ISSN: 0973-7510
E-ISSN: 2581-690X
Methicillin-Resistant Staphylococcus aureus (MRSA) is an important cause of healthcare associated infections globally. New mecA homologue (mecC), was first reported in the UK and Denmark. The mecC mediated MRSA is resistant only to Β-lactams antibiotics and is sensitive to other antibiotics. Detecting the prevalence of mecC MRSA provides more options in treatment of MRSA infections. The aim of this study was to prevalence of mecC gene in clinical isolates of MRSA in Ain-Shams university hospitals & to correlate Minimal Inhibitory concentration (MIC) of Oxacillin with the mecC gene expression in MRSA isolates. Fifty MRSA isolates were collected from different intensive care units (ICUs) of Ain-Shams university hospital from April-December 2018. Methicillin resistance was detected by Cefoxitin disc, and antimicrobial susceptibility testing was done for all isolates and its results were interpreted according to Clinical & Laboratory Standards Institute (CLSI) guidelines 2018. Minimal Inhibitory Concentration of Oxacillin was detected using Oxacillin E-test and the results were interpreted according to the manufacturer’s instructions, then Polymerase Chain Reaction was done to detect mecA and mecC genes among MRSA isolates. Fifty isolates were identified as MRSA by Cefoxitin disc out of 163 samples. Twelve isolates were sensitive to Oxacillin while 38 isolates were resistant to Oxacillin. All isolates were positive to mecA gene while only 3 isolates were positive to both mecA and mecC genes. MecC is a new emerging gene responsible for methicillin resistance in staphylococci and was detected in 6 % of the isolates in this study.
MRSA, mecC gene, mecA gene, PCR
Methicillin-Resistant Staphylococcus aureus (MRSA) is considered as one of the most common causes of community and hospital-acquired infections, leading to high morbidity and mortality. MRSA poses a serious problem in hospitals and its detection is crucial for infection prevention and control1.
Resistance to beta-lactams antibiotics is conferred by the presence of mecA gene, which encodes a penicillin-binding protein (PBP2’). mecA is part of a mobile genetic element called the “Staphylococcal cassette chromosome (SCC) mec.”2
The report of MRSA carrying a new variant of the mecA gene in 2011 in humans was highly significant. This new variant was named mecC gene. The presence of this gene poses diagnostic problems due to the probability of misdiagnosis of isolates as methicillin-sensitive S. aureus which may affect the results of MRSA surveillance3.
The mecC gene shared only 70% DNA identity with the mecA gene. MRSA isolates carrying mecC gene have been reported in different European countries and from several host species4.
This mecC MRSA shows a characteristic antimicrobial susceptibility pattern compared to mecA MRSA. Where mecA MRSA displays resistance to both Oxacillin and Cefoxitin while most of mecC MRSA shows resistance to Cefoxitin and susceptibility to Oxacillin5.
There is limited available data about the prevalence of MRSA that carry mecC gene in Egypt, hence, this study was performed.
Collection of clinical samples
Different clinical samples were obtained from patients admitted to different ICUs of Ain-Shams university hospitals according to the regulations of scientific research Ethical Committee Faculty of Medicine-Ain Shams University during the period from April 2018 to December 2018. The collected specimens include blood, pus, sputum and swabs from burn and surgical wound.
Identification and antimicrobial susceptibility testing of the isolated organism
Sample collection and identification of the isolated organism were performed by conventional bacteriological methods according to Colle et al, 19966 and Cheesbrough, 20097.
For identification of Staphylococcus aureus isolates, the samples were inoculated on Blood agar and Mannitol salt agar, the colonies were identified based on morphology of the isolated colonies, microscopic examination of Gram-stained films, the β-hemolytic effect on blood agar and yellow colonies on mannitol Salt agar.
Then detection of MRSA strains was done by Cefoxitin discs 30µg (FOX) supplied by (Oxoid, Basingstoke, UK) using Kirbey-Bauer disc diffusion method and the results were interpreted according to CLSI guidelines 20188.
