ISSN: 0973-7510
E-ISSN: 2581-690X
With an aim to isolate a tannase positive organism, the microbial mat growing on the stored areca extract leachate surface was screened. Once the tannase positive organism was isolated, it was identified by ITS/18S rRNA gene sequencing. Further, the enzyme was purified and examined for its biochemical properties. A potent extracellular tannase-producing yeast was isolated and was identified as Geotrichum cucujoidarum. After the shake flask studies, the enzyme activity of 4.42 U/ml and specific activity of 29.86 U/mg were achieved in a medium with tannic acid as an inducer. Later, ethanol (70%) precipitation followed by purification through FPLC using SEC 650 column resulted in 166.37 U/mg specific activity and a recovery of 50.54%. The purified enzyme was a monomer with a molecular weight of 63 kDa. The optimum pH and the temperature of the enzyme were found to be 5.0 and 30°C, respectively. The Michaelis-Menten constant (Km) was found to be 2.9 mM, and the turn over number (kcat) and catalytic efficiency (kcat/km) of the purified tannase were 102 S-1 and 35.17 mM-1S-1 respectively. Temperature and pH stability profiles of the enzyme, influence of various metal ions, chelators and surfactants on enzyme activity and kinetic constants of enzyme shows that the tannase produced from Geotrichum cucujoidarum is unique and is a potential candidate for further studies.
Geotrichum cucujoidarum, Isolation, Production, Tannase
Tannin acyl hydrolase (E.C.3.1.1.20), commonly known as tannase is an important hydrolytic enzyme belongs to the esterase super family.1 It is an attractive biocatalyst and an inducible enzyme which is widely utilized for tannin degradation. It helps in the hydrolysis of the ester bond (galloyl ester of alcohol moiety) and depside bond (galloyl ester of gallic acid) present in hydrolysable tannins thereby releasing simple phenolic molecules like gallic acid and some gallic acid esters along with glucose molecule.2,3 It also hydrolyzes various substrates like epicatechin gallate, epigallocatechin gallate, and various gallic acid esters such as digallic acid, methyl gallate, ethyl gallate, n-propyl gallate, isoamyl gallate, to release gallic acid.4 Tannase being one of the industrially important hydrolases, it finds an immense range of applications in various industries such as food, animal feed, cosmetics, pharmaceutical, chemical, and leather industries.2,5,6 It is extensively used for the clarification of polyphenolic compounds structure, animal feed preparation, nutritional improvement of legume flours and instantaneous tea elaboration, bioremediation of tannin-contaminated wastewaters.7,8 It is extensively used as a clarifying agent in the production of beverages such as instant tea, corn liquors, beers, and fruit juices.9 Gallic acid, the primary product of tannin hydrolysis finds immense applications in food industry, agriculture sector, dye industry, photography industry, cosmetic industry and pharmaceutical industry.10
The major source for the industrial production of tannase were microorganisms because of their biochemical heterogenicity, facile cultivation, better control over process parameters, and amenability to genetic engineering.11-14 Numerous reports on production and characterization of Tannases from bacteria and molds have been published.2,15 Studies dealing with yeast tannases are relatively less in number.16 In the present investigation, a novel tannase positive yeast strain has been isolated, identified and the enhanced tannase titer has been achieved through media engineering. Further, the tannase enzyme has been purified and its biochemical properties have been studied.
Tannic acid, rhodanine, potassium hydroxide, Microbial media namely potato dextrose agar (PDA), and Potato dextrose broth (PDB) were purchased from Himedia, India. Analytical grade chemicals and have in the study for all experiments and analysis.
Isolation and Identification
The microbial mat growing on the one-year-old aqueous areca extract was used as a source of inoculum. The sample collected was soaked in sterile distilled water and was spread on potato dextrose agar (PDA) and incubated at 30°C for 72 h17,18. The isolated pure cultures were further stored at 4°C. The screening of isolated pure cultures was carried out in 50mL diluted areca extract (1:3) mixed with 2.4% (w/v) of potato dextrose broth in 250 mL conical flasks. The initial pH of the medium was 4.9. The areca nut extract was collected from areca nut farmers of Sagar Taluk, Shimoga district, Karnataka state, India. The sterile flasks were inoculated by transferring the colonies growing on PDA plate culture using a sterilized inoculation needle. The shake flask studies were carried out at 25°C at 120 rpm.
