary to ensure that the patient receives adequate treatment.81 It was extensively stated that teicoplanin, a naturally found antibacterial, is equally effective as vancomycin but has fewer negative consequences. Teicoplanin kills gram-positive bacteria by blocking the cell membrane synthesis.82 The consumption rate of teicoplanin has traditionally been administered, based on the patient’s condition and their weight. While commencing the concentration could reach >10mg/l/day. The average unsuccessful rate was shown to be related to 4mg/kg rather than 6mg/kg.83 Recommended teicoplanin concentrations in infected people are frequently evasive because the drug is mainly peptide-binding. According to the literature on multidrug pharmacology, the teicoplanin dosage should be loaded at 6mg/kg twice daily for 2 days against MRSA.84,85 The treatment monitoring of antibiotic recommendations is lacking due to the absence of data indicating sedative-related adverse reactions. For specific individuals, optimizing treatment could be aided by measuring the levels of plasma.83 Teicoplanin is among the scarce drugs still available for preventing diseases brought on by Multidrug microorganisms. Teicoplanin’s unique property as a commercialized medicine is that its composition is a blend of five main chemical components (A2-1-A2-5), each of which differs in the overall length and bifurcation of a saturated fatty acyl chain which further demonstrates the teicoplanin’s multiplex chemical structure as well as the difficulties of manufacturing and purifying it in a repeatable manner. As a basis for creating the future of antimicrobial agents, GPAs as a whole and Teicoplanin, specifically, are drawing growing attention. Teicoplanin, for example, is currently being synthesized with micron-sized particles, which were demonstrated to have improved effectiveness towards bacteria that produce biofilms.86
Mechanism of action Teicoplanin
As per the literature available, the recreation of the binding site was necessary for the research on the chemical reaction of teicoplanin with the binding site. The synthesis of glycopeptide chains is divided into three phases. Teicoplanin involves glycosylation and cross-linking, which acts as a predecessor and prevents the last step of glycopeptide synthesis by bonding with acyl-D-alanyl-D-alanine of the nascent glycopeptide.87 In the first stage, in which UDP-MurNAc-pentapeptide transforms UDP-GlcNAc. In the second stage, the predecessor is coupled with a nanostructured lipid carrier for drug delivery and then transferred to the exoplasmic phase by an amphipathic molecule. In the third stage, Transacylation and glycosidic bond formation by the reticulum accumulation between the predecessor chaperoned by the lipid and the reprocess inside the cell membrane. Teicoplanin treatment for nascent bacteria aggregates by UDP-MurNAc-pentapeptide inhibits glycopeptide predecessor in the cytoplasmic phase.88 Teicoplanin interferes with glycosidic bond formation, particularly the building of glycopeptide. Teicoplanin is firmly bound to the D-alanyl-D-alanine on the polynucleotide ladder, further emulating the glycosidic bond formation and transacylation building blocks without affecting any molecule.88-91 Glycopeptides create a snug niche in which the N-acyl-D-alanyl-D-alanine group can join. This reactant is created by establishing several hydrogen-bonded chains connecting the glycopeptide chain’s terminus and the D-Ala-D-Ala. The exact positioning of glycosidic bond formation along with its target is chemically hampered with the development of the above intricate, which precludes any extra manufacturing of peptidoglycan. Transacylation interactions are likewise prevented by affecting the end D-alanine leftovers.91 Teicoplanin can bind to gram-positive cell walls and may enhance the snugness of its connection with the D-Ala-D-Ala motif, positioning the antibiotic next to glycopeptide. In contrast, vancomycin repeatedly creates complexes that give it molecular stiffness.92-95
Telavancin
Antimicrobial agents known as lipoglycopeptides, such as dalbavancin and telavancin, are derived from their older antimicrobial agents equivalents (vancomycin and teicoplanin), but with the inclusion of new parameters features known as lipid-soluble chain reactions.96 Vancomycin was altered quasi-synthetically to produce telavancin. It was introduced by a cutting-edge treatment against MRSA and other antibiotic-resistant bacteria by “Theravance.” Telavancin was approved against MRSA and complex skin disease in 2009 by USFDA.97-99 The non-polar chain (decyl aminoethyl ) binds with the aminosugars giving rise to Telavancin, whereas on the 4 location of an amino acid (heptapeptide) is coupled to a polar group of phosphomethyl. These several distinctive pharmacological characteristics of telavancin help understand the Polar and non-polar components.100 Telavancin interacts with the bacterial transmembrane and interferes by affecting the protective layer, thereby triggering lipid two, which is the bacterial transmembrane predecessor. Telavancin has in vitro antibacterial activity against Staphylococcus bacteria that is quantity-dependent. It also contains a variety of medical strains that resist antimicrobial activity.99,101,102 The two analogous created, unpredictable or independent, address telavancin’s effectiveness and safety. Patients of all ages with severe skin diseases that are adequately characterized.99,102 As per the literature, the seven to fourteen days experiments conducted compared to vancomycin, treated with telavancin hydrochloride on the patients with skin infections have more success rates than vancomycin.99
Telavancin possesses several unique qualities
Telavacin showed an antibacterial effect on gram-positive microorganisms that is immediate and quantity-dependent. Maximum human serum values exceed the specified minimal inhibiting limits for Staphylococcus by multiple logarithmic.97,103 Being crucial in the small percentage of severe skin infection people, only 2-5% of people acquire bacterial infections. Due to the two active mechanisms in telavancin, tolerance will arise more slowly.97 Telavancin research studies haven’t revealed the emergence of susceptibility. Telavancin is a promising treatment for pneumonic illnesses because it has not been confined to the alveoli region.104
Mechanism of resistance of telavancin
There have been 2 suggested modes of therapeutic action of telavancin. Telavancin has antibacterial properties by interacting with the d-alanyl-d-alanine sequence of peptide g predecessors transmembrane, just like vancomycin does. This relationship significantly alters the phases of transmembrane which include the synthesis of transglycosylation and successive peptide cross-link formationTelavancin is more effective than vancomycin at inhibiting the glycosaminoglycan chains linked with peptide production in undamaged Staphylococcus bacteria because it strongly reduces peptide g manufacture at the glycosyltransferase stage. Since telavancin compromises microbial reliability. In in vitro, the bacteria-killing effect is observed within minutes.97,99 There has been discussion of an additional mechanism of activation. Hypo-polarization across the bacterial cell transmembrane occurs, affecting the membrane’s function. As not many additional glycopeptides are thought to function similarly, this double procedure that works in this special significance.105 Telavancin has the propensity for lipids 2, a molecule found in bacterial cellular membranes, which is facilitated by its lipophilicity. Since vancomycin bonds more strongly to the transmembrane of the bacterial cell in which transpeptidase occurs. Telavancin can easily enter the transmembrane of bacterial cells and interfere with the glycosyltransferase procedure of cell membrane composite.106 According to the literature, lipid-2 interaction is necessary to activate telavancin to cause cell wall hypopolarization during MRSA facs analysis. This may not represent the crucial phase of microbial transmembrane disruption.107 The reduction of potassium (+) and cytosolic (ATP) may also be related to cell wall hypopolarization. Telavancin’s quicker antibacterial impact against MRSA compared to vancomycin might be caused by this alternative method of operation, which only affects the transmembrane of microorganisms as opposed to vertebrate cells.
The chemical reactions that occur in lipophilicity and constituent of telavancin and the double layer of lipids of transmembrane of microorganism are believed to be the basis for the rupture of transmembrane of microorganisms with telavancin. However, this procedure of action remains not fully comprehended.108,109 In contrast with the antibiotic vancomycin, telavancin is believed to have a more significant attraction to the microorganism’s septal area. Septum adherence to the medication was found in sixty-one percent of microorganisms exposed to telavancin. Whereas Just thirteen percent of the cells that received vancomycin have been shown to have septum attached with medication. Vancomycin had a more significant attraction for the area of adherence with the bacterium (exterior part of the transmembrane) than the other two antibiotics.110 Fewer quantities of telavancin result in less attachment of the transmembrane of microorganisms because its ability to attach to the bacterial membrane is quantity-dependent. Whereas, bonding to the septal area has an impact unaffected by dose absorption.111 The quick antibacterial telavancin effect is not triggered by microbial cell death. It is conceivable that telavancin disrupts the transmembrane perspective by allowing the influx of ions like K+ utilizing ATP to pass through and ultimately causing the death of cells. According to the research conducted with Staphylococcus aureus (n = 8), telavancin sustains its antibacterial activity in vitro whether the pathogen is outside the cell or internalized.112
Oxazolidinone
Oxazolidinones are an emerging category of synthetic antibacterial drugs that is effective towards various Gram-positive microbes, including (VRE), MRSA, and (Mtb). Oxazolidinones suppress the production of pathogenic bacteria’s proteins by attaching to the 50S ribosome unit. LNZ was introduced in 1996 and in 2000, authorized for medicinal purposes by FDA.113,114 The curative properties enforce specific illnesses of plants. The curative properties enforce specific illnesses of plants. Oxazolidinones were first synthesized by EI DuPont de Nemours & Co.Inc. in 1978. The 2 effective antibiotics clinically isolated from Oxazolidinones (DuP721 & DuP105) in 1987. Fortunately, the production of such antibiotics was halted due to their harmful effects. Later on, in 1996, Pfizer studied oxazolidinones and created 2 harmless products Linezolid and eperezolid.115 LNZ is frequently used to treat gram-positive bacterial illnesses and is regarded as an effective medication for operational diseases and Tuberculosis and pneumonia. Oxazolidinones (Linezolid, Tedizolid) are the solely antibacterial drug approved for TB; furthermore, they also get approved for the cure of cSSSI.113,116 The oxazolidine-2-one ring’s stereocentre at position C-5 is connected to its antibacterial activity. LNZ and TZD emerged as intriguing drugs due to the formation of STD with acetamide methyl (2-oxazolidine), Fluorophenyl, and thiomorpholine. The C-5 is connected to antimicrobial activity due to the stereocenter in oxazolidine.117
Additionally, by utilizing suitable DDSs, we can get around the primary barriers that hinder the therapeutic application of oxazolidinones, such as poor bioavailability and negative systemic ramifications, by incorporating the medication or covalently conjugating it. A number of novel substances, notably oxazolidine, are being created and examined in the laboratory for their potency as antibiotics against TB agents.118 The analysis of microbial susceptibility to antibiotics in the bodily fluid has drawn growing curiosity because of its unsettling global expansion. As antimicrobial agents escape into water bodies and wind up in the earth’s soil, endangering wellness, scientific oxazolidinone tenacity techniques are also pertinent for ecological uses.119
Linezolid
Linezolid was first found in E. faecium, gram-positive pathogenic microbes by the periodic susceptibility.120-122 The synthesized bactericidal drug linezolid inhibits enterococci, staphylococci, and most streptococcus strains. It is primarily used to treat sepsis and pneumonia.123,124 Modern therapies, particularly linezolid (oxazolidines) or tedizolid have emerged as crucial drugs for combating disease triggered by bactericides due to the rise of vancomycin-resistant.113 The changes involve the receptor, due to the insertion of upright variation, e.g. modification occurs in ribosomal genes or proteins (like 23S rRNA or L3, L4 & L22). In the bacterium, there is an accumulation of plasmid-borne ARGs which were found to include the 23S RNA (methyltransferases cfr and cfr(B) ), efflux-pump genes (optrA and poxtA).125-130 They may disrupt the 50S & 30S ribosomal subunits and impede the development of the 70s integrated cluster, further interfering with the synthesis in bacterium by binding the 23s region to 50s ribosomal subunits.131 The 52 freely accessible genomic sequences of E. faecium isolates were added to the 41 transcribed sequences from USA and 8 from Pakistan for the data analysis. The analysis of data shows that the linezolid resistance mechanism is correlated with the geographical regions not with the adaptation mechanism, as offered in susceptible variants from the USA retaining the G2576T SMP in 23S rRNA genes, where Pakistan inputting multiple variants of poxtA, OptrA, and cfr like ARGs.