Antimicrobial susceptibility testing for the following antibiotics was done using Muller Hinton agar: Ciprofloxacin 5µg (CIP), Cefoxitin 30µg (FOX), Chloramphenicol 30µg (C), Penicillin 0.6µg (P), Linezolid 10µg (LZO), Rifampicin 5µg (RA), Doxycycline 30µg (DO), Erythromycin 15µg (E), Clindamycin 2µg (DA), Levofloxacin 5µg (LEV), Gentamycin 10µg (CN), Trimethoprim/ sulfamethoxazole 1.25 + 23.75µg (SXT), all antibiotic discs were supplied by (Oxoid, Basingstoke, UK), and the results were interpreted according to CLSI 20188.
Detection of oxacillin minimal inhibitory concentration (MIC)
MIC was done for the Fifty MRSA isolates by using Oxacillin E-test supplied by LIOFILCHEM company, and the interpretation was done according to CLSI guidelines,2018.
Detection of mecA gene and mecC gene by Polymerase chain reaction (PCR)
All MRSA strains were subjected to PCR for detection of mecA and mecC gene, first DNA extraction was done using ultraclean microbial DNA extraction kit supplied by MoBio by QIAGEN, then amplification of mecA gene9 was performed using the primers forward 5’-AAA ATC GAT GGT AAA GGT TGGC-3’ and reverse 5’-AGT TCT GCA GTA CCG GAT TTG C-3’ and for mecC gene4 forward 5′-GAA AAA AAG GCT TAG AAC GCC TC-3′ reverse 5′ GAA GAT CTT TTC CGT TTT CAG C-3′ .
PCR conditions for mecA gene9
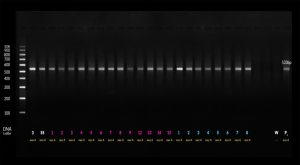
Thermal cycler was adjusted to 2 minutes at 95°C for primary denaturation followed by 40 cycles of 45 seconds denaturation at 95°C then 45 seconds at 50°C for annealing, and 1 min at 72°C for extension. The amplified product was identified at 530 base pair (bp) by electrophoresis on 1.5% agarose gels and using 100 bp DNA ladder.
PCR conditions for mecC gene4
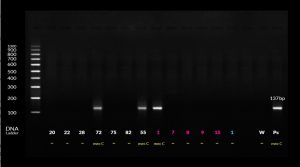
Thermal cycler was adjusted to 2 minutes at 95°C for primary denaturation followed by 40 cycles of 45 seconds denaturation at 95°C then 45 seconds at 60°C for annealing, and 1 min at 72°C for extension. The amplified product was identified at 137 bp by electrophoresis on 1.5% agarose gels and using 100 bp DNA ladder (Fig. 1 & 2).
Data analysis
All statistical procedures were carried out using SPSS version 22.0 for Windows (SPSS Inc, Chicago, IL, USA).
In this work, different clinical samples were collected from 163 patients admitted to different ICUs. From these samples, fifty isolates were identified as MRSA in which 25 isolates were collected from males (50%) and 25 from females (50%). Their age ranged from 18-75, and the mean age was 49.6 ± 13.1 years and all patients were under antibiotic therapy.
Most of MRSA isolates were obtained from blood samples (25 isolates (50%)) followed by 9 (18%) sputum and 9 (18%) swabs from surgical and burn wounds and 7(14%) pus samples.
The results of antimicrobial susceptibility of MRSA revealed that the highest sensitivity values were found with Trimethoprim/ sulfamethoxazole (SXT), linezolid (LZD), and Chloramphenicol (C) (68% for each antimicrobial agent) while lower sensitivity (66%) was found with Rifampin (RA) followed by Levofloxacin (LEV) (62%). Sensitivity values dropped to 38% with Clindamycin (DA) followed by Doxycycline (DO) and Ciprofloxacin (CIP) with sensitivity in 36% of the isolates. The highest resistance pattern was to Erythromycin (E) and Penicillin (P) as 41 isolates (82%) were resistant to erythromycin and 48 isolates (98%) were resistant to Penicillin (P) as shown in Table (1).