The isolated organisms were examined under microscope (Olympus India) after lactophenol cotton blue staining. To evaluate the presence of tannase enzyme, the isolated cultures were plated on the PDA plates containing tannic acid. The clear zones indicate the hydrolysis of tannic acid indicates the presence of tannase enzyme in the isolated yeast culture. 19 The culture was preserved on PDA slants at 4°C and glycerol stock at -80°C. Taxonomic characterization of the organisms was performed by ITS/18S rRNA gene sequencing at National Collection of Industrial Microorganisms (NCIM), Pune.
Tannase Assay and Protein Estimation
The enzyme activity of tannase was estimated spectrophotometrically based on the rhodamine method proposed by Sharma et al.20 The amount of gallic acid produced was determined based on the chromogen (pink color) developed by the gallic acid and the methanolic rhodanine (2-thio-ketothiazolidine) during the hydrolysis of tannic acid, at room temperature (≈30°C) & pH 5.0. The absorbance was measured using a spectrometer (LABINDIA®Analytical) at 520 nm. One unit of tannase activity was defined as 1µmole of gallic acid released per minute under the assay conditions. All the determinations were performed in triplicates. The amount of soluble protein at various stages of enzyme production was quantitatively determined by the Bradford method (1976).21
Shake-flask Studies
For seed culture, a loop full of culture was taken from the PDA plate and inoculated into a 100ml conical flask containing 50 ml of sterile broth (PDB) incubated at 27°C at 150 rpm for 24 h in an incubator shaker (Scigenics, India). The fermentation medium was prepared as given in Table 1. Tannic acid was always filter-sterilized and added to the sterile medium in the flask. The growth rate of the yeast was monitored by measuring the turbidity (absorbance) of the fermentation broth at 600 nm using a spectrophotometer (Labomed, India). Fermentation was performed in Erlenmeyer flasks, containing 100 ml of culture media at 27°C in an incubator shaker for 48 h. The fermented broth was centrifuged at 10,000 rpm for 20 min at 4°C and the cell-free supernatant (crude) was assayed for the tannase activity. The sediment was dried for 24 h at 80°C to find the dry weight of biomass. All the experiments were done in duplicates.
Table (1):
Various media used for the production of tannase.
Media |
Composition (g/L) |
|---|---|
Medium 1 |
K2HPO4-0.5, KH2PO4-0.5, MgSO4-2.0, CaCl2-1.0,NH4Cl2-3.0, Tannic acid-10.0. |
Medium 2 |
Czapek Dox medium, Tannic acid-10.0. |
Medium 3 |
Potato Dextrose Broth, Tannic acid-10.0. |
Medium 4 |
Sucrose-1.0, NaCl-0.5, Yeast extract-1.0, Tannic acid-10.0. |
Purification of Tannase
Tannase enzyme produced by the yeast isolate was purified by fractional precipitation employing salt (Ammonium sulphate) and solvent (Ethanol, Isopropanol and acetone individually) followed by size exclusion Chromatography. In order to achieve 10-100% saturation, calculated amount of ammonium sulphate was added to the cell-free broth. At different degrees of saturation, the precipitated protein was collected by centrifugation (10,000 rpm for 15 min at 4°C). The citrate buffer (50mM, pH 5.0) was used to resuspend precipitate. The enzyme activity and protein concentrations of various saturations were measured. Prechilled solvents (ethanol, isopropanol and acetone) were slowly added to the cell-free broth until the required saturation (10-90 %) is attained. The solution was incubated at -20°C for about 2 hr. The precipitated protein was separated by centrifugation at 10,000 rpm for 10 min at 4°C. The supernatant was discarded and the precipitate was air-dried, washed once with double distilled water. Citrate buffer (50 mM, pH 5) was added to dissolve the precipitate and tested for enzyme activity and protein concentration.
Further purification was performed using a fast protein liquid chromatography (FPLC) system (BIO-RAD NGC Chromatography System). The precipitated enzyme samples were applied to SEC 650 column in FPLC of dimensions (10 mm × 300 mm) pre-equilibrated with citrate buffer (50mM, pH 5) and the constant flow rate of 0.5 mL/min was maintained to perform the elution. The eluted fractions were measured at A280 and collected by an automated fraction collector as 1mL fractions and each fraction was subjected to the standard assay for tannase activity. The fractions showing tannase activity were pooled together and used for further analysis.
