ISSN: 0973-7510
E-ISSN: 2581-690X
The current research demonstrates the biotechnological economization of accumulated and inefficiently used agro-industrial orange peel wastes to generate amylase, endoglucanase, exoglucanase, pectinase, and xylanase, industrially essential enzymes with growing demands in enzyme markets, from three Cladosporium isolates. In submerged fermentation (SmF) at 10°C, the isolate AUMC 10865 produced the highest level of amylase (4164 IU/gram dry substrate). Endoglucanase, exoglucanase and xylanase had development peaks (923 IU/gds, 2280 IU/gds, and 1646 IU/gds, respectively in case of Cladosporium sp. AUMC 11366. Pectinase produced the most (7840 IU/gds) in the strain AUMC 11340. At 30°C, the strain AUMC 11340 secretes the most amylase (4120 IU/gds), endoglucanase (2700 IU/gds) and xylanase (3220 IU/gds). Exoglucanase development reached the peak (8750 IU/gds) in the isolate AUMC 10865. The overall production (5570 IU/gds) was instead enhanced by pectinase in the AUMC 11366 isolate. In solid-state fermentation (SSF) at 10°C, the isolate AUMC 10865 outperformed the other two isolates producing 640.0 IU/gds amylase, 763.3 IU/gds endoglucanase, 771.0 IU/gds exoglucanase, 1273.23 IU/gds pectinase and 1062.0 IU/gds xylanase, while the isolate AUMC 11366 produced the least amount of 399.7 IU/gds, 410.0 IU/gds, 413.3 IU/gds, 558.7 IU/gds, and 548.0 IU/gds, respectively. At 30°C, the isolate AUMC 11340 was superiorly producing higher levels of amylase (973.3 IU/gds), endoglucanase (746.0 IU/gds), exoglucanase (1052.0 IU/gds), pectinase (1685.3 IU/gds) and xylanase (1340.0 IU/gds), whereas isolate AUMC 10865 generated the least amounts of amylase (556.7 IU/gds) and exoglucanase (452.7 IU/gfs), and the isolate AUMC 11366 produced the least endoglucanase (256.3 IU/gds), pectinase (857.7 IU/gfs) and xylanase (436.3 IU/gds) amounts.
Agro-industrial, Bioconversion, Cladosporium, Enzymes, Fermentation
Cladosporium is amongst the largest and diverse hyphomycetes, with more than 772 names.1 Cladosporium was previously used to refer to all unrelated dematiaceous hyphomycetes with amero-to-phragmosporous conidia produced in acropetal chains. Cladosporium species are well suited to disperse in vast numbers over long distances, so they are cosmopolitan and commonly found in all forms of plants, and other debris, often isolated from air, soil, crops, grains, fruit, paint, textiles, and other organic matter.1-9 Some species of this genus are plant pathogenic causing leaf spots and other lesions,10 or they occur as hyper parasites on other fungi.11
Active research on converting agricultural by-products into value-added products has been purposed to produce different enzymes of great economic value. Orange juice is now one of the best drinks in the world12,13 and orange is one of Egypt’s important commercial crops.14 The production of orange juice is made with a large percentage of citrus fruits, of which around 50% to 60% is converted into citrus peel wastes,15,16 which leads to the accumulation of large quantities of peel waste in the citrus processing industries. Important quantities have been collected of orange peel waste along with environmental issues to ensure that health risks arising from unsatisfactory treatment practices meet the essential needs for identifying alternative biotechnological waste recycling options.17,18
In accordance with existing law on the environment, any wastes can be used as an ingredient for recycling.19 The use of orange peel as a potentially useful low-cost resource will provide high-quality goods.12,18,20 The literature extensively describes a variety of promising approaches for effective use of orange peel wastes. Consequently, three Cladosporium isolates were used for biosynthesis of some important enzymes in submerged fermentation (SmF) and their enzymatic ability was used through solid-state (SSF) methods in order to bio-convert orange peel wastes as a low-cost substrate into valuable enzymes.