Table (1):
Frequencies (n) and percentages (%) for antimicrobial susceptibility of MRSA arranged from highest to lowest sensitivity.
| Antimicrobial agent | Sensitive | Intermediate | Resistant | |||
|---|---|---|---|---|---|---|
| n | % | N | % | N | % | |
| Trimethoprim/ sulfamethoxazole | 34 | 68 | 6 | 12 | 10 | 20 |
| linezolid | 34 | 68 | 1 | 2 | 15 | 30 |
| Chloramphenicol | 34 | 68 | 1 | 2 | 15 | 30 |
| Rifampin | 33 | 66 | 2 | 4 | 15 | 30 |
| Levofloxacin | 31 | 62 | 1 | 2 | 18 | 36 |
| Clindamycin | 19 | 38 | 15 | 30 | 16 | 32 |
| Doxycycline | 18 | 36 | 6 | 12 | 26 | 52 |
| Ciprofloxacin | 18 | 36 | 17 | 34 | 15 | 30 |
| Gentamycin | 16 | 32 | 4 | 8 | 30 | 60 |
| Erythromycin | 9 | 18 | 14 | 28 | 27 | 54 |
| Penicillin | 2 | 4 | 0 | 0 | 48 | 96 |
| Cefoxitin | 0 | 0 | 0 | 0 | 50 | 100 |
Minimal inhibitory concentration (MIC) of MRSA isolates to Oxacillin was done by using E Test and 12 isolates (24%) were sensitive to Oxacillin while there were 38 isolates (76%) resistant to Oxacillin.
As regards PCR results of determining the presesnce of mecA and mecC genes, all isolates (100%) were positive to mecA gene while only 3 isolates (6%) were positive to both mecA and mecC genes and the isolates that carrying both genes were isolated from blood samples
The three isolates carrying both mecA and mecC, two of them were resistant to Oxacillin E-test and one was sensitive, while 47 MRSA isolates carrying only mecA gene, 11 (92%) were sensitive to Oxacillin E-test and 36 (95%) were resistant to Oxacillin E-test.
The results of antimicrobial susceptibility of MRSA carrying both mecA and mecC genes revealed that the highest sensitivity values were found with (CIP), (SXT), (LZD) and (C) agents (66.7% for each antimicrobial agent). Lower sensitivity was found with (LEV), (RA) and (DA) which was 33.3% for each. There was no sensitivity towards other antimicrobial agents as shown in Table (2).
Table (2):
Frequencies (n) and percentages (%) for antimicrobial susceptibility of MRSA in mecC positive cases (n = 3) arranged from highest to lowest sensitivity.
| Antimicrobial agent | Sensitive | Intermediate | Resistant | |||
|---|---|---|---|---|---|---|
| n | % | n | % | n | % | |
| Ciprofloxacin | 2 | 66.7 | 0 | 0 | 1 | 33.3 |
| Trimethoprim/ sulfamethoxazole | 2 | 66.7 | 0 | 0 | 1 | 33.3 |
| linezolid | 2 | 66.7 | 0 | 0 | 1 | 33.3 |
| Chloramphenicol | 2 | 66.7 | 0 | 0 | 1 | 33.3 |
| Levofloxacin | 1 | 33.3 | 0 | 0 | 2 | 66.7 |
| Rifampin | 1 | 33.3 | 0 | 0 | 2 | 66.7 |
| Clindamycin | 1 | 33.3 | 0 | 0 | 2 | 66.7 |
| Erythromycin | 0 | 0 | 1 | 33.3 | 2 | 66.7 |
| Deoxycycline | 0 | 0 | 0 | 0 | 3 | 100 |
| Penicillin | 0 | 0 | 0 | 0 | 3 | 100 |
| Cefoxitin | 0 | 0 | 0 | 0 | 3 | 100 |
| Gentamycin | 0 | 0 | 0 | 0 | 3 | 100 |
MRSA infections are well established in both the healthcare setting and in the community. MecC is a new emerging gene responsible for methicillin resistance in staphylococci. The mecC MRSA produces a characteristic antimicrobial susceptibility pattern different from mecA MRSA as mecA MRSA typically displays resistance to both oxacillin and cefoxitin antibiotic. On the other hand, most of mecC MRSA show resistance to Cefoxitin and are therefore reported as MRSA, although it shows susceptibility to Oxacillin 5.