Mechanism of linezolid
The molecular signature of linezolid susceptibility was investigated via two Linezolid resistance variants MRSA strains from French Hospitals (2017 & 2019) recovered from the cystic fibrosis victim. To address the antibacterial resistance, evaluation of the utilizing antibiotic susceptibility by diluting the broth and transition strips through PCR (cfr, cfr(B)) encode portA & oprA genomic sequence was performed. The identification and genotyping of 23S rRNA by nanopores technology evaluate the factor influencing the genomic linezolid basis of resistance by practicing PCR, applying significant markers in PCR to the existence of 23S rRNA variation. The amplified fragments were then sequenced by Sanger., and matched to those of RP62A benchmark S. epidermidis variants. Underlying G2576t alternation has been detected in every MRSA strain subjected to the cfr alteration. While L4 and L22 translational proteins were barbaric-type, the 12 MRSE strains with the G2576T alteration also had C280G and A437C alterations in the genome encoding the L3 translational protein, leading to L94V and H146P, etc. The lone MRSE strain that lacked the G2576T alteration displayed an L94V, L3 rearrangement and persisted in being tedizolid-susceptible. Elevated MIC (256 mg/L) was shown by colonies with the G256T genome alteration and bacterial cfr trait. In contrast, none of the three S. aureus strains deemed cfr-positive included supplementary pathways for linezolid rejection, and the MICs for both linezolid (16-24mh/ml) and tedizolid (0.75-1mg/ml) were racing.132
Tedizolid
Tedizolid, also known as ”torezolid,” is a subsequent generations antibiotic belonging to the class oxazolidinone that Cubist Pharmaceuticals is developing to manage life-threatening disease (S. aureus). A further investigation against tedizolid in therapeutic management (cSSSI, ABSSSI, HA-MRSA, VAP).133 Plasma phosphate empirically metabolizes the ineffective prodrug tedizolid phosphatase (TR-701) into active therapeutics. Tedizolid, concerning gram-positive bacterial infection, functions identically with linezolid by bonding with the 50S subunits with 23S ribosomal RNA, preventing the assembly of integrating 70S units and impeding translation. There is a hydroxyl group in place of the CH3CONH2 compound at the fifth carbon and a CH3NH5 a substituent that linezolid lacks, forming the main molecular distinctions. These alterations could enhance the relationship of tedizolid with aminomethyltransferase of the attachment site and, in certain instances, improve the efficacy about two-to eight times against sensitive microbial strains.133 The D-ring structure is thought to increase the number of bonds with hydrogen and stabilize relationships with the area of interest. Surprisingly tedizolid’s geometry allowed for an antibacterial property that proved several times greater than linezolid’s. Through the help of the phosphorylation of prodrug tedizolid phosphates, the C-5 hydroxymethylation is protected from reactions with MAO, or monoamine oxidase, and has a greatly enhanced mobility in liquid.134 Patients subjected to cSSSI responded to each of the dosages investigated in stage 2 prescription-ranging results in having similar effectiveness rates. For stage 3 studies in subjects having (ABSSSI), a minimal feasible dosage of 200 milligrams was chosen; in ESTABLISH-1, 200 milligrams per day (QD), as It was found that 600 milligrams of the linezolid each for 10 days weren’t superior to tedizolid phosphatase for six days.135 Tedizolid had 90% MIC values that were roughly fourfold lesser concerning linezolid. Increased MIC % is frequently observed in microorganisms with reduced linezolid sensitivity. Teizolid’s minimum inhibitory concentration (MIC), which ranged from zero point five to eight milligrams per milliliter among strains resistant to linezolid, was 8–16 times lesser than that of linezolid, depending on the individual susceptibility pathway.136 Medication-resistant or resistant to drug characteristics like MRSA (VRE) and LR-MRSA. Additionally, tedizolid exhibits activity against LR-MRSA strains carrying the cfr genetic material even in the lack of specific RNA-binding alterations resulting in decreased tedizolid susceptibility. No dosing modifications are necessary for tedizolid in individuals with any impairment of the kidneys or liver. Research on animals has shown that the pharmacokinetic factor strongly linked to tedizolid’s effectiveness is fAUC0-24h/MIC. Tedizolid showed a dosing modifications increase in pseudo-neutropenia organisms, and less administration was needed than in neutropenia groups. A total of two stages of experimental studies.137
Mechanism of resistance of Tedizolid
Tedizolid mechanism of action concerning gram-positive bacterial infection functions identically with linezolid by bonding with the 50S subunits with 23S ribosomal RNA, preventing the assembly of integrating 70S units and impeding translation.138,139 According to in vitro investigations, unexpected susceptibility to tedizolid seems rare.G2576T, T2500A, G2505A, and G2447TA are the four primary mutational sites. The susceptibility of Tedizolid against staphylococcus aureus strains consisted of the T2500A alteration in the 23sRNA, the average levels of transient alteration that reduced tolerance to Tedizolid were approximately 1.1 10-10 and 1.9 10-10, respectively; these factor rates are 16–18 times fewer than those observed using linezolid. At the same time, the emergence of unplanned susceptibility to linezolid engaged either G2576T and T2500A alterations along with the RB amino acids L3 variations His146, Met169Leu, and DPhe127.140 Regarding gram-negative bacteria carrying the G2576T alteration, tedizolid proved more than four times more successful than linezolid, blocking over eighty percent of bacterial infections. The CFR gene is responsible for the diminished tolerance antibacterial response, such as specific mutation concerning the RB subunit. The earliest generally accepted methods of decreased sensitivity were specific alteration of genes within the 23S rRNA or RB amino acids L3 & L4.141 Such genomic heterogeneity, maybe not completely, could be the cause of oxazolidinone’s persistent potency. With repeated administration of linezolid, further changes and a quicker generation of drug-resistant stains are feasible due to specific gene alteration as demonstrated in moderate stability.142 The lateral transferability of the cfr gene, formerly most notorious for causing animal chemical-resistant illnesses, was recently demonstrated.143 cfr gene conferred in both gram-positive and gram-negative microorganisms.
The ermB methylating enzymes were present in CM05, the first diagnostic strain that had Cfr-mediated linezolid susceptibility. The mlr gene, a collection of multiple genes controlled by a specific enhancer, turns all translational antagonists currently used in hospitals inactive.144,145 Due to the CFR-interceded (A2503) methylate enzyme coinciding concerning the C-5 position, linezolid MIC increases by 2-4 in animals, whereas tedizolid.146 On the other hand, tedizolid Minimum Inhibitory Concentration is often constant when the Cfr gene (methylated enzymes) is present.147 The HOCH2 C-5 blockchain of tedizolid is known to be compact & elastic, in contrast with the acetamido methyl blockchain of linezolid, as a primary cause of this. The conserved efficacy of tedizolid with cfr genes shows it could continue to be effective versus certain LR-MRSA. The therapeutic practice has not yet confirmed the medical importance of this in vitro benefit.148
Tetracyclines
Tetracyclines are a group of wide-ranging bacteriostatic drugs prescribed for treating a variety of illnesses, such as chlamydiae, acne, MRSA PID, and a number of infections like zoonosis. These medications prevent tRNA ligase molecules from attaching to the translational receiver site, which inhibits the production of RB proteins in bacteria. Tetracycline drugs, especially minocycline, and doxycycline, a preferred class of prescribed medication, are linked to a higher risk of PTC. However, these conditions are uncommon. Tetracyclines are a class of drugs that are frequently used, particularly by youths, for the treatment of pimples, thus, doctors must be aware of the server complications that can cause death and disability.149 Tetracyclines, a medication class of antibiotics found in 1940 from streptomyces strains.