Isolation and Maintenance of Cladosporium Isolates
Three Cladosporium isolates involved in the current study, of which two were isolated from air of Beni Suef and Qena cities and one from grapevine fruits in Sohag city, Egypt. Exposure method21 was employed for isolation of Cladosporium from air and direct plating technique22 for isolation from grapevine fruits. Czapek’s Dox agar was used as an isolation medium. The medium is composed of (g/L): Sucrose, 30; Na2NO3, 2; K2HPO4, 1; KCl, 0.5; MgSO4.7H2O, 0.5; FeSO4, 0.01; ZnSO4, 0.01; CuSO4, 0.005; Rose Bengal, 0.05; chloramphenicol, 0.25; agar, 15 and the final pH 7.3. Interesting isolates obtained were preserved as frozen and lyophilized cultures, as well as on cotton balls23 in the culture collection of Assiut University Mycological Centre as AUMC 10865 (air, Beni-Suef), AUMC 11340 (grapevine fruits, Sohag) and AUMC 11366 (air, Qena).
Molecular Identification of the Cladosporium Isolates
DNA Extraction, PCR and Sequencing of Internal Transcribed Spacer (ITS) Gene
For DNA isolation, a small portion from fungal growth of 7-day-old colonies of Cladosporium isolates AUMC 10865, AUMC 11340 and AUMC 11366 grown on potato dextrose agar (PDA) at 25°C were collected and transferred individually to 2 ml-Eppendorf tube. The DNA extraction was performed following CTAB method described in Moubasher et al24. The universal primers ITS1 and ITS425 were used for amplification of the internal transcribed spacer (ITS) region.26,9
Alignment and Phylogenetic Analysis
The sequences of Cladosporium isolates in this study were compared to the closely related sequences in GenBank as well as sequences of the type and ex-type Cladosporium species. Sequences of Cladosporium isolates in this study and those retrieved from GenBank were aligned together using MAFFT (version 6.861b) with the default options.27 Alignment gaps and parsimony uninformative characters were cleaned using BMGE.28 Maximum-likelihood (ML) and Maximum parsimony (MP) phylogenetic analyses were performed using PhyML 3.0.29 The robustness of the most parsimonious trees was evaluated by 100 bootstrap replications.30,31 The best optimal model of nucleotide substitution for the ML analyses was determined using Smart Model Selection (SMS) version 1.8.1.32 The phylogenetic tree was drawn using MEGA X,33 and edited using Microsoft Power Point (2016) and saved as TIF file.9
Assessment of Enzymatic Potential in Submerged Fermentation (SmF)
Cladosporium AUMC 10865, AUMC 11340 and AUMC 11366 were cultivated, separately in 250 ml Erlenmeyer conical flasks containing 50 ml sucrose-free Czapek’s broth medium supplemented with 1% soluble starch (for amylase production) or 1% pectin (for pectinase) or 1% xylan (for endoglucanase, exoglucanase and xylanase production). The flasks were inoculated, individually with 1 ml containing 1.8 x 108 spore/ml of spore suspension obtained from 7-day-old cultures of the Cladosporium isolates tested. The flasks were then incubated for 7 days at 10°C and 30°C in shacking condition at 120 rpm. After incubation, the fermented media were filtered through Whatman No.1 filter paper and the cell-free supernatants obtained by centrifugation (10000 xg at 4°C for 10 min) were used as enzyme sources.
Enzymes Assay and Protein Estimation
Starch, carboxymethyl cellulose (CMC), microcrystalline cellulose (avicel), pectin, and oat spelts xylan were used for assays of amylase, endoglucanase, exoglucanase, pectinase and xylanase respectively. The reaction mixture consists of 0.5 ml of the filtered crude enzyme and 0.5 ml of previously-mentioned substrates at a concentration of 1% (prepared in 50 mM Na-citrate buffer, pH 5.0). The reaction was carried out in water bath at 50°C for 20 min. Afterwards, the process was stopped by introducing 2 ml of 3, 5-dinitrosalicylic acid (DNS) and the tube contents were placed in a boiling water bath for 10 min.34 After cooling, the color absorbance was measured at 540 nm using UV-Visible spectrophotometer (T80+; UK). The amount of reducing sugar liberated was quantified using standard curves of glucose (for amylase, endoglucanase, exoglucanase, and pectinase), or xylose (for xylanase). One unit of the enzyme is defined as the amount of enzyme that liberates 1 µmol of the reducing sugar (glucose or xylose) equivalent per minute under the standard assay conditions.35 Total protein content was measured by using bovine serum albumin (BSA) as the standard.36 Calculations of enzyme activities were done.37,38