Characterization of Purified Tannase
Determination of Molecular Weight of the Enzyme
The molecular weight of the purified tannase was determined by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) employing mini gel system (Bio-Rad). The SDS gel comprises of 12.5% separating and 4% stacking gels according to the method of Laemmli (1970),22 with a wide range of protein ladder. The samples for SDS-PAGE were prepared by incubating 20µl of the purified enzyme with 5µl of buffer containing 10% β- mercaptoethanol (β- ME) at 100°C for 5 min. The prepared enzyme samples were loaded and run under reducing and denaturing conditions with a constant voltage of 70V. The gel was stained overnight by Coomassie blue and de-stained using destaining solution after which protein bands were detected. The standard protein marker was used to compared and estimate the molecular weight of the resultant bands produced by the purified enzyme.
Effect of pH on the Enzyme Activity and Stability
To examine the effect of pH on tannase activity the reactions were carried out at various pHs of 0.1M buffers ranging from pH 2 to pH 10 at 30°C. Various buffers used to maintain different pH were glycine buffer of pH 2.0-3.0; citrate buffer of pH 3.0-6.0; phosphate buffer of pH 6.0-8.0 and glycine-NaOH buffer of pH 8.0-10.0. The enzyme tannase was pre incubated at different buffers of pH values ranging from 2.0 to 8.0 at 30°C for 12 h to determine the pH stability profile. The residual enzyme activity was measured under the standard assay conditions and compared with the activity determined before the incubation.
Effect of Temperature on the Enzyme Activity and Stability
The enzyme activity was measured at different temperatures (25 – 100ºC) in 0.1M citrate buffer (pH 5) for 30 min to determine the optimum temperature. The temperature stability profile of the enzyme tannase was estimated by incubating the enzyme solution at varying temperatures (25-100°C) in 0.1M citrate buffer (pH 5) for a maximum of 12h and the aliquots were taken periodically. After the incubation, the temperature of enzyme solution was rapidly brought down to room temperature and the residual activity was estimated under standard assay conditions. The Activity of the enzyme determined before the incubation was considered as 100%.
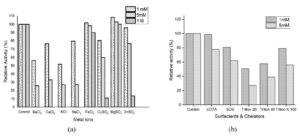
Effect of Different Metal Ions, Chelators and Surfactants
The tannase activity was studied in presence of various metal ions (Mg2+, Fe3+, Cu2+, Ca2+, Zn2+, Ba2+, Na+, and K+). Equal quantity of the enzyme and metal ions with different concentrations (1mM, 5mM, and 1M) were mixed and incubated at 30°C for 30min. Desired concentrations of metal ions were prepared by dissolving different metal salts MgSO4.7H2O, FeCl3, CuSO4.5H2O, CaCl2, ZnSO4.7H2O, BaCl2, NaCl2, and KCl in the buffer to get different concentration. Then the enzyme samples were assayed for tannase activity as stated in the standard assay method. The activity devoid of metal ions was considered 100%. The surfactants and chelator of different concentrations were evaluated for their influence on tannase activity. The chelator like, ethylene diamine tetraacetic acid disodium salt (EDTA) and the surfactants namely Tween 20, Tween 80, and Triton X-100 at two different concentrations (1mM and 5mM) were added to the purified enzyme and incubated for 30 min at 30°C and then the enzyme activity was estimated as per the protocol. Enzyme activity without the chemical agents was considered as 100%.
Determination of Kinetic Parameters
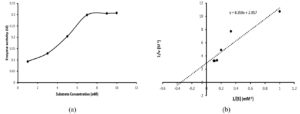
The kinetic parameters like Km and Vmax were determined from the Lineweaver-Burk double reciprocal plot. The turnover number (Kcat) and catalytic efficiency (kcat/km) was also calculated. Tannic acid with varying concentrations from 1-10 mM prepared in 0.1 M citrate buffer (pH 5.0) was used as the substrate.
Isolation and Identification of Tannase Positive Organism
Only two types of colonies were visible on the PDA plates. Based on the colony morphology, pigmentation, cell shape, color of mycelia/cells and microscopic observation, it was concluded that one is a filamentous fungi capable of sporulation and another one is a yeast. The amplification and sequencing of the variable region of the 18s rRNA of the test organism showed 99% pairwise similarity with AB250164.1 Rhizopus oryzae. The sequences of the isolate were submitted to the GenBank with Accession No. MW538932. Similarly, the amplification and sequencing of the hypervariable region of the 28s rRNA of yeast showed 90% pairwise similarity with KY107748.1 Geotrichum cucujoidarum. Hence, the isolate was identified as Geotrichum cucujoidarum. The phylogenetic tree was constructed and analysed. The sequences of the isolate were submitted to the GenBank with Accession No. MW538971.
