ISSN: 0973-7510
E-ISSN: 2581-690X
The goal of this study was to isolate, count, and identify coagulase-negative Staphylococci from meat and dairy products collected in Riyadh, Saudi Arabia, as well as to perform molecular identification of the mecA gene. In addition, the prevalence of these bacteria among the examined food products was determined. Ninety samples of both meat and dairy products were collected and examined between February 2018 and August 2019. Mannitol salt agar and VITEK 2 system were carried out and VITEK 2 system was used to identify all bacterial isolates. Also, the molecular technique was used to detect the target gene of mecA among CoNS. The proportion of samples in which Staphylococcus species isolated is 13.33% (Camel meat), 6.66% (Beef mortadella), 6.66% (Turkish labneh), 33.33% (Cows cheese), 6.66% (Goat labneh), 13.33% (Nabulsy cheese), 13.33% (Haloumi goat cheese) and 6.66% (Akawy white cheese). Counts of coagulase-negative Staphylococcus species (as cfu/gm) of sample were around 11×104, 10×104, 9×104, 12×104, 4×105, 11×104 (Nabulsy cheese), 14×104 and 12×104, respectively. The Prevalence of species in both products was 3.30% (Staphylococcus vitulinus), 53.30% (Staphylococcus saprophyticus), 16.66% (Staphylococcus hominis), 6.66%, (Staphylococcus equorum and Staphylococcus lentus) and 13.33% (Staphylococcus sciuri). Furthermore, results showed methicillin specific mecA gene was harbored in 40 % of the CoNS.
Coagulase-negative staphylococci, meats, dairy products
Staphylococcus spp. could cause food poisoning and a production of enterotoxin in nourishment products.1 Staphylococcus has different species and divided in two main groups on the basis of coagulase enzyme production; the first group is coagulase-positive Staphylococci (CoPS) such as S. aureus, S. intermedius and S. hyicus.2 The other group is coagulase negative Staphylococci (CoNS) that consist of various species habitat human body.3 The Staphylococci have been frequently isolated from human, animal, and food (diary and meat products).4
Staphylococci are foodborne pathogen that are associated with raw unpasteurized milk of cattle suffering from mastitis.4,5 Meat and dairy products are abundant sources of nutrition such as proteins, fat, minerals, vitamins; and play a significant role in meet nutrient requirements of human.6 Bacteria causing food poisoning have the ability to habitat animals and might be transmitted to human through the consumption of animal products.7 Selvaggi study have focused on the resistance of bacteria in domestic animals. Its effect on human health is mostly linked to the misuse of antibiotics.8
Recently, CoNS is a typical opportunist and represents one of the main nosocomial pathogens that have a crucial impact on human health9,10 and associated with high resistance to many antibiotics.11 The CoNS are important reservoirs of antibiotic resistance genes and associated mobile genetic elements and are believed to contribute to the emergence of successful methicillin resistant Staphylococcus aureus (MRSA) clones. Also, multiple antibiotic resistance CoNS were isolated from food and other sources.9,10 Therefore, the present study aimed to isolate, count and identify CoNS and molecular detection of mecA gene from meat and dairy products in Riyadh city, Saudi Arabia.
Sample collection
To assess the identification of CoNS species and the prevalence rate in meats and dairy products, 45 samples of meat products and 45 samples of dairy products were randomly collected from three local hypermarkets of different three major chains in Riyadh City. All samples were collected in clean, dry and sterile polythene bags and then transported to the laboratory (under cold condition) then refrigerated at 4°C for maximum of 2 hours (hrs) for microbiological analysis and analysis.
Serial dilution preparation of samples
Five gram of meats and dairy products were homogenized with 95 ml sterile distilled water for 2 minutes (Ningbo Sklon, China). Subsequently, 1 ml of the total amount was transferred to 9 ml of sterilized distilled water under sterilized conditions until 10-5 dilution.
Enumeration and isolation of Staphylococci spp.
Total Staphylococci spp. were performed using mannitol salt agar medium. The plates were inoculated with 1 ml-1 of homogenate dilutions using surface plating technique. Then, plates were incubated at 37°C for 24-48 hrs. Typical yellow colonies on mannitol salt agar considered as mannitol fermenter Staphylococci spp.
Purification of isolated strains
A typical colony of selected Staphylococcus spp. heavily streaked on mannitol salt agar and then incubated at 37°C for 24 hrs. Bacterial isolates were enriched in nutrient broth containing 30% (v/v) glycerol (BDH Chemicals Ltd. Pool, England) and then stored at −80°C for further analysis.
Inoculum preparation
Each organism suspension was prepared from growth of pure cultures of bacteria cultivated on plates containing nutrient agar. Then they were incubated overnight at 37°C. Bacterial cells were suspended in 2.5 ml of a 0.45% sodium chloride solution. The suspension used in the VITEK 2 system (bioMerieux) was adjusted to a McFarland standard of 0.5 by using a Denscheck. Suspensions used for the comparative identification method were made according to the manufacturer’s instructions.
Biochemical tests of isolated Staphylococcus spp. with the VITEK 2 System
The VITEK 2 system was used according to the manufacturer’s instructions; ID-Gram Positive Cocci cards (ID-GPC cards; bioMérieux) were used for identification. The ID-GPC card is a 64-well plastic card containing 18 empty wells and 46 wells for fluorescent and inhibitory tests. Final results were obtained automatically. All used cards were automatically discarded into a waste container.
Molecular detection of mecA gene in the recovered coagulase-negative Staphylococci
To determine the targeted gene in CoNS, as a reservoir for antibiotics resistance genes, two primer sets previously reported Moussa and Shibl 200912 The primers were used for the detection of the mecA gene, that amplifies 1319 bp fragments specific for mecA gene.