Bioconversion of Orange Peel Wastes into Cocktail Enzyme
Determination of Moisture Content (MC) of Orange Peels
Twenty grams of orange peels were weighed and dried in an oven at 105°C over-night, then cooled in a desiccator for 30 minutes and reweighed. The moisture content was calculated as follows:
% MC = [wt.of wet sample-wt.of dry sample / wt.of wet sample ] x 100
Solid-State Fermentation
Cladosporium isolates were grown separately in Erlenmeyer flasks containing 10 g of fresh orange peel wastes and 10 mL of fermentation medium. The fermentation medium contained (g/L): pectin from citrus peel, 1.0; sodium nitrate, 2.0; magnesium sulphate, 0.5; di-potassium hydrogen orthophosphate, 1.0; potassium chloride, 0.5; zinc sulphate, 0.01; and copper sulphate, 0.005. After autoclaving at 121°C for 20 minutes, each flask was inoculated with 1% spore suspension obtained from a 7-day-old cultures of the Cladosporium isolates. Two sets of flasks were incubated in stagnant conditions at 10°C and 30°C for 10 days. Following the incubation period, the contents of each flask were extracted with 100 ml of 50 mM citrate buffer (pH 5.0), and cell-free supernatants were obtained through centrifugation at 10,000 xg for 10 min. The transparent supernatant was used as a source of cocktail enzymes. As previously stated, the amount of reducing sugar liberated, as well as enzyme activity and soluble protein were calculated.
Phylogenetic Analyses
The ITS sequences obtained from 32 strains produced 566 characters of which 494 were complete characters (no gaps, no N), 114 variable characters (23.1% of the complete characters) and 29 characters were parsimony informative (5.9% of complete). Tamura-Nei (TN93) was the best nucleotide substitution model. The dataset for maximal parsimony yielded 2 best parsimony trees with a tree length of 136 steps. The strongest ML tree score with a final ML probability optimization value of -1662.15949 and a tree size of 0.40191 was monitored to explain and explore the phylogenetic relationship between taxa (Fig. 1). The ITS sequences of Cladosporium isolates AUMC 10865, AUMC 11340, and AUMC 11366 from this study were uploaded to GenBank as MN826828, MN826919 and MN826822, respectively.
Fig. 1. Phylogenetic tree generated from MP analysis based on alignment of ITS sequences of Cladosporium isolates AUMC 10865, AUMC 11340 and AUMC 11366 with the most similar sequences belonging to Cladosporium in GenBank database. Sequences from this study are in blue color. Bootstrap support values (100 replications) for ML/MP combination ≥50% are indicated at the respective nodes. The tree was rooted to Cercospora beticola CBS 116456 as outgroup.
Enzymatic Capability of Cladosporium Isolates in SmF
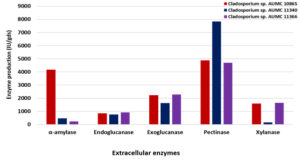
Using SmF at 10°C and 30°C, the three Cladosporium isolates could develop amylase, endoglucanase, exoglucanase, chitinase, pectinase and xylanase enzymes in varied levels of production and activity. At 10°C, the isolate AUMC 10865 produced the most amylase (4164 IU/gds) and had the highest specific activity (62.5 IU/mg). Endoglucanase and exoglucanase had the development peak (923 IU/gds and 2280 IU/gds, respectively) in the strain AUMC 11366, while both enzymes had the highest specific activity (5.85 IU/mg and 12.43 IU/mg, respectively) in the strain AUMC 11340. Pectinase produced the most (7840 IU/gds) in the strain AUMC 11340 and had the highest specific activity (8.0 IU/mg) in the strain AUMC 10865. Xylanase reached the maximum production (1646 IU/gds) and specific activity (6.065 IU/mg) in the strain AUMC 11366 (Table 1; Fig. 2).