Only two microbes were isolated from the stored liquid surface of the areca nut extract. Usually tender areca nut is peeled and boiled till a considerable amount of polyphenols are leached out and gives a red colouration to the boiling liquid and then areca nuts are drained and sun dried to get “kaliadeke or kalipak” in several districts of Karnataka state, India.23 The extract is enriched with large quantity of areca tannins, procyanidins, arecoline etc.24 Perhaps, due to the lack of fermentable sugars and the presence of large amount of polyphenols, most of the microorganisms cannot grow in this extract. Hence it is a popular practice to store it and reuse it in the next season for boiling the areca nuts. Isolation of only two organisms corroborate that it is an ecological niche, where only limited number of organisms can grow. Subsequent culturing of these two pure cultures in the screening medium showed that the yeast isolate, Geotrichum cucujoidarum could produce extracellular tannase. Whereas the other isolate Rhizopus oryzae failed to show any extracellular tannase production. Thus Geotrichum cucujoidarum was taken for all the further studies.
Growth Studies and Enzyme Production
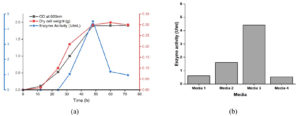
Maximum tannase was produced in the early stationary phase of growth (Figure 1). Culturing of this yeast in a medium which is devoid of tannic acid showed growth but there was no tannase activity indicating that it is an inducible enzyme. Its peak production in the early stationary phase and inducible nature of production witnessed here corroborate the findings of numerous authors.2,25 Among the different media studied, Medium 3 (PDB+1% tannic acid) was found to be a good producer of tannase enzyme.
Figure 1. Growth studies. (a) Time course study of biomass and enzyme production, (b) Enzyme production in different nutrient media
Production of tannase using microbial source received wide attention by the research groups world over due to the importance of the enzyme, tannase. Numerous reports were available on tannase production by bacterial fermentation and mycelial fermentation.10 Marquez-Lopez et al.,16 have reported the detection of tannase activity in Debaryomyces hansenii, Debaryomyces hansenii, Candida parapsilosis, Candida utilis, Pichia pastoris, Pichia kluyveri, Issatchenkia terricola. Kanpiengjai et al.26 have reported the isolation of three extracellular tannase positive yeasts, Debaryomyces hansenii, Cyberlindnera rhodanensis, and Sporidiobolus ruineniae from Miang, a fermented food product from Thailand. However, reports on tannase production from yeasts are limited. Only three reports were available on tannase production from yeasts, Arxula adeninivorans, Kluyveromyces marxianus and Rhodosporidium diobovatum.27-29 This is the first report of extracellular tannase production from Geotrichum cucujoidarum.
Purification of Tannase
The tannase was partially purified from a 48h cell-free broth using salt and solvent precipitation followed by FPLC. The results of different stages of tannase purification were summarized in the Table 2. The crude cell-free broth having total activity of 442 U and specific activity of 29.86 U/mg was subjected to purification.
Table (2):
Purification of tannase from Geotrichum cucujoidarum.
Purification steps |
Volume (ml) |
Total Activity (U) |
Total Proteins (mg) |
Specific Activity (U/mg) |
Purification Fold |
Recovery (%) |
|---|---|---|---|---|---|---|
Crude Extract |
100 |
442 |
14.8 |
29.86 |
1 |
100 |
Ammonium Sulphate precipitation |
54 |
106 |
9.85 |
10.76 |
0.36 |
23.98 |
Iso propanol
precipitation |
54 |
403 |
4.9 |
82.24 |
2.75 |
91.17 |
Acetone precipitation |
54 |
323.5 |
6.05 |
53.47 |
1.79 |
73.19 |
Ethanol precipitation |
54 |
460.6 |
5.08 |
90.67 |
3.03 |
96.49 |
FPLC |
24 |
223.4 |
1.34 |
166.37 |
5.57 |
50.54 |
Initially, an attempt was made to precipitate tannase using ammonium sulphate at different saturations (10-100%). At 20% saturation, the yield was 23.28%. As the salt concentration increased, yield of enzyme was progressively decreasing perhaps due to enzyme inactivation. This led to the exploration of precipitation using various solvents and found that acetone, ethanol and isopropanol can precipitate tannase enzyme. Among all the solvents and their concentration tested, 70% ethanol could give an enzyme yield of 96.49% with a specific activity of 90.67 U/mg. The precipitated enzyme was further purified by FPLC. The elution profile of tannase from FPLC was shown in Figure 2. Upon purification, the specific activity had increased by 5.57 folds with 50.54% recovery.