In this study, the highest susceptibility of MRSA strains was found to Trimethoprim/ Sulfamethoxazole, Linezolid and Chloramphenicol antibiotic (68% for each antimicrobial agent) While lower susceptibility was found with Clindamycin (38%) followed by Doxycycline and Ciprofloxacin (36% for each). The highest resistance pattern was to Erythromycin (82%) and penicillin (98%). This goes in accordance with a study done in Egypt by Abdel-Maksoud et al. in 201610 who reported that 100% of all tested isolates were resistant to Penicillin and 70.5 % were resistant to Ciprofloxacin and 64.4% were resistant to Erythromycin and highest susceptibility was to Trimethoprim/ Sulfamethoxazole, and Rifampicin.
Also, Al-Zoubi et al in 201711, reported that the highest susceptibility of MRSA isolates was found to Linezolid (96.5%) and Chloramphenicol (86.7 %) and medium sensitivity percentages were found to Clindamycin (54.9%), Gentamicin (47%) and Levofloxacin (42.5%) and highest resistance was to Erythromycin and Penicillin.
Regarding the minimal inhibitory concentration (MIC) using E-test, we found that 12 (24%) isolates were sensitive to Oxacillin and there were 38 (76%) isolates resistant to Oxacillin .on the other hand Saeed et al.in 201412 reported (1.2%) strains were sensitive to oxacillin using phenotypic methods in the United Kingdom and Cikman et al. in 201913 reported that all tested MRSA isolates were phenotypically resistant to oxacillin and Cefoxitin.
In the current study results of PCR showed that all cases (100%) were positive to mecA gene while only three cases (6%) were positive to both mecA and mecC gene, one of the mecC cases was sensitive to Oxacillin E-test with MIC ≤ 2 and two mecC were resistant to Oxacillin E-test with MIC≥ 4.
One of the first papers published to discuss the emergence of mecC gene in MRSA isolates versus mecA gene was conducted in Denmark 2011 by Garcia-Alvarez4 who found that mecC gene represents 2.8% of MRSA isolates while in the current study we found 6% mecC gene of MRSA isolates, the author of the previous study contributed this result to rural areas as the detection of mecC gene was linked to livestock.
On the other hand, Rania et al. in 201714 in Egypt conducted a similar study showed that of 150 isolates (110 MRSA + 40 MR-CoNS) all were subjected to PCR where no MRSA isolates carrying mecC gene were reported detected. This was similar to Ganesan15 who also did not detect mecC gene in his study. Another study done in Egypt by Khairalla et al. in 201716 reported that all the MRSA isolates included in the study contain the mecA gene (n = 34, 100%), while mecC was not identified.
On the other hand Khan and co-workers17 reported that the prevalence of mecA gene was 54% which is less than reported in this study, they also found 3% mecA negative isolates carrying mecC, while only one MRSA isolate carrying both mecA and mecC genes. also, Aklilu and Hui Ying18 reported first mecC and mecA positive livestock-associated MRSA in Malaysia, they concluded that out of the 15 mecC positive isolates, 12 were positive for both mecA and mecC genes this concedes with results of the current study concerning the 3 mecC positive were mecA positive, so we can assume that we can find both mecA and mecC genes in same isolate which needs further studies for confirmation of this finding .
In conclusion, this work is one of first report of presence of mecC gene in MRSA isolates in Egypt, mecC gene was detected in 6% of MRSA isolates included in this study and this isolates also carry mecA gene, although this percentage is not alarming, further studies testing for mecC gene in larger scale using different primer sets for mecC, may increase the probability to detect mecC gene especially in rural areas as the presence of mecC is linked to livestock.
ACKNOWLEDGMENTS
The authors are grateful to members of intensive care units of Ain Shams University hospitals, for their support to carry out this study.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors designed the experiments. HR performed the experiments.HR and SA drafted the manuscript, compiled information from the literature, and designed the figures and tables. WK and AN supervised and reviewed the manuscript. All authors read and approved the manuscript for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Lakhundi S, Zhang K. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev. 2018;31(4).