Table (1):
Production and specific activity of amylase, endoglucanase, exoglucanase, pectinase and xylanase enzymes produced by Cladosporium isolates AUMC 10865, AUMC 11340 and AUMC 11366 in SmF at 10°C.
| Extracellular enzymes | Cladosporium sp. AUMC 10865 | Cladosporium sp. AUMC 11340 | Cladosporium sp. AUMC 11366 | |||
|---|---|---|---|---|---|---|
| Production IU/gds |
Specific activity IU/mg |
Production IU/gds |
Specific activity IU/mg |
Production IU/gds |
Specific activity IU/mg |
|
| α-amylase | 4164 | 62.5 | 458 | 0.375 | 235 | 3.2 |
| Endoglucanase | 846 | 2.0 | 760 | 5.85 | 923 | 4.053 |
| Exoglucanase | 2220 | 5.4 | 1620 | 12.43 | 2280 | 10.9 |
| Pectinase | 4870 | 8.0 | 7840 | 1.78 | 4690 | 7.52 |
| Xylanase | 1600 | 3.26 | 150 | 0.971 | 1646 | 6.065 |
Fig. 2. Enzymes production (IU/gds) by Cladosporium isolates AUMC 10865, AUMC 11340 and AUMC 11366 at 10°C in SmF.
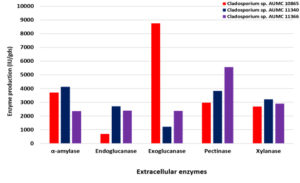
On the other hand, using SmF at 30°C, the strain AUMC 11340 secretes the most amylase (4120 IU/gds), endoglucanase (2700 IU/gds) and xylanase (3220 IU/gds) with the peak of specific activity of endoglucanase (27.8 IU/mg) and xylanase (28.1 IU/mg), while the maximum specific activity of amylase (43.7 IU/mg) was recorded in the strain AUMC 10865. Exoglucanase development reached the peak (8750 IU/gds) in the strain AUMC 10865 and specific activity (16.8 IU/mg) in the strain AUMC 11366. The overall production (2860 IU/gds and 5570 IU/gds) was instead enhanced by chitinase and pectinase in the AUMC 11366 strain with the specific activity maximum (28.4 IU/mg and 62.0 IU/mg, respectively) in the AUMC 10865 strain (Table 2; Fig. 3).
Table (2):
Production and specific activity of amylase, endoglucanase, exoglucanase, pectinase and xylanase enzymes produced by Cladosporium isolates AUMC 10865, AUMC 11340 and AUMC 11366 in SmF at 30°C.
| Extracellular enzymes | Cladosporium sp. AUMC 10865 | Cladosporium sp. AUMC 11340 | Cladosporium sp. AUMC 11366 | |||
|---|---|---|---|---|---|---|
| Production IU/gds |
Specific activity IU/mg |
Production IU/gds |
Specific activity IU/mg |
Production IU/gds |
Specific activity IU/mg |
|
| α-amylase | 3700 | 43.7 | 4120 | 11.9 | 2350 | 6.34 |
| Endoglucanase | 700 | 1.63 | 2700 | 27.8 | 2400 | 17.0 |
| Exoglucanase | 8750 | 1.86 | 1220 | 12.53 | 2370 | 16.8 |
| Pectinase | 2970 | 62.0 | 3820 | 10.64 | 5570 | 2.64 |
| Xylanase | 2690 | 5.32 | 3220 | 28.1 | 2900 | 17.34 |
Fig. 3. Enzymes production (IU/gds) by Cladosporium isolates AUMC 10865, AUMC 11340 and AUMC 11366 at 30°C in SmF.
Bioconversion of Orange peel wastes into Cocktail Enzymes in SSF
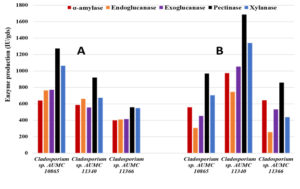
The moisture content of the orange peel wastes was found to be 70%. Based on the MC percent, the enzymatic activity was calculated as IU/gram dry substrate (IU/gds). In SSF at 10°C, the isolate AUMC 10865 outperformed the other two isolates producing 640.0 IU/gds amylase, 763.3 IU/gds endoglucanase, 771.0 IU/gds exoglucanase, 1273.23 IU/gds pectinase and 1062.0 IU/gds xylanase, while the isolate AUMC 11366 produced the least amount of 399.7 IU/gds, 410.0 IU/gds, 413.3 IU/gds, 558.7 IU/gds, and 548.0 IU/gds, respectively (Table 3; Fig. 4).