The second generation antibiotic of Aureomycin received authorization in 1948, whereas Doxycycline and minocycline got consent in 1967, 1971.150 There are increasingly more infectious, proactive, and parasitic microorganisms susceptible to tetracycline. The existence of susceptibility of microbes constrains these drugs from curing illness. Exposure to tetracycline can result from developing novel strains that encode RB proteins and shield the damaging effects of tetracyclines from potential energy extrusion. and can be identified using molecular techniques because they are linked to transposon, While it is unclear how new tetracycline variations will be used in therapy. They show resistance to both gram-positive and gram-negative microbes. Tetracycline-resistant bacteria are becoming more common, leading to research into the molecular basis of mechanisms of resilience and the transformation of genes. Additionally, ongoing studies are recognizing methods for creating antibiotic resistance-inhibiting agents that could be combined with older Tetracycline antibiotics to reactivate them as antimicrobial drugs.151,152
Doxycycline
Doxycycline is a second-generation antibiotic derived from the first-generation tetracycline from a soil-based microorganism. The only distinctive feature of doxycycline is the positioning of the OH group, which differentiates it from minocycline. Its many advantageous features make it distinct from other tetracyclines as they have high lipophilicity value which exposes them to cure skin disease.153-155 A ribosomal (30s) targeting drug blocks the translation process. Doxycycline has been shown to have as an oral drug broad-spectrum bioaccumulation of more than 80%. Doxycycline also has anti-inflammatory properties.156,157
Additionally, it is a helpful antibacterial drug for preventing and curing several possible severe weapons of mass destruction. As a result, it is used extensively worldwide, particularly for treating certain invertebrate-borne rickettsial diseases and for preventing malaria, pneumonia, and STIs.158 Compared with other tetracyclines, doxycycline invades the transmembrane of bacterial cells more quickly.159
Doxycycline is an oral drug. In the digestive tract and the intestine, it gets completely absorbed; in contrast to other tetracyclines, the effects of meals or milk-based items on intake are minimal, with plasma levels just twenty percent lower. Doxycycline reaches its destination in the small intestine as a free medication because it creates compounds with metal particles found in meals that are transient in the acrylic environs of the abdomen. A small amount of doxycycline cannot be taken in due to strong complexes of metals that are generated in the intestines, though. The intake of doxycycline will be hampered by ion polyvalency.160,161 Doxycycline has been known as the “secret arsenal treatment for viral illnesses” due to its broad spectrum of therapeutic applications, particularly treating other uncommon and challenging-to-diagnose disorders. While the persistent macrolide azithromycin has expanded its applications, it still holds a key position in the arsenal of antibiotics. It is also a helpful antimicrobial for the prevention and therapy of many significant possible weapons of mass destruction. Beta-lactam or other antibacterial drugs having more vital resistance against streptococcus pneumonia potency should be combined with doxycycline. Surprisingly satisfactory results were found in CAP treatment with doxycycline in combination with potent antibacterial drugs (ceftriaxone, Amoxycillin, or benzylpenicillin) and effects were terrific for all proven cases of legionellosis. There aren’t any current randomized analyses contrasting doxycycline with other medications.162,163
Mechanism evolved by Doxycycline
Incorporating genes (tet, otr) leads to doxycycline susceptibility, which often comes from the transposition of bacterial DNA,164 a genetic engineering tool(plasmid), and stockpiling by bacterial conjugation. The bulk of such descent produces outflow proteins, which use power to transport doxycycline out of the cell’s membrane. Doxycycline cannot associate with ribosomes because inevitable decline produces an Rb protective layer that causes ribosome mutation. By recursively bonding to the ribosomal unit (the 30s) and the hindrance in mitochondria by bonding of (70s) Rb and blocking the connection of tRNA with aminoacylation to ribosomes of bacterial cells, doxycycline reduces the production of Ubiquitous, as a bacteriostatic drug. Through polar porins in the transmembrane of the bacterial cells and a pH-determined transport (active) in the inner plasmalemma selective barrier, doxycycline break into the cell.
Additionally, it prevents the production of apicomplexa Rb in plassmodium falciparum and impairs haeme biosynthesized and fatty acid synthesis in the postmalarial morphological life stages.150,165,166 Further, it promotes gingivitis cell types connection, inhibits lymphangiogenesis and cell death, and speeds up wound healing, among many other things. Several MMPs, which are proteinase or peptidase produced by cell inflammation, inhibit it. Due to this, potent applications in numerous antibacterial and anti-cancer duties have emerged. Doxycycline in sub-antibacterial intake prevents gingival MMP expression and elastin deterioration in.152-155,158,
167-173 The use of doxycycline as a treatment for several species, including Enterobacteriaceae, S. aureus, S. pneumoniae (including penicillin-resistant pneumococci), and Bacteroides, doxycycline has been restricted due to the rising incidence of doxycycline resistance in different disease (S.aus, S.pneumoniae) in distinct geographical areas. However, doxycycline is still effective in various particular conditions e.g. (penicillin-allergic patients).29
Minocycline
Minocycline(C23H27N3O7) is a semi-synthesized tetracycline, MW -457.5 g/mol. It is a second-generation antibiotic from the tetracycline class, as it lacks CH2OH at the 5 positions and has a (CH3)2NH group at position seven, having high lipophilicity and having the best penetration into the cells. 161,171,174-179 In the early 1960s, it made its appearance. Minocycline binds to the 30S Rb unit of the bacterial cell and stops translation, which is the main principle underlying their antibacterial effectiveness.198 After that, the effectiveness of tetracycline(minocycline) is due to a mutation in C-7 to 9 of the D-ring.177,178 Minocycline is similar to doxycycline but distinct as more prolonged than the first-generation Tetracycline; minocycline has an elimination of 15 to 23 hours or a 76% PPB (plasma protein binding) capability compared to other tetracyclines. The circulatory amount of the minocycline varies between 67.5 – 115 L.
The bile duct, prostate glands, organs of the reproductive system, and urinary system all have tissue-to-serum ratios that are greater than one. Additionally, the trachea plasma concentration is 3.8.174,175, 178,181 Constitutes concentrating 3-8.75 mg/L of minocycline equivalent to other tetracyclines. Minocycline is an oral drug having strong perforation. Followed by i.v. (200mg) the infusion of minocycline. The synergic approach was used to treat gram-negative disease, and the outcomes were comparable to those obtained with first-generation tetracyclines. Minocycline is a promising treatment for severe SSI since it can be administered orally and has suitable perforation. Minocycline is the second most or maybe the only effective treatment for a disease like A. baumannii in vitro. In contrast, using minocycline either by solus or as synergic with other medications is supported by the drug’s therapeutic involvement. Due to the limited available information, minocycline should be an ultimate choice to cure additional multi-drug resistance illnesses. Minocycline retains its antibacterial properties contrary to both MSSA and MRSA, as well as many gram-negative bacteria.175 Initially, tetracycline-resistant UTIs caused by staphylococcus and gram-negative microorganisms were among the conditions for which minocycline was prescribed.
Mechanism evolved Minocycline
Minocycline shares the same methodology as tetracycline compared to the tetracycline antibacterial class by bonding 30s Rb retroactively at the H34 position, further inhibiting protein synthesis. That prevents amino acid-peptides incorporation, which hinders the development of bacteria.182,183 Minocycline susceptibility is caused by several processes involving the antibacterial focus region’s discharge and alteration or shielding. Tet genes are potent ones that can lessen the intransigence of minocycline, the membrane-associated genes, and also can enable the minocycline to leave microorganisms and lower the MICs in the protection of Rb, The Tet genes are of fives types Tet(S), Tet(B), Tet(M), Tet(A)and Tet(K), among them Tet(B), Connected by the leading promoter group outflow mechanism provides minocycline susceptibility.184-190 However, it was demonstrated that the secretion of tetB in tetB-positive, A. baumannii microbes increases the minimal inhibitory concentration (MIC) of minocycline (1-8 g/mL) or drastically decreases tolerance.191 AdeFGH and AdeIJK are two examples of RND-type outflow mechanisms that have been hypothesized as a second outflow system for tetracycline resistance.
Additionally, evidence suggests a clear link between minocycline sensitivity and the absence of the Tet(B) efflux pump. In studies, it was discovered that isolated A. baumannii had the Tet(B) efflux pump and shows 93.3% sensitivity to tetracycline. As a result, it has been suggested that TetB could be a potent option for quick screening genetics to determine minocycline susceptibility. Changes to the Tet(A) gene’s looping process section increase minocycline extrusion.188,189,192,193 According to the researcher, all of the strains of the bacteria A. baumanni that were susceptible to minocycline have the tet(B) gene, which is found on a flanking region of an ISCR2 migratory component. The tet(B) DNA structure supports an innovative process through which A. baumannii strains can communicate with one another. The inhibitory strategy to minocycline is shown,194 The Tet(M) genes produce an RPP in minocycline. It has been suggested that this locus was acquired through an inherited horizontal transmission from microorganisms to S. aureus. A clinical nasal Bacteria infant strain was found to have Tet(S), which is carried by a novel, tiny, reduced replica plasmid resistant to minocycline.186,195 The RPPs encoded by the tet proteins provide a diverse antimicrobial susceptibility. Sensitivity to tetracycline is caused by Ribosomal protein protection, which is protoplasmic with GTPase activation.85,196 The antimicrobial protein changes its shape when it binds to the Rb. Tet(O)-GTP compound was formed by combining GTP and Tet(O). The antimicrobial protein continues to travel once this structure connects to the Rb. The GTP component is subsequently broken down to create a Tet(O)-GDP composite, which is then released from the Rb to ensure it can resume its standard configuration. On the other hand, the drug is thought to be unbound from the protein by the GTP breakdown, and the drug remains undisturbed. 164,185
Miscellaneous drugs
Significance of clindamycin
Clindamycin, an artificially produced bacteriostatic, belongs to the class of lincosamide it is frequently used as a therapeutic to prevent susceptible microbes from synthesizing peptides. At greater dosages, though, they might be bactericidal. When applied with different antibacterial or non-antimicrobials drugs, clindamycin is typically far more successful than other lincosamides at diagnosing fungal infections, especially those brought on by oxygen-deprived organisms. In addition, it may be utilized to combat crucial protozoal illnesses, such as malaria-related illnesses.197 Clindamycin is a sulfonamide, chlortetracycline only attaches to 50 s Rb unit of the microbes that inhibits cytoplasmic protein synthesis.179 The enhanced antimicrobial capacity of clindamycin. Both clindamycin and lincomycin have effective anti-streptolysin O and Penicillin resistance S. aureus properties. Research conducted using in vitro methods showed that in minimal amounts, clindamycin also inhibits the discharge of toxins by producing isolates. Additionally, it has been demonstrated that these drugs have beneficial properties in managing illnesses brought on by fragilis bacterium and a few other anaerobioses. The range of possible behaviors does not include P. aeruginosa.
Compared to most cephalosporins, it covers a broader range of anaerobiosis microbes. However, it seldom affects gram-negative aerobics. Clindamycin plays a part in the medical management of the skull, torso, respiration, skeletal and skin, stomach, and urinary tract illness because of its great action over anaerobic bacteria. Douglas, R. G. (1995). Mandell, Douglas and Bennett’s Principles and Practice of Infectious Diseases. United Kingdom: Churchill Livingstone. Clindamycin reportedly has an excellent rate of absorption (90%) through the GI tract and is present in large amounts in the majority of structures, particularly macrophages, bones (60%), and connections (85%), however, it is not in the brain or spinal cord. According to new studies, clindamycin retention could be up to 50%, despite curiously greater amounts being attained among individuals with severe HIV I (75% retention), probably due to a reduced liver metabolism.198 The FDA (US) has approved the antibacterial drug clindamycin for aerobic management, an anti-streptolysin O and Penicillin resistance S. aureus. Its tendency to trigger antibacterial drug diarrhea, particularly C. difficile colitis, is one of its main drawbacks. Clindamycin seems capable of suppressing toxic generation in poison-elaborating isolates of microorganisms, and it obtains significant internalized concentrations in cells that eat and high concentrations in bones, both of which have raised curiosity in its application.199
Mechanism of resistance of clindamycin
Clindamycin mainly impacts microbes by attaching to their 50s Rb unit. This substance prevents the translation process by inhibiting the peptide formation steps and preventing synthesis. In addition to acting on the 50s Rb unit, Macrolides might contend with one another for bonding at this location. Although they aren’t biologically connected, clindamycin and the accompanying medication lincomycin are frequently mentioned in conjunction with macrolides.199 Despite nonlethal amounts, clindamycin may enhance bacterial interaction and autophagy. Clindamycin alters the outermost layer of bacteria by interfering with peptide production, thereby decreasing the adhesion of microbes to their host cells and boosting the intrinsic death of microorganisms.