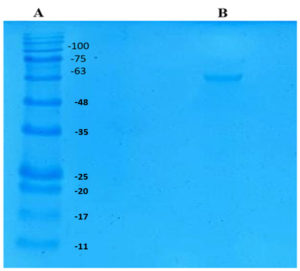
Molecular Weight of Tannase
SDS-PAGE analysis was performed to determine the molecular weight of the purified tannase. As seen in Figure 2. the purified enzyme showed a single band of 63 kDa indicating the monomeric nature of the enzyme. Molecular weight of tannases studied by various authors were found to be in the range of 50–320 kDa30 and the result obtained here concur with those findings. Most of the authors have reported that tannases of microbial origin consists of two or more subunits30 which is contrary to the result obtained here. The tannase enzyme produced by a yeast, Arxula adeninivorans was found to be a homotetramer with the molecular weight of 80 kDa.31 The tannase produced by another yeast, Kluyveromyces marxianus NRRL Y-8281 was about 65 kDa and was monomeric.28 Similarly, the molecular weight of tannase produced by Rhodosporidium diobovatum Q95 was 75.1 kDa.29 The molecular weight of tannase produced in this study is in same range of other yeasts studied so far. However, the number of subunits present in the native enzyme might be more than one and it could be confirmed by analytical gel filtration.
Effect of pH on Enzyme Activity and Stability
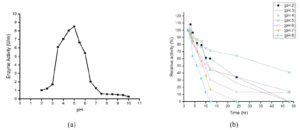
Enzymes are greatly influenced by the change of pH and function best over a finite extent, with a definite optimal pH. The influence of pH on the activity of the tannase enzyme was shown in Figure 3a. The maximal activity was observed at pH 5.0 (8.52 U/ml), which is found to be the optimal pH for tannase from Geotrichum cucujoidarum. As the pH proceeds towards the alkaline range the enzyme activity started decreasing and lost most of its activity (0.26 U/ml) at the pH of 10. This shows that enzyme tannase was more active in an acidic environment and also proves that the tannase from Geotrichum cucujoidarum is an acidic protein.32,33 This is in agreement with the general observation that the optimal pH values of most fungal tannases were detected around 6.0, whereas those of bacterial tannases predominantly lie between 7.0 and 9.0.29
The pH stability profile of Geotrichum cucujoidarum tannase was investigated by varying the pH from 2.0 to 8.0 as shown in Figure 3b. Enzyme activity without 12h of incubation was considered as 100%. Furthermore, it possessed at least 60% of the activity in a pH range of 2.5–6.5. The enzyme had retained 71% of its activity at pH 5 after 12h of incubation and seemed to be quite stable in the pH range of 3-6. This is in agreement with general observation that most of the fungal tannases were ascertained to be stable in the pH range of 3 to 6.34
Figure 3. Effect of pH on the (a) activity of the enzyme and (b) stability of tannase produced by Geotrichum cucujoidarum
Effect of Temperature on Enzyme Activity and Stability
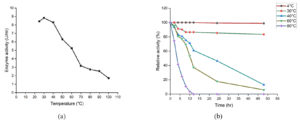
Enzyme activity of Geotrichum cucujoidarum tannase at different functional temperatures (25-100°C) was examined. The markable increase in tannase activity was observed and the optimal activity of 8.83 U/ml was recorded at 30°C (Figure 4a). Escalation in temperature beyond the optimal temperature results in decreased enzyme activity. This is in agreement with all the three yeast tannases studied so far exhibited temperature optima in the range of 30-40°C.20,31 This may be due to the thermal inactivation of enzymes resulting in structural loss/change.28,29 Many fungal species which produce tannase were reported to have 30-40°C as their temperature optima.30
The thermal stability profile of tannase shows around 37% of enzyme activity was retained after 12h of incubation at 60°C and it also revealed that the enzyme was stable at 30-60°C (Figure 4b). The loss of activity was negligible at 4°C and the loss of activity was close to 20% after 48 h of storage at 30°C.