Table (3):
Production and specific activity of amylase, endoglucanase, exoglucanase, pectinase and xylanase enzymes produced by Cladosporium isolates AUMC 10865, AUMC 11340 and AUMC 11366 from untreated orange peels in SSF at 10 °C.
| Extracellular enzymes | Cladosporium sp. AUMC 10865 | Cladosporium sp. AUMC 11340 | Cladosporium sp. AUMC 11366 | |||
|---|---|---|---|---|---|---|
| Production IU/gds |
Specific activity IU/mg |
Production IU/gds |
Specific activity IU/mg |
Production IU/gds |
Specific activity IU/mg |
|
| Amylase | 640.0 | 1.45 | 586.7 | 0.766 | 399.7 | 0.788 |
| Endoglucanase | 763.3 | 1.73 | 661.3 | 0.862 | 410.0 | 0.8 |
| Exoglucanase | 771.0 | 1.75 | 555.3 | 0.724 | 413.3 | 0.814 |
| Pectinase | 1273.23 | 2.88 | 919.7 | 1.2 | 558.7 | 1.1 |
| Xylanase | 1062.0 | 2.0 | 672.0 | 0.87 | 548.0 | 0.9 |
Fig. 4. Enzyme production by Cladosporium isolates AUMC 10865, AUMC 11340 and AUMC 11366 from fresh orange peels in SSF at 10°C (A) and 30°C (B).
On the other hand, at 30°C the isolate AUMC 11340 was superiorly producing higher levels of amylase (973.3 IU/gds), endoglucanase (746.0 IU/gds), exoglucanase (1052.0 IU/gds), pectinase (1685.3 IU/gds) and xylanase (1340.0 IU/gds), whereas isolate AUMC 10865 generated the least amounts of amylase (556.7 IU/gds) and exoglucanase (452.7 IU/gfs), and the isolate AUMC 11366 produced the least endoglucanase (256.3 IU/gds), pectinase (857.7 IU/gfs) and xylanase (436.3 IU/gds) amounts (Table 4; Fig. 4).
Table (4):
Production and specific activity of amylase, endoglucanase, exoglucanase, pectinase and xylanase enzymes produced by Cladosporium isolates AUMC 10865, AUMC 11340 and AUMC 11366 from untreated orange peels in SSF at 30°C.
| Extracellular enzymes | Cladosporium sp. AUMC 10865 | Cladosporium sp. AUMC 11340 | Cladosporium sp. AUMC 11366 | |||
|---|---|---|---|---|---|---|
| Production IU/gds |
Specific activity IU/mg |
Production IU/gds |
Specific activity IU/mg |
Production IU/gds |
Specific activity IU/mg |
|
| Amylase | 556.7 | 1.264 | 973.3 | 1.268 | 642.7 | 1.267 |
| Endoglucanase | 306.0 | 1.386 | 746.0 | 2.73 | 256.3 | 1.0 |
| Exoglucanase | 452.7 | 2.0546 | 1052.0 | 2.74 | 532.3 | 2.1 |
| Pectinase | 969.0 | 4.4 | 1685.3 | 4.385 | 857.7 | 3.38 |
| Xylanase | 703.3 | 2.66 | 1340.0 | 2.89 | 436.3 | 1.43 |
All living systems contain enzymes, which act as biological catalysts.39,40 Enzymes are proteins in origin and catalyse a wide range of biological processes. Enzymes have been employed in several industries such as wine, cheese, bread, beer, vinegar, leather and linen.41,42 In this study, three Cladosporium species were isolated from air and grapevine fruits. It was determined that the three isolates could not be identified based just on ITS sequencing, and that they may require sequencing of other genes such as actin (ACT), elongation factor 1-alpha (EF1), and large subunit (LSU) in order to be positively identified.
The enzymatic activity of the three Cladosporium species was assessed. With the aid of SmF, all isolates were able to produce different quantities of amylase, endoglucanase, exoglucanase, pectinase, and xylanase enzymes. In this context, numerous studies have shown that Cladosporium species can generate a variety of enzymes, including glucoamylase by Cladosporium gossypiicola,43 laccase, cellulases, and hemicellulases by C. cladosporioides,44,45 alpha-amylase, cellulase, and xylanase from C. cladosporioides,46 and amylase, endoglucanase, exoglucanase and xylanase by C. allicinum AUMC 14511.9
Cladosporium isolates in this study were used in SSF to convert orange peel wastes into valuable extracellular enzymes by fermenting the wastes. At 10°C, the isolate AUMC 10865 was superior to the other two isolates in the ability to produce the five extracellular enzymes from orange peels in SSF, while the isolate AUMC 11340 was the strongest at 30°C for all enzymes assayed. Orange peel waste has been shown to contain 16.9% soluble sugar, 9.21% cellulose, 10.5% hemicellulose, and 42.5% pectin,18 which may be the explanation for the high enzyme production utilizing orange peels.