Additionally, the medication has a prolonged PAE (post-antibiotic effects) on specific microbial isolates, which might be explained by the medication’s ability to remain at the ribosome interaction region.178,201 The physiological process through which clindamycin hinders the translation process appears to be connected to the reality that clindamycin’s three-dimensional configuration is similar to the initial-Proline-Methionine tRNA and the d-ribose ring of adenosine taking place near each other of initial-Proline-Methionine-tRNA and aminoacylated tRNA for an extended period after the development of the peptide bond among l-Pro-tRNA and l-Met-tRNA at three ends. Clindamycin & lincosamide may potentially function in the first stage of pre-translocation in the protein synthesis process as molecular analogs of the three-carbon ends of initial-Proline-Methionine-tRNA and amino acylated-tRNA. Whereas macrolides, lincosamides, and streptogramins have quite distinct molecular structures, their modes of administration are comparable. Macrolides inhibit translation by bonding to the 3S Rb unit, which causes a sudden release of aminotransferase.202 Furthermore, the list of current options for treating life-threatening MRSA is in Table.
Table:
Listing the antibiotics active against MRSA
Antibiotics |
Classes |
Dosage |
Ref. |
|---|---|---|---|
Zyvox |
Oxazolidinone |
600mg |
203 |
Cubicin |
Miscellaneous antibiotics |
350mg |
204 |
Vancocin |
Glycopeptide |
500mg |
205 |
Floxin |
Quinolones |
20mg |
206 |
Ofloxacin |
Quinolones |
200mg |
207 |
Vancocin HCL |
Glycopeptide |
125mg |
208 |
Vancocin HCL Pelvolus |
Glycopeptide |
500mg |
209 |
Dalfopristin |
Strepto- graminis |
350-150mg |
210 |
Cubicin RF |
Daptomycin |
500mg |
211 |
Dazpura RT |
Daptomycin |
10mg |
212 |
Synercid |
Streptogramins |
150mg |
213 |
Linezolid |
Oxazolidinone |
600mg |
214 |
Daptomycin |
Miscellaneous drug |
500mg |
215 |
Vancomycin |
Glycopeptides |
500mg |
216 |
Delafloxacin |
Fluoroquinolone |
300mg |
217 |
Omadacycline |
Amino-methylcycline |
200mg |
218 |
Oritavancin |
Glycopeptide |
1200mg |
219 |
Sulfo- methoxazole |
Sulfonamides |
200mg |
220 |
Ceftobiprole |
Cephalosporins |
500mg |
221 |
Mupirocin |
Miscellaneous |
500mg |
222 |
Fusidic acid |
Miscellaneous |
600mg |
200 |
MRSA is a dangerous and challenging “Anti-Infective” that may alter its genes’ gene regulation and function to produce variations with increased lethality and colonization ability. The CU currently views MRSA as an important pathogen to the public’s well-being due to its extraordinary adaptability as a bacterium and its shown capacity to build tolerance. Chandigarh University considers it one of the infections with the most significant potential for antibiotic resistance. Considering the ongoing appearance of New clones, which frequently cause long-lasting outbreaks, it might be viewed as a constantly developing marvel. Once just a nosocomial disease encountered in healthcare facilities, the infectious agent is setting up a base in the neighborhood and discovering an innovative animal biological niche. Determining determinants thus represents a vital endeavor for contemporary MRSA studies.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- De Oliveira DMP, Forde BM, Kidd TJ, et al. Antimicrobial Resistance in ESKAPE Pathogens. Clin Microbiol Rev. 2020;33(3):e00181-19.
Crossref - Ogston A. Report upon Micro-Organisms in Surgical Diseases. Br Med J. 1881;1(1054):369.b2-369.b375.
Crossref - Ogston A. Micrococcus Poisoning. J Anat Physiol. 1882;16(Pt 4):526-567.
- Newstead LL, Harris J, Goodbrand S, Varjonen K, Nuttall T, Paterson GK. Staphylococcus caledonicus sp. nov. and Staphylococcus canis sp. nov. isolated from healthy domestic dogs. Int J Syst Evol Microbiol. 2021;71(7).
Crossref - Haag AF, Fitzgerald JR, Penadיs JR. Staphylococcus aureus in Animals. Microbiol Spectr. 2019;7(3).
Crossref - Lakhundi S, Zhang K. Methicillin-Resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clin Microbiol Rev. 2018;31(4):e00020-18.
Crossref - Diekema DJ, Pfaller MA, Schmitz FJ, et al. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin Infect Dis Off Publ Infect Dis Soc Am. 2001;32 Suppl 2:S114-132.
Crossref - Lindsay JA, Holden MTG. Staphylococcus aureus: superbug, super genome? Trends Microbiol. 2004;12(8):378-385.
Crossref - Wertheim HFL, Melles DC, Vos MC, et al. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005;5(12):751-762.
Crossref - Borg MA, de Kraker M, Scicluna E, et al. Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in invasive isolates from southern and eastern Mediterranean countries. J Antimicrob Chemother. 2007;60(6):1310-1315.
Crossref - Kaur DC, Chate SS. Study of Antibiotic Resistance Pattern in Methicillin Resistant Staphylococcus Aureus with Special Reference to Newer Antibiotic.
J Glob Infect Dis. 2015;7(2):78-84.
Crossref - Mlynarczyk A, Mlynarczyk B, Kmera-Muszynska M, Majewski S, Mlynarczyk G. Mechanisms of the resistance and tolerance to beta-lactam and glycopeptide antibiotics in pathogenic gram-positive cocci. Mini Rev Med Chem. 2009;9(13):1527-1537.
Crossref - Hryniewicz W. Epidemiology of MRSA. Infection. 1999;27 Suppl 2:S13-S16.
Crossref - Poole K. Resistance to β-lactam antibiotics. Cell Mol Life Sci CMLS. 2004;61(17):2200-2223.
Crossref - Acred P, Brown DM, Knudsen ET, Rolinson GN, Sutherland R. New semi-synthetic penicillin active against Pseudomonas pyocyanea. Nature. 1967;215(5096):25-30.
Crossref - Abraham EP, Newton GGF. The structure of cephalosporin C. Biochem J. 1961;79(2):377-393.
Crossref - Matono T, Nagashima M, Mezaki K, et al. Molecular epidemiology of β-lactamase production in penicillin-susceptible Staphylococcus aureus under high-susceptibility conditions. J Infect Chemother. 2018;24(2):153-155.
Crossref - Trends in Microbiology, 1994;2(10):341-425. Accessed May 8, 2023. https://www.sciencedirect.com/journal/trends-in-microbiology/vol/2/issue/10
- Goffin C, Ghuysen JM. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol Mol Biol Rev. 1998;62(4):1079-1093.
Crossref - McGuinness WA, Malachowa N, DeLeo FR. Vancomycin Resistance in Staphylococcus aureus . Yale J Biol Med. 2017;90(2):269-281. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5482303/#
- Lee YD, Park JH. Phage Conversion for β-Lactam Antibiotic Resistance of Staphylococcus aureus from Foods. 2016;26(2):263-269.
Crossref - Harada Y, Chong Y, Shimono N, et al. Nosocomial spread of methicillin-resistant Staphylococcus aureus with β-lactam-inducible arbekacin resistance.
J Med Microbiol. 2014;63(5):710-714.
Crossref - Hashizume H, Takahashi Y, Masuda T, et al. In vivo efficacy of β-lactam/tripropeptin C in a mouse septicemia model and the mechanism of reverse β-lactam resistance in methicillin-resistant Staphylococcus aureus mediated by tripropeptin C. J Antibiot (Tokyo). 2018;71(1):79-85.
Crossref - Heidary M, Khosravi AD, Khoshnood S, Nasiri MJ, Soleimani S, Goudarzi M. Daptomycin. J Antimicrob Chemother. 2018;73(1):1-11.
Crossref - Miller WR, Bayer AS, Arias CA. Mechanism of Action and Resistance to Daptomycin in Staphylococcus aureus and Enterococci. Cold Spring Harb Perspect Med. 2016;6(11):a026997.
Crossref - Bayer AS, Schneider T, Sahl HG. Mechanisms of daptomycin resistance in Staphylococcus aureus: role of the cell membrane and cell wall. Annals of the New York Academy of Sciences – Wiley Online Library. 2013. Accessed May 8, 2023. https://nyaspubs.onlinelibrary.wiley.com/doi/10.1111/j.1749-6632.2012.06819.x
- Gómez Casanova N, Siller Ruiz M, Muñoz Bellido JL. Mechanisms of resistance to daptomycin in Staphylococcus aureus. Rev Espanola Quimioter Publicacion Of Soc Espanola Quimioter. 2017;30(6):391-396.
- Moore CL, Osaki-Kiyan P, Haque NZ, Perri MB, Donabedian S, Zervos MJ. Daptomycin Versus Vancomycin for Bloodstream Infections Due to Methicillin-Resistant Staphylococcus aureus With a High Vancomycin Minimum Inhibitory Concentration: A Case-Control Study. Clin Infect Dis. 2012;54(1):51-58.
Crossref - Jung SY, Kim BG, Kwon D, et al. An outbreak of joint and cutaneous infections caused by non-tuberculous mycobacteria after corticosteroid injection. Int J Infect Dis. 2015;36:62-69.
Crossref - Berti AD, Theisen E, Sauer JD, et al. Penicillin Binding Protein 1 Is Important in the Compensatory Response of Staphylococcus aureus to Daptomycin-Induced Membrane Damage and Is a Potential Target for β-Lactam-Daptomycin Synergy. Antimicrob Agents Chemother. 2015;60(1):451-458.
Crossref - Pereira SF, Henriques AO, Pinho MG, de Lencastre H, Tomasz A. Evidence for a dual role of PBP1 in the cell division and cell separation of Staphylococcus aureus. Mol Microbiol. 2009;72(4):895-904.
Crossref - Peacock SJ, Paterson GK. Mechanisms of Methicillin Resistance in Staphylococcus aureus. Annu Rev Biochem. 2015;84(1):577-601.