Figure 4. Effect of temperature on the (a) activity of the enzyme and (b) stability of tannase produced by Geotrichum cucujoidarum
Effect of Metal Ions, Chelators and Surfactants on Enzyme Activity
Three fourth of all the industrially important enzymes require metal ion activation to exhibit its maximum catalytic potential.31 Therefore, the influence of various metal ions on tannase was shown in Figure 5a. Among the various metal ions studied, Mg2+ increased the tannase activity. The other metal ions like Ba2+, Ca2+, Na+, and K+ completely inhibited the tannase activity as the concentration increases to 1M. Fe3+ slightly stimulated the tannase activity at 1mM whereas Cu2+ and Zn2+ mildly inhibited the tannase enzyme activity, but these metal ions have a staunch hindering effect on the activity as the concentration increases to 1mM. Many reports on the effects of metal ions on tannases concur with the results presented here. Magnesium was found to stimulate tannases produced by Aspergillus nomius GWA5,32 Rhizopus oryzae, Aspergillus foetidus,33 Verticillium sp. P934 and Rhizopus oryzae.35 Interestingly Mg2+, Mn2+, Ca2+ and Zn2+ had no effect on the tannase produced by the yeast Arxula adeninivorans.31 In case of tannase produced by a yeast Kluyveromyces marxianus, Fe2+, Cu2+ and Ca2+ enhanced the activity whereas all the other metal ions exhibited weak inhibition.28 Fe2+ also found to stimulate the tannase produced by a yeast Rhodosporidium diobovatum Q95.29 The inhibition in enzyme activity might be due to the binding of metal ions to sulfhydryl groups, tryptophan residues, and/or carboxyl groups at the active site of an enzyme.36
Surfactants were reported to have some role in the catalytic activity of enzymes.33 In this study surfactants like SDS, Triton X-100, Tween 20, Tween 80, and chelators like EDTA have been investigated and the effect on tannase activity was shown in Figure 5b. The results depict that the tannase activity was inhibited by the surfactant and EDTA. It was observed that tannase activity was slightly inhibited by the chelator EDTA (1mM) and a significant loss was observed as the concentration increased to 5mM. The inhibition by surfactants could be due to the some of the combined effect of factors involving reduction in the hydrophobic interactions that possess a major part in holding the protein tertiary structure and the direct interactions with the protein molecule.37 It was noticed that SDS and Triton X-100 inhibited the enzyme activity by 19.23% and 21.02% at low concentration (1%) as the concentration increases (5%) the inhibition was found to be 38.27% and 44.11%.
Kinetic Studies
The kinetic constants were studied by employing standard assay conditions (pH 5.0 and ≈30°C) using tannic acid as substrate. Determination of enzyme activity with varying substrate concentration revealed that the reaction obeys Michaelis-Menten kinetics (Figure 6a). Using Origin pro-2019, the constructed Lineweaver-Burk double reciprocal plot was shown in Figure 6b. The Michaelis-Menten constant (Km) was found to be 2.9 mM and maximum velocity of the enzyme reaction (Vmax) was appraised to be 0.34 U/ml. The turn over number (kcat) and catalytic effieciency (kcat/km) of the produced tannase were 102 S-1 and 35.17 mM-1S-1 respectively. Reports on the kinetic constants of yeast are rather scarce. Mahmoud et al.28 had reported a Km and Vmax value of 0.77 mM and 263.20 U/ml respectively using tannic acid as substrate for the tannase enzyme produced from Kluyveromyces marxianus. Tannase from Arxula adeninivorans has Km of 0.14 mM for endogenous enzyme and 0.17 mM for recombinant tannase and the catalytic efficiency was found to be 78 μM/s for endogenous enzyme and 64 μM/s for recombinant enzyme.31
In the present study, an extracellular tannase producing yeast Geotrichum cucujoidarum was isolated from an ecological niche. The titer of the extra cellular tannase has been enhanced by media engineering. Further, tannase has been purified and its important characteristics have been elucidated. Temperature and pH stability profiles of enzyme, effect of various metal ions, chelators and surfactants on Tannase activity and kinetic constants of enzyme shows that the tannase enzyme produced from Geotrichum cucujoidarum is unique and is a potential candidate for industrial exploitation.