Microorganisms that have evolved to cold temperatures have a lot of promise in biotechnological applications, such as waste treatment and bioremediation at lower temperatures, as well as the textile and food sectors.47-49 Cold-adaptive enzymes produced by psychrophilic microorganisms such as Cladosporium species have significant goals in bioconversion processes because of their high activity at low and moderate temperatures, they have the potential to economize processes by saving energy, and thus offer potential economic and environmental benefits such as reducing heat-sensitive substrate alteration and the production of harmful by-products due to mild industrial conditions at low temperatures, and using psychrophilic enzymes. The aforementioned will make industrial operations more easy and safe, as in the case of conventional industry.48,50
Using Cladosporium species as enzyme developers, the current work demonstrated a low-cost substrate that does not require pretreatment. The expense of enzymes in industrial processes is a major problem. In addition, the usage of enzymes in diverse industrial processes indirectly influences the final product’s price. Plant equipment and installation expenses account for a substantial percentage of an enzyme manufacturing facility’s annual operating costs.51
As a means of producing industrially significant enzymes, SSF offers numerous economic benefits. For the synthesis of fungal enzymes52 and53 have indicated that the SSF method is the most suited due to its high productivity, ease of enzyme recovery, and cost efficiency. Due to their cheap cost of collection, agro-industrial wastes are not only inexpensive solid substrates, but they are also ideal for extracellular enzyme synthesis.54 The current study, on the other hand, revealed that SmF was the best for the synthesis of all enzymes examined. The increased productivity of submerged fermentation over solid state fermentation may be ascribed to more substrate availability in SmF due to improved aeration, and the surface of the substrate particles is routinely more susceptible to enzyme action than in solid state. Furthermore, the heat transmission and homogeneity of the submerged fermentation media make it preferable to the non-homogeneous solid state.
As part of this study, three Cladosporium isolates from air and grapevine fruits in Egypt were evaluated for their enzyme activity, as well as the bioconversion of orange peel wastes as a low-cost substrate for SSF’s extracellular enzymes. Amylase, endoglucanase, exoglucanase, pectinase and xylanase were all produced at varying quantities by the isolates when grown in SmF and SSF.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest
AUTHORS’ CONTRIBUTION
AMM and AAZ conceived this project. MAM isolated the fungi. AEH and OAMSA-B performed phylogenetic analysis. OAMSA-B and MAM performed the enzymatic activity and bioconversion experiment. All authors have read and agreed to the published version of the manuscript.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Bensch K, Braun U, Groenewald JZ, Crous PW. The genus Cladosporium. Studies in Mycology. 2012;72:1-401.
Crossref - Ellis MB. More dematiaceous hyphomycetes. Commonwealth Mycological Institute, Kew, Surrey, England. 1976;507.
- Yehia RS, Osman GH, Assaggaf H, Salem R, Mohamed MS. Isolation of potential antimicrobial metabolites from endophytic fungus Cladosporium cladosporioides from endemic plant Zygophyllum mandavillei. South African Journal of Botany. 2020;134:296-302.
Crossref - Moubasher A. Soil fungi in Qatar and other Arab countries. The Centre for Scientific and Applied Research, University of Qatar; 1993.
- Ismail M, Moubasher A, Mohamed R, Al-Bedak O. Extremophilic fungi and chemical analysis of hypersaline, alkaline lakes of Wadi-El-Natrun, Egypt. International Journal of Technical Research and Science. 2017;1(10):345-363.
- Farr DF, Bills GF. Chamuris GP, Rossman AY. Fungi on plants and plant products in the United States. APS press; 1989.
Crossref - Flannigan B, Samson RA, Miller JD. Microorganisms in home and indoorwork environments: diversity, health impacts,investigation and control. 1st edition, CRC Press; 2002.
- Flannigan B, Samson RA, Miller JD. Microorganisms in home and indoor work environments: diversity, health impacts, investigation and control. CRC Press; 2011.
Crossref - Al-Bedak OA, Teama EA, Ali E, Said M, Shalaby E, Moharram ZA. Impact of fumigation with phosphine on viability of wheat grains stored for six months at two levels of moisture content, in addition to description of four new records of associated fungi and assessment of their potential for enzymatic production. Journal of Basic & Applied Mycology (Egypt). 2020;11:77-97.