Crossref - Chuang YC, Lin HY, Chen PY, Lin CY, Wang JT, Chang SC. Daptomycin versus linezolid for the treatment of vancomycin-resistant enterococcal bacteraemia: implications of daptomycin dose. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2016;22(10):890.e1-890.e7.
Crossref - Mediavilla JR, Chen L, Mathema B, Kreiswirth BN. Global epidemiology of community-associated methicillin-resistant Staphylococcus aureus (CAMRSA). Curr Opin Microbiol. 2012;15(5):588-595.
Crossref - Stoll H, Dengjel J, Nerz C, Götz F. Staphylococcus aureus deficient in lipidation of prelipoproteins is attenuated in growth and immune activation. Infect Immun. 2005;73(4):2411-2423.
Crossref - Dhand A, Bayer AS, Pogliano J, Yang SJ, Bolaris M, Nizet V, Wang G, Sakoulas G. Use of Antistaphylococcal β-Lactams to Increase Daptomycin Activity in Eradicating Persistent Bacteremia Due to Methicillin-Resistant Staphylococcus aureus: Role of Enhanced Daptomycin Binding. Clin Infect Dis. 2011;53(2):158-163.
Crossref - Moise PA, Amodio-Groton M, Rashid M, et al. Multicenter evaluation of the clinical outcomes of daptomycin with and without concomitant β-lactams in patients with Staphylococcus aureus bacteremia and mild to moderate renal impairment. Antimicrob Agents Chemother. 2013;57(3):1192-1200.
Crossref - Barna JC, Williams DH. The structure and mode of action of glycopeptide antibiotics of the vancomycin group. Annu Rev Microbiol. 1984;38:339-357.
Crossref - Damiati SA. Digital Pharmaceutical Sciences. AAPS PharmSciTech. 2020;21(6):206.
Crossref - Harris CM, Thomas MH. Structure of the glycopeptide antibiotic vancomycin. Evidence for an asparagine residue in the peptide. Journal of the American Chemical Society. 1982;104(15):4293-4295.
Crossref - Allen NE, LeTourneau DL, Hobbs JN Jr. The role of hydrophobic side chains as determinants of antibacterial activity of semisynthetic glycopeptide antibiotics. J Antibiot (Tokyo). 1997;50(8):677-684.
Crossref - Jakaria SM, Budil DE, Murtagh J. Glycopeptide antibiotic drug stability in aqueous solution. AAPS Open. 2022;8(1):20.
Crossref - Wenzler E, Rodvold KA. Telavancin: the long and winding road from discovery to food and drug administration approvals and future directions. Clin Infect Dis. 2015;61 Suppl 2:S38-S47.
Crossref - Zeng D, Debabov D, Hartsell TL, et al. Approved Glycopeptide Antibacterial Drugs: Mechanism of Action and Resistance. Cold Spring Harb Perspect Med. 2016;6(12):a026989.
Crossref - Damodaran SE, Madhan S. Telavancin: A novel lipoglycopeptide antibiotic. J Pharmacol Pharmacother. 2011;2(2):135-137.
Crossref - Wilhelm MP. Vancomycin. Mayo Clin Proc. 1991;66(11):1165-1170.
Crossref - Okano A, Isley NA, Boger DL. Peripheral modifications of [Ψ[CH2NH]Tpg4]vancomycin with added synergistic mechanisms of action provide durable and potent antibiotics. Proc Natl Acad Sci U S A. 2017;114(26):E5052-E5061.
Crossref - Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover FC. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility.
J Antimicrob Chemother. 1997;40(1):135-136.
Crossref - Smith TL, Pearson ML, Wilcox KR, et al. Emergence of vancomycin resistance in Staphylococcus aureus. Glycopeptide-Intermediate Staphylococcus aureus Working Group. N Engl J Med. 1999;340(7):493-501.
Crossref - Shah P, Kalra S, Yadav Y, et al. Management of Glucocorticoid-Induced Hyperglycemia. Diabetes Metab Syndr Obes Targets Ther. 2022;15:1577-1588.
Crossref - Limper M, de Leeuw K, Lely AT, et al. Diagnosing and treating antiphospholipid syndrome: a consensus paper. Neth J Med. 2019;77(3):98-108.
- M³ynarczyk G, M³ynarczyk A, Zabicka D, Jeljaszewicz J. Lysogenic conversion as a factor influencing the vancomycin tolerance phenomenon in Staphylococcus aureus. J Antimicrob Chemother. 1997;40(1):136-137.
Crossref - Smith DL, Dushoff J, Morris JG. Agricultural antibiotics and human health. PLoS Med. 2005;2(8):e232.
Crossref - Aslam B, Wang W, Arshad MI, et al. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist. 2018;11:1645-1658.
Crossref - Hastings PJ, Rosenberg SM, Slack A. Antibiotic-induced lateral transfer of antibiotic resistance. Trends Microbiol. 2004;12(9):401-404.
Crossref - Beaber J, Hochhut B, Waldor M. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature, 2004;427;72-74.
Crossref - National Agricultural Library. Accessed May 8, 2023. https://www.nal.usda.gov/
- Wilkie T. The Lancet, 1997;350(9092): 1641-1716.
Crossref - Weigel LM, Clewell DB, Gill SR, et al. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science. 2003;302(5650):1569-1571.
Crossref - Noble WC, Virani Z, Rosemary GAC. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiology Letters, 1992;93(2):195-198.
Crossref - Arthur M, Molinas C, Depardieu F, Courvalin P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1993;175(1):117-127.
Crossref - Courvalin P. Vancomycin resistance in gram-positive cocci. Clin Infect Dis Off Publ Infect Dis Soc Am. 2006;42 Suppl 1:S25-34.
Crossref - Dmitriev BA, Toukach FV, Holst O, Rietschel ET, Ehlers S. Tertiary structure of Staphylococcus aureus cell wall murein. J Bacteriol. 2004;186(21):7141-7148.
Crossref - Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G. Pili in gram-positive pathogens. Nat Rev Microbiol. 2006;4(7):509-519.
Crossref - Bugg TD, Wright GD, Dutka-Malen S, Arthur M, Courvalin P, Walsh CT. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry. 1991;30(43):10408-10415.
Crossref - Weigel LM, Donlan RM, Shin DH, et al. High-level vancomycin-resistant Staphylococcus aureus isolates associated with a polymicrobial biofilm. Antimicrob Agents Chemother. 2007;51(1):231-238.
Crossref - Klinker KP, Borgert SJ. Beyond Vancomycin: The Tail of the Lipoglycopeptides. Clin Ther. 2015;37(12):2619-2636.
Crossref - Zhanel GG, Calic D, Schweizer F. et al. New Lipoglycopeptides. Drugs, 2010;70;859-886.
Crossref - Population Pharmacokinetic Analysis of Dalbavancin, a Novel Lipoglycopeptide – Buckwalter – 2005 – The Journal of Clinical Pharmacology – Wiley Online Library. Accessed May 8, 2023.
Crossref - Marbury T, Dowell JA, Seltzer E, Buckwalter M. Pharmacokinetics of Dalbavancin in Patients With Renal or Hepatic Impairment. J Clin Pharmacol. 2009;49(4):465-476.
Crossref - Glenney JR, Soppet D. Sequence and expression of caveolin, a protein component of caveolae plasma membrane domains phosphorylated on tyrosine in Rous sarcoma virus-transformed fibroblasts. Proc Natl Acad Sci U S A. 1992;89(21):10517-10521.
Crossref - Xhemali X, Smith JR, Kebriaei R, et al. Evaluation of dalbavancin alone and in combination with β-lactam antibiotics against resistant phenotypes of Staphylococcus aureus. J Antimicrob Chemother. 2019;74(1):82-86.
Crossref - Campanile F, Bongiorno D, Perez M, et al. Epidemiology of Staphylococcus aureus in Italy: First nationwide survey, 2012. J Glob Antimicrob Resist. 2015;3(4):247-254.
Crossref - Bongiorno D, Mongelli G, Stefani S, Campanile F. Burden of Rifampicin- and Methicillin-Resistant Staphylococcus aureus in Italy. Microb Drug Resist. 2018;24(6):732-738.
Crossref - Malabarba A, Goldstein BP. Origin, structure, and activity in vitro and in vivo of dalbavancin. J Antimicrob Chemother. 2005;55(suppl_2):ii15-ii20.
Crossref - Smith JR, Roberts KD, Rybak MJ. Dalbavancin: A Novel Lipoglycopeptide Antibiotic with Extended Activity Against Gram-Positive Infections. Infect Dis Ther. 2015;4(3):245-258.
Crossref - Bardone MR, Paternoster M, Coronelli C. Teichomycins, new antibiotics from Actinoplanes teichomyceticus nov. sp. II. Extraction and chemical characterization. J Antibiot. 1978;31(3):170-177.
Crossref - Parenti F. Structure and mechanism of action of teicoplanin. J Hosp Infect. 1986;7:79-83.
Crossref - Cavalcanti AB, Goncalves AR, Almeida CS, Bugano DD, Silva E. Teicoplanin versus vancomycin for proven or suspected infection. Cochrane Database Syst Rev. 2010;(6):CD007022.
Crossref - Ramos-Martín V, Johnson A, McEntee L, et al. Pharmacodynamics of teicoplanin against MRSA. J Antimicrob Chemother. 2017;72(12):3382-3389.
Crossref - Liu C, Bayer A, Cosgrove SE, et al. Clinical practice guidelines by the infectious diseases society of america for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children Clin Infect Dis. 2011;52(3):e18-e55. https://academic.oup.com/cid/article/52/3/e18/306145
- Svetitsky S, Leibovici L, Paul M. Comparative efficacy and safety of vancomycin versus teicoplanin: systematic review and meta-analysis. Antimicrob Agents Chemother. 2009;53(10):4069-4079.
Crossref - Harding I, MacGowan AP, White LO, Darley ESR, Reed V. Teicoplanin therapy for Staphylococcus aureus septicaemia: relationship between pre-dose serum concentrations and outcome. Journal of Antimicrobial Chemotherapy. 2000;45(6):835-841.
Crossref - Bernareggi A, Borghi A, Borgonovi M, et al. Teicoplanin metabolism in humans. Antimicrobial Agents and Chemotherapy. 1992;36(8):1744-1749.
Crossref - Esposito S, Tagliabue C, Bosis S, Principi N. Levofloxacin for the treatment of Mycoplasma pneumoniae-associated meningoencephalitis in childhood. International Journal of Antimicrobial Agents. 2011;37(5):472-475.