ACKNOWLEDGMENTS
The authors would like to thank the National Institute of Technology Karnataka (NITK), Surathkal, India, for providing a fellowship to pursue doctoral research work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
PDB conceptualized the study, applied methodology, acquired funding, and performed supervision. NT data curation. NT and PH performed investigation and wrote the manuscript. PDB reviewed, edited and approved the manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
ETHICS STATEMENT
Not applicable.
- Kumar M, Singh A, Beniwal V, Salar RK. Improved production of tannase by Klebsiella pneumoniae using Indian gooseberry leaves under submerged fermentation using Taguchi approach. AMB Express. 2016;6(1).
Crossref - Dhiman S, Mukherjee G, Kumar A, Mukherjee P, Verekar SA, Deshmukh SK. Fungal tannase: Recent advances and industrial applications. Dev Fungal Biol Appl Mycol. 2017:295-313.
Crossref - Mondal KC, Banerjee D, Jana M, Pati BR. Colorimetric assay method for determination of the tannin acyl hydrolase (EC 3.1.1.20) activity. Anal Biochem. 2001;295(2).
Crossref - Azzaz HH, Kholif AE, Abd El Tawab AM, Khattab MSA, Murad HA, Olafadehan OA. A newly developed tannase enzyme from Aspergillus terreus versus commercial tannase in the diet of lactating Damascus goats fed diet containing pomegranate peel. Livest Sci. 2020;241.
Crossref - Al-mraai STY, Al-fekaiki DF, Al-manhel AJA. Purification and characterization of tannase from the local isolate of Aspergillus niger. J Appl Biol Biotechnol. 2019;7(1):29-34.
Crossref - Jana A, Halder SK, Ghosh K, et al. Tannase Immobilization by Chitin-Alginate Based Adsorption-Entrapment Technique and Its Exploitation in Fruit Juice Clarification. Food Bioprocess Technol. 2015;8(11).
Crossref - Albuquerque KKSA, Albuquerque WWC, Costa RMPB, et al. Biotechnological potential of a novel tannase-acyl hydrolase from Aspergillus sydowii using waste coir residue: Aqueous two-phase system and chromatographic techniques. Biocatal Agric Biotechnol. 2020;23.
Crossref - Samanta S, Giri S, Parua S, Nandi DK, Pati BR, Mondal KC. Impact of tannic acid on the gastrointestinal microflora. Microb Ecol Health Dis. 2004;16(1).
Crossref - Wu C, Zhang F, Li L, Jiang Z, Ni H, Xiao A. Novel optimization strategy for tannase production through a modified solid-state fermentation system. Biotechnol Biofuels. 2018;11(1):1-15.
Crossref - Dhiman S, Mukherjee G, Singh AK. Recent trends and advancements in microbial tannase-catalyzed biotransformation of tannins: a review. Int Microbiol. 2018;21(4):175-195.
Crossref - Beniwal V, Kumar A, Sharma J, Chhokar V. Recent Advances in Industrial Application of Tannases:
A Review. Recent Pat Biotechnol. 2013;7(3):228-233.
Crossref - Banerjee A, Jana A, Pati BR, Mondal KC, Das Mohapatra PK. Characterization of tannase protein sequences of bacteria and fungi: An in silico study. Protein J. 2012;31(4).
Crossref - Jana A, Halder SK, Banerjee A, et al. Biosynthesis, structural architecture and biotechnological potential of bacterial tannase: A molecular advancement. Bioresour Technol. 2014;157.
Crossref - Dayana AT, Dhrithi PM, Nomuk K, Sharadamma N. Purification and Characterization of Tannase: A review. Int J Sci Eng Res. 2021;12(3):941-951.
- de las Rivas B, Rodríguez H, Anguita J, Muñoz R. Bacterial tannases: classification and biochemical properties. Appl Microbiol Biotechnol. 2019;103(2):603-623.
Crossref - Márquez-López A, Ramírez-Conejo JD, Chávez-Parga MDC, et al. Comparative analysis of enzymatic activity of tannase in non-conventional yeasts to produce ellagic acid. Food Sci Technol. 2020;40(3):557-563.
Crossref - Jana A, Maity C, Halder SK, Mondal KC, Pati BR, Mohapatra PK Das. Tannase production by Penicillium purpurogenum PAF6 in solid state fermentation of tannin-rich plant residues following OVAT and RSM. In: Applied Biochemistry and Biotechnology. 167; 2012.