- Schubert K, Braun U. Taxonomic revision of the genus Cladosporium s. lat. 2. Cladosporium species occurring on hosts of the families Bignoniaceae and Orchidaceae. Sydowia. 2004;56(2):296-317.
- Heuchert B, Braun U, Schubert K. Morphotaxonomic revision of fungicolous Cladosporium species (hyphomycetes). Schlechtendalia. 2005;13:1-78.
- Martin M, Siles J, Chica A, Martin A. Biomethanization of orange peel waste. Bioresource Technology. 2010;101(23):8993-8999.
Crossref - Embaby AM, Masoud AA, Marey HS, Shaban NZ, Ghonaim TM. Raw agro-industrial orange peel waste as a low cost effective inducer for alkaline polygalacturonase production from Bacillus licheniformis SHG10. Springer Plus. 2014;3(1):327.
Crossref - Mohamed SA, Drees EA, El-Badry MO, Fahmy AS. Biochemical Properties of α-Amylase from Peel of Citrus sinensis cv. Abosora. Appl Biochem Biotechnol. 2010;160(7):2054-2065.
Crossref - Wilkins MR, Suryawati L, Maness NO, Chrz D. Ethanol production by Saccharomyces cerevisiae and Kluyveromyces marxianus in the presence of orange-peel oil. World J Microbiol Biotechnol. 2007;23(8):1161-1168.
Crossref - Li P-J, Xia J-L, Shan Y, et al. Optimizing production of pectinase from orange peel by Penicillium oxalicum PJ02 using response surface methodology. Waste and Biomass Valorization. 2015;6(1):13-22.
Crossref - Martin M, Fernandez R, Serrano A, Siles J. Semi-continuous anaerobic co-digestion of orange peel waste and residual glycerol derived from biodiesel manufacturing. Waste Management. 2013;33(7):1633-1639.
Crossref - Rivas B, Torrado A, Torre P, Converti A, Dominguez JM. Submerged citric acid fermentation on orange peel autohydrolysate. J Agric Food Chem. 2008;56(7):2380-2387.
Crossref - Moller M, Schmitz P, Thiele H, Wronka T. Economically and ecologically integrated valuation of land-use in less favoured areas. Ber Landwirtsch. 2001;79(1):19-48.
- Balu AM, Budarin V, Shuttleworth PS, et al. Valorisation of orange peel residues: waste to biochemicals and nanoporous materials. Chem Sus Chem. 2012;5(9):1694-1697.
Crossref - Flannigan B. Air sampling for fungi in indoor environments. Journal of Aerosol Science. 1997;28(3):381-392.
Crossref - Berry ED, Wells JE. A direct plating method for estimating populations of Escherichia coli O157 in bovine manure and manure-based materials. Journal of Food Protection. 2008;71(11):2233-2238.
Crossref - Al-Bedak OA, Sayed RM, Hassan SH. A new low-cost method for long-term preservation of filamentous fungi. Biocatal Agric Biotechnol. 2019;22:101417.
Crossref - Moubasher A, Ismail M, Al-Bedak O, Mohamed R. Ramophialophora chlamydospora, a new species from an alkaline lake of Wadi-El-Natron, Egypt. Asian J Mycol. 2019;2(1):110-117.
Crossref - White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications. 1990;18(1):315-322
Crossref - Al-Bedak O, Moubasher A. Aspergillus gaarensis, a new addition to section Circumdati from soil of Lake El-Gaar in Wadi-El-Natron, Egypt. Studies in Fungi. 2020;5(1):59-65.
Crossref - Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772-780.
Crossref - Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10(1):210.
Crossref - Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology. 2010;59(3):307-321.
Crossref - Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783-791.
Crossref - Hesham AEL. New safety and rapid method for extraction of genomic DNA from bacteria and yeast strains suitable for PCR amplifications. J Pure Appl Microbiol. 2014;8(1):383-388.
- Lefort V, Longueville J-E, Gascuel O. SMS: smart model selection in PhyML. Mol Biol Evol. 2017;34(9):2422-2424.
Crossref - Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547-1549.
Crossref - Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical Chemistry. 1959;31(3):426-428.
Crossref - Ghose T, Bisaria VS. Measurement of hemicellulase activities: Part I Xylanases. Pure Appl Chem. 1987;59(12):1739-1751.