Crossref - Armenia I, Marcone GL, Berini F, et al. Magnetic Nanoconjugated Teicoplanin: A Novel Tool for Bacterial Infection Site Targeting. Frontiers in Microbiology. 2018;9.
Crossref - Kahne D, Leimkuhler C, Lu W, Walsh C. Glycopeptide and Lipoglycopeptide Antibiotics. Chemical Reviews. 2005;105(2):425-448.
Crossref - Somma S, Gastaldo L, Corti A. Teicoplanin, a new antibiotic from Actinoplanes teichomyceticus nov. sp. Antimicrob Agents Chemother. 1984;26(6):917-923.
Crossref - PETERS G, PULVERER G. Antibacterial activity of teichomycin, a new glycopeptide antibiotic, in comparison to vancomycin. Journal of Antimicrobial Chemotherapy. 1983;11(1):94-95.
Crossref - Li L, Li X, Xia Y, et al. Recommendation of Antimicrobial Dosing Optimization During Continuous Renal Replacement Therapy. Frontiers in Pharmacology. 2020;11:786.
Crossref - Reynolds PE. Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur J Clin Microbiol Infect Dis. 1989;8(11):943-950.
Crossref - Beauregard DA, Williams DH, Gwynn MN, Knowles DJ. Dimerization and membrane anchors in extracellular targeting of vancomycin group antibiotics. Antimicrob Agents Chemother. 1995;39(3):781-785.
Crossref - Mackay JP, Gerhard U, Beauregard DA, Williams DH, Westwell MS, Searle MS. Glycopeptide antibiotic activity and the possible role of dimerization: a model for biological signaling. J Am Chem Soc. 1994;116(11):4581-4590.
Crossref - Schäfer M, Schneider TR, Sheldrick GM. Crystal structure of vancomycin. Structure. 1996;4(12):1509-1515.
Crossref - Jung HM, Jeya M, Kim SY, et al. Biosynthesis, biotechnological production, and application of teicoplanin: current state and perspectives. Appl Microbiol Biotechnol. 2009;84(3):417-428.
Crossref - Terrin N, Schmid CH, Lau J. In an empirical evaluation of the funnel plot, researchers could not visually identify publication bias. Journal of Clinical Epidemiology. 2005;58(9):894-901.
Crossref - Plotkin P, Patel K, Uminski A, Marzella N. Telavancin (vibativ), a new option for the treatment of gram-positive infections. PT. 2011;36(3):127-138.
- Draghi DC, Benton BM, Krause KM, Thornsberry C, Pillar C, Sahm DF. Comparative surveillance study of telavancin activity against recently collected gram-positive clinical isolates from across the United States. Antimicrob Agents Chemother. 2008;52(7):2383-2388.
Crossref - Label of Vibatv. Published online 2009. http://www.accessdata.fda.gov/drugsatfda_docs/label/2009/022110s000lbl.pdf
- Leadbetter MR, Adams SM, Bazzini B, et al. Hydrophobic vancomycin derivatives with improved ADME properties: discovery of telavancin (TD-6424). J Antibiot (Tokyo). 2004;57(5):326-336.
Crossref - Dunbar LM, Tang DM, Manausa RM. A review of telavancin in the treatment of complicated skin and skin structure infections (cSSSI). Ther Clin Risk Manag. 2008;4(1):235-244.
Crossref - Krause KM, Renelli M, Difuntorum S, Wu TX, Debabov DV, Benton BM. In vitro activity of telavancin against resistant gram-positive bacteria. Antimicrobial Agents and Chemotherapy. 2008;52(7):2647-2652.
Crossref - Sun HK, Duchin K, Nightingale CH, Shaw JP, Seroogy J, Nicolau DP. Tissue penetration of telavancin after intravenous administration in healthy subjects. Antimicrobial Agents and Chemotherapy. 2006;50(2):788-790.
Crossref - Corey GR, Stryjewski ME, Weyenberg W, Yasothan U, Kirkpatrick P. Telavancin. Nat Rev Drug Discov. 2009;8(12):929-930.
Crossref - Sharma B, Moghimianavval H, Hwang SW, Liu AP. Synthetic Cell as a Platform for Understanding Membrane-Membrane Interactions. Membranes (Basel). 2021;11(12):912.
Crossref - Benton B, Breukink E, Visscher I, et al. Telavancin inhibits peptidoglycan biosynthesis through preferential targeting of transglycosylation: evidence for a multivalent interaction between telavancin and lipid II. Int J Antimicrob Agents. 2007;29(Suppl):S51-S52.
Crossref - Lunde CS, Hartouni SR, Janc JW, Mammen M, Humphrey PP, Benton BM. Telavancin disrupts the functional integrity of the bacterial membrane through targeted interaction with the cell wall precursor lipid II. Antimicrob Agents Chemother. 2009;53(8):3375-3383.
Crossref - Higgins DL, Chang R, Debabov DV, et al. Telavancin, a multifunctional lipoglycopeptide, disrupts both cell wall synthesis and cell membrane integrity in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2005;49(3):1127-1134.
Crossref - Das B, Sarkar C, Das D, Gupta A, Kalra A, Sahni S. Telavancin: a novel semisynthetic lipoglycopeptide agent to counter the challenge of resistant Gram-positive pathogens [published correction appears in Ther Adv Infect Dis. 2017 Nov;4(6):193]. Ther Adv Infect Dis. 2017;4(2):49-73.
Crossref - Bambeke FV. Glycopeptides in clinical development: pharmacological profile and clinical perspectives. Current Opinion in Pharmacology. 2004;4(5):471-478.
Crossref - Lunde CS, Rexer CH, Hartouni SR, Axt S, Benton BM. Fluorescence Microscopy Demonstrates Enhanced Targeting of Telavancin to the Division Septum of Staphylococcus aureus. Antimicrobial Agents and Chemotherapy. 2010;54(5):2198-2200.
Crossref - Barcia-Macay M, Lemaire S, Mingeot-Leclercq MP, Tulkens PM, Van Bambeke F. Evaluation of the extracellular and intracellular activities (human THP-1 macrophages) of telavancin versus vancomycin against methicillin-susceptible, methicillin-resistant, vancomycin-intermediate and vancomycin-resistant Staphylococcus aureus. J Antimicrob Chemother. 2006;58(6):1177-1184.
Crossref - Bozdogan B, Appelbaum PC. Oxazolidinones: activity, mode of action, and mechanism of resistance. International Journal of Antimicrobial Agents. 2004;23(2):113-119.
Crossref - Jiang J, Hou Y, Duan M, et al. Design, synthesis and antibacterial evaluation of novel oxazolidinone derivatives nitrogen-containing fused heterocyclic moiety. Bioorganic & Medicinal Chemistry Letters. 2021;32:127660.
Crossref - Bolmstrom A, Ballow CH, Qwarnstrom A, Biedenbach DJ, Jones RN. Multicentre assessment of linezolid antimicrobial activity and spectrum in Europe: report from the Zyvox® antimicrobial potency study (ZAPS-Europe). Clinical Microbiology and Infection. 2002;8(12):791-800.
Crossref - Kokilambigai KS, Lakshmi KS, Sai Susmitha A, Seetharaman R, Kavitha J. Linezolid-A Review of Analytical Methods in Pharmaceuticals and Biological Matrices. Crit Rev Anal Chem. 2020;50(2):179-188.
Crossref - Michalska K, Lewandowska K, Mizera M, Bocian W, Pałys B, Cielecka-Piontek J. Spectroscopic identification of intermediates and final products of the chiral pool synthesis of sutezolid. Journal of Molecular Structure. 2020;1217:128396.
Crossref - Ang W, Ye W, Sang Z, et al. Discovery of novel bisoxazolidinone compounds as potential potent and selective antitubercular agents. Bioorganic & Medicinal Chemistry Letters. 2014;24(6):1496-1501.
Crossref - Pinar PT, Erturk Z. Electrochemical and analytical performance of cathodically pretreated boron-doped diamond electrode for the determination of oxazolidinone antibiotic linezolid in cationic surfactant media. Journal of Electroanalytical Chemistry. 2020;878:114681.
Crossref - Auckland C, Teare L, Cooke F, et al. Linezolid-resistant enterococci: report of the first isolates in the United Kingdom. Journal of Antimicrobial Chemotherapy, 2002;50(5):743-746.
Crossref - Bi R, Qin T, Fan W, Ma P, Gu B. The emerging problem of linezolid-resistant enterococci. Journal of Global Antimicrobial Resistance. 2018;13:11-19.
Crossref - Kumar S, Bandyoapdhyay M, Chatterjee M, Mukhopadhyay P, Poddar S, Banerjee P. The first linezolid-resistant Enterococcus faecium in India: High level resistance in a patient with no previous antibiotic exposure. Avicenna J Med. 2014;4(1):13-16. https://pubmed.ncbi.nlm.nih.gov/24678466/.
- Sazdanovic P, Jankovic SM, Kostic M, Dimitrijevic A, Stefanovic S. Pharmacokinetics of linezolid in critically ill patients. Expert Opin Drug Metab Toxicol. 2016;12(6):595-600.
Crossref - Krueger WA, Unertl KE. Neue Therapieoption für Intensivpatienten mit Infektionen durch Gram-positive Bakterien – Uberblick über Linezolid. Anasthesiol Intensivmed Notfallmed Schmerzther. 2002;37(4):199-204. https://pubmed.ncbi.nlm.nih.gov/11967745/.
- Stefani S, Bongiorno D, Mongelli G, Campanile F. Linezolid Resistance in Staphylococci. Pharmaceuticals (Basel). 2010;3(7):1988-2006. https://pubmed.ncbi.nlm.nih.gov/27713338/.
- Dong W, Chochua S, McGee L, Jackson D, Klugman KP, Vidal JE. Mutations within the rplD Gene of Linezolid-Nonsusceptible Streptococcus pneumoniae Strains Isolated in the United States. Antimicrob Agents Chemother. 2014;58(4):2459-2462. https://pubmed.ncbi.nlm.nih.gov/24492357/.
- Ikonomidis A, Grapsa A, Pavlioglou C, Demiri A, Batarli A, Panopoulou M. Accumulation of multiple mutations in linezolid-resistant Staphylococcus epidermidis causing bloodstream infections; in silico analysis of L3 amino acid substitutions that might confer high-level linezolid resistance. J Chemother. 2016;28(6):465-468. https://pubmed.ncbi.nlm.nih.gov/27077930/
- Morales G, Picazo JJ, Baos E, et al. Resistance to linezolid is mediated by the cfr gene in the first report of an outbreak of linezolid-resistant Staphylococcus aureus. Clin Infect Dis. 2010;50(6):821-825. https://pubmed.ncbi.nlm.nih.gov/20144045/.