Crossref - Jana A, Maity C, Halder SK, Pati BR, Mondal KC, Mohapatra PK Das. Rapid screening of tannase producing microbes by using natural tannin. Brazilian J Microbiol. 2012;43(3).
Crossref - Bradoo S, Gupta R, Saxena RK. Screening of extracellular tannase-producing fungi: Development of a rapid and simple plate assay. J Gen Appl Microbiol. 1996;42(4).
Crossref - Sharma S, Bhat TK, Dawra RK. A spectrophotometric method for assay of tannase using rhodanine. Anal Biochem. 2000;279(1).
Crossref - Bradford M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal Biochem. 1976;72(1-2).
Crossref - Laemmli,U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nat Publ Gr. 1970;228:726-734.
- Mathew AG, Govindarajan VS. Studies on Arecanut: Part I – Changes in Chemical Composition & Physical Characteristics of Nuts with Maturity, 1964;2(3):90-96.
- Peng W, Liu YJ, Wu N, et al. Areca catechu L. (Arecaceae): A review of its traditional uses, botany, phytochemistry, pharmacology and toxicology.
J Ethnopharmacol. 2015;164:340-356.
Crossref - Iibuchi S, Minoda Y, Yamada K. Studies on tannin acyl hydrolase of microorganisms. Agric Biol Chem. 1968;32(7):803-809.
Crossref - Kanpiengjai A, Chui-Chai N, Chaikaew S, Khanongnuch C. Distribution of tannin-tolerant yeasts isolated from Miang, a traditional fermented tea leaf (Camellia sinensis var. assamica) in northern Thailand. Int J Food Microbiol. 2016;238:121-131.
Crossref - Böer E, Breuer FS, Weniger M, Denter S, Piontek M, Kunze G. Large-scale production of tannase using the yeast Arxula adeninivorans. Appl Microbiol Biotechnol. 2011;92(1):105-114.
Crossref - Mahmoud AE, Fathy SA, Rashad MM, Ezz MK, Mohammed AT. Purification and characterization of a novel tannase produced by Kluyveromyces marxianus using olive pomace as solid support, and its promising role in gallic acid production. Int J Biol Macromol. 2018;107:2342-2350.
Crossref - Pan J, Wang NN, Yin XJ, Liang XL, Wang ZP. Characterization of a robust and pH-stable tannase from mangrove-derived yeast Rhodosporidium diobovatum Q95. Mar Drugs. 2020;18(11):1-11.
Crossref - Yao J, Guo GS, Ren GH, Liu YH. Production, characterization and applications of tannase. J Mol Catal B Enzym. 2014;101:137-147.
Crossref - Böer E, BOde R, Mock H peter, Piontek M, Kunze G. Atan1p — an extracellular tannase from the dimorphic yeast Arxula adeninivorans:molecular cloning of the ATAN1 gene and characterization of the recombinant enzyme. Yeast. 2009;26(6):323-37.
Crossref - Mahapatra K, Nanda RK, Bag SS, Banerjee R, Pandey A, Szakacs G. Purification, characterization and some studies on secondary structure of tannase from Aspergillus awamori nakazawa. Process Biochem. 2005;40(10):3251-3254.
Crossref - Mehta M, Muddapur M, Shanmuga Priya VG. Fungal Production of Tannase:A Review. Int J Sci Eng Technol. 2013;755(28):2277-1581.
- Ordoñez RM, Colombo I, Alberto MR, Isla MI. Production of tannase from wood-degrading fungus using as substrate plant residues: Purification and characterization. World J Microbiol Biotechnol. 2011;27(10):2325-2333.
Crossref - Mukherjee G, Banerjee R. Effects of temperature, pH and additives on the activity of tannase produced by a co-culture of Rhizopus oryzae and Aspergillus foetidus. World J Microbiol Biotechnol. 2006;22(3).
Crossref - Gayen S, Ghosh U. Purification and characterization of tannin acyl hydrolase produced by mixed solid state fermentation of wheat bran and marigold flower by Penicillium notatum NCIM 923. Biomed Res Int. 2013;2013.
Crossref - Nandi S, Chatterjee A. Extraction , partial purification and application of tannase from Aspergillus niger MTCC 2425. Int J Food Sci Nutr. 2016;1(3):20-23.
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.