Crossref - Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. Journal of Biological Chemistry. 1951;193:265-275.
Crossref - Moubasher A, Ismail M, Mohamed RA, Al-Bedak O. Xylanase and cellulase production under extreme conditions in submerged fermentation by some fungi isolated from hypersaline, alkaline lakes of Wadi-El-Natrun, Egypt. J Basic Appl Mycol. 2016;7:19-32.
- AL-Kolaibe AM, Moharram AM, Al-Bedak OA. Worthwhile enzyme production and eco-friendly bioconversion of three agricultural residues by Aspergillus curvatus and Aspergillus gaarensis, promising enzyme-producers isolated from extreme environment. Journal of Basic & Applied Mycology (Egypt). 2021;12:1-14.
- Hesham AEL, Alrumman SA, Al-Dayel MA, Salah HA. Screening and genetic identification of acidic and neutral protease-producing yeasts strains by 26S rRNA gene sequencing. Cytology and Genetics. 2017;51(3):221-229.
Crossref - Salah HA, Temerk HA, Salah NA, et al. Production and Optimization of Xylanase and α-Amylase from Non-Saccharomyces Yeasts (Pichia membranifaciens). J Pure Appl Microbiol. 2021;15(1):452-461.
Crossref - Ravindran R, Jaiswal AK. Microbial enzyme production using lignocellulosic food industry wastes as feedstock: a review. Bioengineering. 2016;3(4):30.
Crossref - Adrio JL, Demain AL. Microbial enzymes: tools for biotechnological processes. Biomolecules. 2014;4(1):117-139.
Crossref - Quigley T, Kelly C, Doyle E, Fogarty W. Patterns of raw starch digestion by the glucoamylase of Cladosporium gossypiicola ATCC 38026. Process Biochemistry. 1998;33(6):677-681.
Crossref - Aslam MS, Aishy A, Samra ZQ, Gull I, Athar MA. Identification, purification and characterization of a novel extracellular laccase from Cladosporium cladosporioides. Biotechnol Biotechnol Equip. 2012;26(6):3345-3350.
Crossref - Halaburgi VM, Sharma S, Sinha M, Singh TP, Karegoudar TB. Purification and characterization of a thermostable laccase from the ascomycetes Cladosporium cladosporioides and its applications. Process Biochemistry. 2011;46(5):1146-1152.
Crossref - Mushimiyimana I. A statistical strategy for the production of cellulase, xylanase and α-amylase by Cladosporium cladosporioides. Fungal Territory. 2019;2(2):16-21.
Crossref - Ji L, Yang J, Fan H, et al. Synergy of crude enzyme cocktail from cold-adapted Cladosporium cladosporioides Ch2-2 with commercial xylanase achieving high sugars yield at low cost. Biotechnology for Biofuels. 2014;7(1):130.
Crossref - Kasana RC, Gulati A. Cellulases from psychrophilic microorganisms: a review. J Basic Microbiol. 2011;51(6):572-579.
Crossref - Margesin R, Schinner F. Properties of cold-adapted microorganisms and their potential role in biotechnology. J Biotechnol. 1994;33(1):1-14.
Crossref - Hamid B, Rana RS, Chauhan D, et al. Psychrophilic yeasts and their biotechnological applications-a review. African J Biotechnol. 2014;13(22):2188-2197.
Crossref - Klein-Marcuschamer D, Oleskowicz-Popiel P, Simmons BA, Blanch HW. The challenge of enzyme cost in the production of lignocellulosic biofuels. Biotechnology and Bioengineering. 2012;109(4):1083-1087.
Crossref - Pandey A, Soccol CR, Nigam P, Soccol VT. Biotechnological potential of agro-industrial residues. I: sugarcane bagasse. Bioresource Technology. 2000;74(1):69-80.
Crossref - Viniegra-Gonzalez G, Favela-Torres E, Aguilar CN, de Jesus Romero-Gomez S, Dıaz-Godınez G, Augur C. Advantages of fungal enzyme production in solid state over liquid fermentation systems. Biochemical Engineering Journal. 2003;13(2-3):157-167.
Crossref - Mahmoodi M, Najafpour G, Mohammadi M. Bioconversion of agroindustrial wastes to pectinases enzyme via solid state fermentation in trays and rotating drum bioreactors. Biocatal Agric Biotechnol. 2019;21:101280.
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.