- Deshpande LM, Ashcraft DS, Kahn HP, et al. Detection of a New cfr-Like Gene, cfr(B), in Enterococcus faecium Isolates Recovered from Human Specimens in the United States as Part of the SENTRY Antimicrobial Surveillance Program. Antimicrob Agents Chemother. 2015;59(10):6256-6261. https://pubmed.ncbi.nlm.nih.gov/26248384/.
- Doern CD, Park JY, Gallegos M, Alspaugh D, Burnham CA. Investigation of Linezolid Resistance in Staphylococci and Enterococci. J Clin Microbiol. 2016;54(5):1289-1294. https://pubmed.ncbi.nlm.nih.gov/26935728/.
- Livermore DM. Linezolid in vitro: mechanism and antibacterial spectrum. J Antimicrob Chemother. 2003;51 Suppl 2:ii9-ii16. https://pubmed.ncbi.nlm.nih.gov/12730138/
- Côrtes MF, André C, Martins Simões P, et al. Persistence of a multidrug-resistant worldwide-disseminated methicillin-resistant Staphylococcus epidermidis clone harbouring the cfr linezolid resistance gene in a French hospital with evidence of interspecies transfer to several Staphylococcus aureus lineages. J Antimicrob Chemother. 2022;77(7):1838-1846.
Crossref - Ferry T, Conrad A, Senneville E, et al. Safety of Tedizolid as Suppressive Antimicrobial Therapy for Patients With Complex Implant-Associated Bone and Joint Infection due to Multidrug-Resistant Gram-Positive Pathogens: Results From the TediSAT Cohort Study. Open Forum Infect Dis. 2021;8(7):1-6.
Crossref - Flanagan S, Bartizal K, Minassian SL, Fang E, Prokocimer P. In vitro, in vivo, and clinical studies of tedizolid to assess the potential for peripheral or central monoamine oxidase interactions. Antimicrob Agents Chemother. 2013;57(7):3060-3066.
Crossref - Prokocimer P, Bien P, Surber J, et al. Phase 2, randomized, double-blind, dose-ranging study evaluating the safety, tolerability, population pharmacokinetics, and efficacy of oral torezolid phosphate in patients with complicated skin and skin structure infections. Antimicrobial Agents and Chemotherapy. 2011;55(2):583-592.
Crossref - Rybak JM, Roberts K. Tedizolid Phosphate: a Next-Generation Oxazolidinone. Infectious Diseases and Therapy. 2015;4(1):1-14.
Crossref - Zhanel GG, Love R, Adam H, et al. Tedizolid: a novel oxazolidinone with potent activity against multidrug-resistant gram-positive pathogens. Drugs. 2015;75(3):253-270. https://link.springer.com/article/10.1007/s40265-015-0352.
- Kisgen JJ, Mansour H, Unger NR, Childs LM. Tedizolid: a new oxazolidinone antimicrobial. Am J Health Syst Pharm. 2014;71(8):621-633.
Crossref - Kanafani ZA, Corey GR. Tedizolid (TR-701): a new oxazolidinone with enhanced potency. Expert Opinion on Investigational Drugs. 2012;21(4):515-522.
Crossref - Locke JB, Hilgers M, Shaw KJ. Novel ribosomal mutations in Staphylococcus aureus strains identified through selection with the oxazolidinones linezolid and torezolid (TR-700). Antimicrobial Agents and Chemotherapy. 2009;53(12):5265-5274.
Crossref - Besier S, Ludwig A, Zander J, Brade V, Wichelhaus TA. Linezolid resistance in Staphylococcus aureus: gene dosage effect, stability, fitness costs, and cross-resistances. Antimicrob Agents Chemother. 2008;52(4):1570-1572.
Crossref - Tsakris A, Pillai SK, Gold HS, et al. Persistence of rRNA operon mutated copies and rapid re-emergence of linezolid resistance in Staphylococcus aureus.
J Antimicrob Chemother. 2007;60(3):649-651.
Crossref - Kehrenberg C, Aarestrup FM, Schwarz S. IS21-558 insertion sequences are involved in the mobility of the multiresistance gene cfr. Antimicrob Agents Chemother. 2007;51(2):483-487.
Crossref - Smith LK, Mankin AS. Transcriptional and translational control of the mlr operon, which confers resistance to seven classes of protein synthesis inhibitors. Antimicrob Agents Chemother. 2008;52(5):1703-1712.
Crossref - Locke JB, Rahawi S, Lamarre J, Mankin AS, Shaw KJ. Genetic environment and stability of cfr in methicillin-resistant Staphylococcus aureus CM05. Antimicrob Agents Chemother. 2012;56(1):332-340.
Crossref - Locke JB, Morales G, Hilgers M, et al. Elevated linezolid resistance in clinical cfr-positive Staphylococcus aureus isolates is associated with co-occurring mutations in ribosomal protein L3. Antimicrob Agents Chemother. 2010;54(12):5352-5355.
Crossref - Issa NC, Rouse MS, Piper KE, Wilson WR, Steckelberg JM, Patel R. In vitro activity of BAL9141 against clinical isolates of gram-negative bacteria. Diagn Microbiol Infect Dis. 2004;48(1):73-75.
Crossref - Shaw KJ, Poppe S, Schaadt R, et al. In Vitro Activity of TR-700, the Antibacterial Moiety of the Prodrug TR-701, against Linezolid-Resistant Strains. Antimicrob Agents Chemother. 2008;52(12):4442-4447.
Crossref - Angelette AL, Rando LL, Wadhwa RD, et al. Tetracycline-, Doxycycline-, Minocycline-Induced Pseudotumor Cerebri and Esophageal Perforation. Adv Ther. 2023;40(4):1366-1378.
Crossref - Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65(2):232-260.
Crossref - Nelson ML, Park BH, Andrews JS, Georgian VA, Thomas RC, Levy SB. Inhibition of the tetracycline efflux antiport protein by 13-thio-substituted 5-hydroxy-6-deoxytetracyclines. J Med Chem. 1993;36(3):370-377.
Crossref - Nelson ML, Levy SB. Reversal of tetracycline resistance mediated by different bacterial tetracycline resistance determinants by an inhibitor of the Tet(B) antiport protein. Antimicrob Agents Chemother. 1999;43(7):1719-1724.
Crossref - Nelson ML, Levy SB. The history of the tetracyclines. Ann N Y Acad Sci. 2011;1241:17-32.
Crossref - Schnappinger D, Hillen W. Tetracyclines: antibiotic action, uptake, and resistance mechanisms. Arch Microbiol. 1996;165(6):359-369.
Crossref - Golub LM, Lee HM, Lehrer G, Nemiroff A, McNamara TF, Kaplan R, Ramamurthy NS. Minocycline reduces gingival collagenolytic activity during diabetes.
J Periodontal Res. 1983;18(5):516-526.
Crossref - Smilack JD. The tetracyclines. Mayo Clin Proc. 1999;74(7):727-729.
Crossref - Agwuh KN, MacGowan A. Pharmacokinetics and pharmacodynamics of the tetracyclines including glycylcyclines. J Antimicrob Chemother. 2006;58(2):256-265.
Crossref - Holmes NE, Charles PGP. Safety and Efficacy Review of Doxycycline. 2009.
Crossref - Nikaido H, Thanassi DG. Penetration of lipophilic agents with multiple protonation sites into bacterial cells: tetracyclines and fluoroquinolones as examples. Antimicrob Agents Chemother. 1993;37(7):1393-1399.
Crossref - Leyden JJ. Absorption of minocycline hydrochloride and tetracycline hydrochloride: Effect of food, milk, and iron. J Am Acad Dermatol. 1985;12(2, Part 1):308-312.
Crossref - Welling PG, Koch PA, Lau CC, Craig WA. Bioavailability of tetracycline and doxycycline in fasted and nonfasted subjects. Antimicrob Agents Chemother. 1977;11(3):462-469.
Crossref - Charles PG, Whitby M, Fuller AJ, et al. The etiology of community-acquired pneumonia in Australia: why penicillin plus doxycycline or a macrolide is the most appropriate therapy. Clin Infect Dis. 2008;46(10):1513-1521.
Crossref - Howden BP, Stuart RL,Tallis G, Bailey M, Johnson PDR. Treatment and outcome of 104 hospitalized patients with legionnaires’ disease. Intern Med J. 2003.
Crossref - Roberts MC. Update on acquired tetracycline resistance genes. FEMS Microbiol Lett. 2005;245(2):195-203.
Crossref - Batty KT, Law AS, Stirling V, Moore BR. Pharmacodynamics of doxycycline in a murine malaria model. Antimicrob Agents Chemother. 2007;51(12):4477-4479.
Crossref - Dahl EL, Shock JL, Shenai BR, Gut J, Derisi JL, Rosenthal PG. Tetracyclines Specifically Target the Apicoplast of the Malaria Parasite Plasmodium falciparum. Antimicrob Agents Chemother. 2006;50(9):3124-3131.
Crossref - Chaidemenos GC. Tetracycline and niacinamide in the treatment of blistering skin diseases. Clin Dermatol. 2001;19(6):781-785.
Crossref - Ramamurthy NS, Rifkin BR, Greenwald RA, et al. Inhibition of Matrix Metalloproteinase Mediated Periodontal Bone Loss in Rats: A Comparison of 6 Chemically Modified Tetracyclines. J Periodontol. 2002;73(7):726-734.
Crossref - Emingil G, Gürkan A, Tervahartiala T, Hernandez M, Özgül S, Sorsa T, Alassiri S. Adjunctive effects of a sub-antimicrobial dose of doxycycline on clinical parameters and potential biomarkers of periodontal tissue catabolism. Dentistry Journal. 2019;7(1):9.
Crossref - Preshaw PM, Hefti AF, Jepsen S, Etienne D, Walker C, Bradshaw MH. Subantimicrobial dose doxycycline as adjunctive treatment for periodontitis. A review. J Clin Periodontol. 2004;31(9):697-707.
Crossref - Pasquale TR, Tan JS. Nonantimicrobial effects of antibacterial agents. Clin Infect Dis. 2005;40(1):127-135.
Crossref - Weinberg JM. The anti-inflammatory effects of tetracyclines. Cutis. 2005;75(4 Suppl):6-11.
- Golub LM, Lee HM, Ryan ME, Giannobile WV, Payne J, Sorsa T. Tetracyclines inhibit connective tissue breakdown by multiple non-antimicrobial mechanisms. Adv Dent Res. 1998;12(2):12-26.