- Kumurya A. A potential diagnostic problem: The Newly Emerging mecC Methicillin- Resistant Staphylococcus aureus strains. IOSR-JPBS. 2015;10(5):84-93.
- Peterson A, Stegger M, Heltberg O, et al. Epidemiology of methicillin-resistant Staphylococcus aureus carrying the novel mecC gene in Denmark corroborates a zoonotic reservoir with transmission to humans. Clin Microbiol Infect. 2013;19(1):E16-22.
- Garcia-Alvarez L, Holden MT, Lindsay H, et al. Methicillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis. 2011;11:595-603.
- Cartwright EJP, Paterson GK, Raven KE, et al. Use of Vitek 2 antimicrobial susceptibility profile to identify mecC in methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2013;51(8):2732-2734.
Crossref - Collee JG, Miles RS and Watt B. Tests for identification of bacteria. In: Mackie and Mc Carteny practical Medical Microbiology. 14th ed. Churchill Livingstone. c1996:131-150.
- Cheesbrough. District Laboratory Practice in Tropical Countries. Cambridge university, UK. 2009;2(2):65-67.
- Clinical and Laboratory Standards Institute (CLSI). https://clsi.org/media/2663/m100ed29_sample.pdf
- Buhlmann M, Bogli-Stuber K, Droz S, et al. Rapid screening for carriage of methicillin-resistant Staphylococcus aureus by PCR and associated costs. J Clin Microbiol. 2008;46(7):2151.
- Abdel-Maksoud M, El-Shokry M, Ismail G, et al. Methicillin-Resistant Staphylococcus aureus Recovered from Healthcare- and Community-Associated Infections in Egypt. Int J Bacteriol. 2016;2016:5751785.
Crossref - Al-Zoubi MS, Al-Tayyar IA, Hussein E, Al Jabali A, et al. Antimicrobial susceptibility pattern of Staphylococcus aureus isolated from clinical specimens in Northern area of Jordan. Iran J Microbiol. 2015;7(5):265.
- Saeed K, Ahmad N, Dryden M, et al. Oxacillin-susceptible methicillin-resistant Staphylococcus aureus (OS-MRSA), a hidden resistant mechanism among clinically significant isolates in the Wessex region/UK. Infection. 2014;42(5):843-847.
Crossref - Cikman A, Aydin M, Gulhan B, et al. Absence of the mecC gene in methicillin-resistant Staphylococcus aureus isolated from various clinical samples: The first multi-centered study in Turkey. Journal of Infection And Public Health. 2019;12(4):528-33.
- Rania AA, Nsreen MK, Rasha HEl, et al. Evaluation for the Novel mecC Methicillin Resistance among Methicillin Resistant Staphylococcal Isolates in two Egyptian University Hospitals. Arch Clin Microbiol. 2017; 9(1):71.
- Ganesan A. Evaluation for a novel methicillin resistance (mecC) homologue in methicillin-resistant Staphylococcus aureus isolates obtained from injured military personnel. J Clin Microbiol. 2013;51(9):3073-3075.
Crossref - Khairalla AS, Wasfi R, Ashour HM. Carriage frequency, phenotypic, and genotypic characteristics of methicillin-resistant Staphylococcus aureus isolated from dental health-care personnel, patients, and environment. Scientific Reports. 2017;7(1):1-6.
- Khan AA, Ali A, Tharmalingam N, Mylonakis E, Zahra R. First report of mecC gene in clinical methicillin resistant S. aureus (MRSA) from tertiary care hospital Islamabad, Pakistan. Journal of Infection And Public Health. 2020;13(10):1501-1507.
Crossref - Aklilu E, Hui Ying C. First mecC and mecA Positive Livestock-Associated Methicillin Resistant Staphylococcus aureus (mecC MRSA/LA-MRSA) from Dairy Cattle in Malaysia. Microorganisms. 2020;8(2):147.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.