Crossref - Macdonald H, Kelly RG, Allen ES, Noble JF, Kanegis LA. Pharmacokinetic studies on minocycline in man. Clin Pharmacol Ther. 1973;14(5):852-861.
Crossref - Colton B, McConeghy KW, Schreckenberger PC, Danziger LH. I.V. minocycline revisited for infections caused by multidrug-resistant organisms. Am J Health Syst Pharm. 2016;73(5):279-285.
Crossref - Hoban DJ, Reinert RR, Bouchillon SK, Dowzicky MJ. Global in vitro activity of tigecycline and comparator agents: Tigecycline Evaluation and Surveillance Trial 2004-2013. Ann Clin Microbiol Antimicrob. 2015;14:27.
Crossref - Yong VW, Wells J, Giuliani F, Casha S, Power C, Metz LM. The promise of minocycline in neurology. Lancet Neurol. 2004;3(12):744-751.
Crossref - Sapadin AN, Fleischmajer R. Tetracyclines: nonantibiotic properties and their clinical implications. J Am Acad Dermatol. 2006;54(2):258-265.
Crossref - Klainer AS. Clindamycin. Med Clin North Am. 1987;71(6):1169-1175.
Crossref - Nelson ML. Chemical and biological dynamics of tetracyclines. Adv Dent Res. 1998;12(2):5-11.
Crossref - Chopra I. Efflux-based antibiotic resistance mechanisms: the evidence for increasing prevalence. J Antimicrob Chemother. 1992;30(6):737-739.
Crossref - Lashinsky JN, Henig O, Pogue JM, Kaye KF. Minocycline for the Treatment of Multidrug and Extensively Drug-Resistant A. baumannii: A Review. SpringerLink. Accessed May 9, 2023. https://link.springer.com/article/10.1007/s40121-017-0153-2.
- Garrido MN, Zarzuelo A, Galvez J. Minocycline: far beyond an antibiotic. Br J Pharmacol. 2013;169(2):337-352. Wiley Online Library. Accessed May 9, 2023. https://bpspubs.onlinelibrary.wiley.com/doi/10.1111/bph.12139.
- McNicholas P, Chopra I, Rothstein DM. Genetic analysis of the tetA(C) gene on plasmid pBR322. J Bacteriol. 1992;174(24).
Crossref - Bishburg E, Bishburg K. Minocycline – an old drug for a new century: emphasis on methicillin-resistant Staphylococcus aureus (MRSA) and Acinetobacter baumannii. Int J Antimicrob Agents. 2009;34(5):395-401.
Crossref - Ciric L, Brouwer MS, Mullany P, Roberts AP. Minocycline resistance in an oral Streptococcus infantis isolate is encoded by tet(S) on a novel small, low copy number plasmid. FEMS Microbiol Lett. 2014;353(2):106-115.
Crossref - Weese JS, Sweetman K, Edson H, Rousseau J. Evaluation of minocycline susceptibility of methicillin-resistant Staphylococcus pseudintermedius. Vet Microbiol. 2013;162(2-4):968-971.
Crossref - Wang P, McElheny CL, Mettus RT, Shanks RMQ, Doi Y. Contribution of the TetB Efflux Pump to Minocycline Susceptibility among Carbapenem-Resistant Acinetobacter baumannii Strains. Antimicrob Agents Chemother. 2017;61(10):e01176-17.
Crossref - Coyne S, Courvalin P, Périchon B. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob Agents Chemother. 2011;55(3):947-953.
Crossref - Roberts M. Environmental Macrolide-Lincosamide-Streptogramin and Tetracycline Resistant Bacteria. Front Microbiol. 2011;2.
Crossref - Huys G, Cnockaert M, Vaneechoutte M, et al. Distribution of tetracycline resistance genes in genotypically related and unrelated multiresistant Acinetobacter baumannii strains from different European hospitals. Res Microbiol. 2005;156(3):348-355.
Crossref - Lomovskaya O, Sun D, Rubio-Aparicio D, et al. Absence of TetB identifies minocycline-susceptible isolates of Acinetobacter baumannii. Int J Antimicrob Agents. 2018;52(3):404-406.
Crossref - Tuckman M, Petersen PJ, Projan SJ. Mutations in the interdomain loop region of the tetA(A) tetracycline resistance gene increase efflux of minocycline and glycylcyclines. Microb Drug Resist. 2000;6(4):277-282.
Crossref - Vilacoba E, Almuzara M, Gulone L, et al. Emergence and spread of plasmid-borne tet(B)::ISCR2 in minocycline-resistant Acinetobacter baumannii isolates. Antimicrob Agents Chemother. 2013;57(1):651-654.
Crossref - Ribera A, Ruiz J, Vila J. Presence of the Tet M determinant in a clinical isolate of Acinetobacter baumannii. Antimicrob Agents Chemother. 2003;47(7):2310-2312.
Crossref - Spahn CM, Blaha G, Agrawal RK, et al. Localization of the ribosomal protection protein Tet(O) on the ribosome and the mechanism of tetracycline resistance. Mol Cell. 2001;7(5):1037-1045.
Crossref - Spirek J, Rezanka T. Lincosamides: Chemical structure, biosynthesis, mechanism of action, resistance, and applications. Biochem Pharmacol. 2017;133:20-28.
Crossref - Gatti G, Flaherty J, Bubp J, White J, Borin M, Gambertoglio J. Comparative study of bioavailabilities and pharmacokinetics of clindamycin in healthy volunteers and patients with AIDS. Antimicrob Agents Chemother. 1993;37(5):1137-1143.
Crossref - Vukadinović D, Samardžić N, Janković S, Tomić SM, Pavlović R, Stefanović S. Factors associated with early treatment failure in adult hospitalized patients with community-acquired pneumonia. Vojnosanit Pregl. 2017;74(9):803-813.
- Fusidic acid. In: Wikipedia. ; 2023. Accessed June 7, 2023.
- Veringa EM, Lambe DW Jr, Ferguson DA Jr, Verhoef J. Enhancement of opsonophagocytosis of Bacteroides spp. by clindamycin in subinhibitory concentrations. J Antimicrob Chemother. 1989;23(4):577-587.
Crossref - Schlünzen F, Zarivach R, Harms J, et al. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413(6858):814-821.
Crossref - Watkins RR, Lemonovich TL, File TM Jr. An evidence-based review of linezolid for the treatment of methicillin-resistant Staphylococcus aureus (MRSA): place in therapy. Core Evid. 2012;7:131-143.
Crossref - Shoemaker DM, Simou J, Roland WE. A review of daptomycin for injection (Cubicin) in the treatment of complicated skin and skin structure infections. Ther Clin Risk Manag. 2006;2(2):169-174.
Crossref - Zar FA, Bakkanagari SR, Moorthi KM, Davis MB. A comparison of vancomycin and metronidazole for the treatment of Clostridium difficile-associated diarrhea, stratified by disease severity. Clin Infect Dis. 2007; 45(3):302–7.
Crossref - Kaatz GW, Seo SM, Barriere SL, Albrecht LM, Rybak MJ. Efficacy of ofloxacin in experimental Staphylococcus aureus endocarditis. Antimicrob Agents Chemother. 1990;34(2):257-260.
Crossref - Kang SL, Rybak MJ, McGrath BJ, Kaatz GW, Seo SM. Pharmacodynamics of levofloxacin, ofloxacin, and ciprofloxacin, alone and in combination with rifampin, against methicillin-susceptible and -resistant Staphylococcus aureus in an in vitro infection model. Antimicrob Agents Chemother. 1994;38(12):2702-2709.
Crossref - Wijesekara PNK, Kumbukgolla WW, Jayaweera JAAS, Rawat D. Review on Usage of Vancomycin in Livestock and Humans: Maintaining Its Efficacy, Prevention of Resistance and Alternative Therapy. Vet Sci. 2017;4(1):6.
Crossref - Brown NM, Goodman AL, Horner C, Jenkins A, Brown EM. Treatment of methicillin-resistant Staphylococcus aureus (MRSA): updated guidelines from the UK. JAC-antimicrobial resistance. 2021; 1;3(1):1-18.
Crossref - Gurk-Turner C. Quinupristin/dalfopristin: the first available macrolide-lincosamide-streptogramin antibiotic. Proc (Bayl Univ Med Cent). 2000;13(1):83-86.
Crossref - Frankenfeld C, Mittal S, Melendez Y, et al. Daptomycin: a comparison of two intravenous formulations. Drug Des Devel Ther. 2018;12:1953-1958.
Crossref - Yamada T, Soda M, Nishida R, et al. Simplified daptomycin dosing regimen for adult patients with methicillin-resistant Staphylococcus aureus infections based on population pharmacokinetic analysis. Drug Metab Pharmacokinet. 2022;44:100444.
Crossref - Synercid Uses, Side Effects & Warnings. Drugs.com. Accessed June 7, 2023. https://www.drugs.com/mtm/synercid.html.
- Stevens DL, Herr D, Lampiris H, Hunt JL, Batts DH, Hafkin B. Linezolid versus vancomycin for the treatment of methicillin-resistant Staphylococcus aureus infections. Clin Infect Dis. 2002;34(11):1481-1490.
Crossref - Daptomycin Uses, Side Effects & Warnings. Drugs.com. Accessed June 7, 2023. https://www.drugs.com/mtm/daptomycin.html.
- Vancomycin Uses, Dosage, Side Effects. Drugs.com. Accessed June 7, 2023. https://www.drugs.com/vancomycin.html.
- Delafloxacin. In: Wikipedia. ; 2023. Accessed June 7, 2023. https://en.wikipedia.org/w/index.php?title=Delafloxacin&oldid=1154032438.
- Omadacycline. In: Wikipedia. ; 2023. Accessed June 7, 2023. https://en.wikipedia.org/w/index.php?title=Omadacycline&oldid=1151638980.
- Oritavancin. In: Wikipedia. ; 2023. Accessed June 7, 2023. https://en.wikipedia.org/w/index.php?title=Oritavancin&oldid=1153705980.
- Trimethoprim/sulfamethoxazole. In: Wikipedia.; 2023. Accessed June 7, 2023. https://en.wikipedia.org/w/index.php?title=Trimethoprim/sulfamethoxazole&oldid=1155976769.
- Ceftobiprole. In: Wikipedia. ; 2023. Accessed June 7, 2023. https://en.wikipedia.org/w/index.php?title=Ceftobiprole&oldid=1158135505.
- Mupirocin. In: Wikipedia. ; 2023. Accessed June 7, 2023. https://en.wikipedia.org/w/index.php?title=Mupirocin&oldid=1158937502.
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.