The prevalence of CoNS
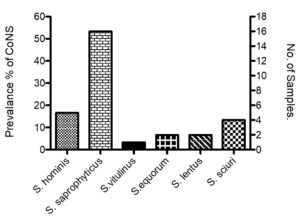
Ninety samples were collected and examined microbiologically to isolate Staphylococcus species. Out of 90 samples, Staphylococci were isolated from 30 samples (33.33%). Of 30 samples, 6 collected from meat products (6.66%) and 24 from dairy products (26.66%). All bacterial isolates were tested for catalase and coagulase activity to confirm isolates belong to Staphylococci group., All bacterial isolates were positive for catalase and negative for coagulase (Table 1). The proportion of CoNS isolates in type of samples and source varied. The predominant species was S. saprophyticus (n=16; 53.33%) followed by S. hominis (n= 5; 16.66%), S. sciuri (n=4; 13.33%), both of S. equorum and S. lentus (n= 2 each; 6.66%), S. vitulinus (n= 1 each; 3.33%) (Table 4). By VITEK 2 equipment, all isolates were identified as species of Staphylococci, named as 99% of Staphylococcus vitulinus, 96.5% of Staphylococcus saprophyticus, 96.9% of Staphylococcus hominis, 99% of Staphylococcus equorum, 97% of Staphylococcus lentus, 98 % of Staphylococcus sciuri (Table 2).
Table (1):
Bacterial isolates in food products that produce catalase enzymes.
| Samples | Bacterial isolates | Catalase test | Coagulase test | |
|---|---|---|---|---|
| Dairy Products | Turkish labneh | Isolate 1 | +ve | -ve |
| Cow cheese | Isolate 2 | +ve | -ve | |
| Goat labneh | Isolate 3 | +ve | -ve | |
| Nabulsy cheese | Isolate 4 | +ve | -ve | |
| Haloumi goat cheese | Isolate 5 | +ve | -ve | |
| Akawy white cheese | Isolate 6 | +ve | -ve | |
| Meat Products | Camel meat | Isolate 7 | +ve | -ve |
| Isolate 8 | +ve | -ve | ||
| Isolate 9 | +ve | -ve | ||
| Beef mortadella | Isolate 10 | +ve | -ve | |
+ve: positive test, -ve: negative test
Table (2):
Identification of bacterial isolates from dairy and meat products with VETIK2 system.
| Samples | Number of isolates | Species | Identification percentage | |
|---|---|---|---|---|
| Dairy Products | Turkish labneh | Isolate 1 | S. saprophyticus | 96.5% |
| Cow cheese | Isolate 2 | S. saprophyticus | 96.5% | |
| Goat labneh | Isolate 3 | S. lentus | 97 % | |
| Nabulsy cheese | Isolate 4 | S. hominis | 96.9% | |
| Haloumi goat cheese | Isolate 5 | S. sciuri | 98 % | |
| Akawy white cheese | Isolate 6 | S. saprophyticus | 96.5% | |
| Meat Products | Camel meat | Isolate 7 | S. vitulinus | 99 % |
| Isolate 8 | S. saprophyticus | 96.5% | ||
| Isolate 9 | S.hominis | 96.9% | ||
| Beef mortadella | Isolate 10 | S. equorum | 99 % | |
CoNS isolates from meat products
Of 45 samples, the proportion of 6 isolates that identified as Staphylococcus species using VITEK 2 were 13.33%. The positive samples of camel meat (n=4) and beef mortadella (n=2) were 8.89% and 4.44%, respectively. Both S. saprophyticus (n=2) and S. equorum (n=2) were predominant (4.44% each) in this product followed by S. hominis and S. Vitulinus (2.22% each). Of 6 isolates, both S. saprophyticus and S. equorum were predominant (33.33% each) in this product followed by S. hominis and S. Vitulinus (16.66% each) (Table 4). Staphylococcus spp. counts in meat products were 9×104 cfu/gm (S. vitulinus), 12×104 cfu/gm (S. saprophyticus), 13×104 cfu/gm (S. hominis) in camel meat. and 10×104 cfu/gm (S. equorum) in beef mortedella (Table 3).
CoNS isolates from dairy products
As stated above in dairy products samples, 53.33% of which (24 out of 45) was positive for the isolation of CoNS isolates. The proportion of different dairy products was 4.44% (Turkish labneh; n=2), 22.22% (Cow cheese; n=10), 4.44% ( Goat labneh; n=2), 8.89% (Nabulsy cheese; n=4), 8.89% (haloumi goat cheese; n=4), 4.44% (Akawy white cheese; n=2). The higher proportion of dairy sample in which Staphylococcus species were isolated was noticed in Cow cheese (33.33%), but Turkish labneh, Goat labneh and Akawy white cheese have lowest proportion of isolates (8.33% each). Of 24 isolates, the higher prevalence of CoNS isolates among diary samples was S. saprophyticus (n=14; 58.33%), followed by S. hominis (n=4; 16.66%) and S. sciuri (n=4; 16.66%), then S. lentus (n=2; 8.88%) (Table 4, Fig. 1). Staphylococcus spp. counts were 9×104 cfu/gm (S. saprophyticus) in Turkish labneh, 12×104 cfu/gm (S. saprophyticus) in Cow Cheese, 4×105 cfu/gm (S. lentus) in Goat labneh, 11×104 cfu/gm (S. hominis) in Nabulsy cheese, 14×104 cfu/gm (S. sciuri) in haloumi goat cheese and 12×104 cfu/gm (S. saprophyticus) in Akawy white cheese (Table 3).
Table (3):
Total counts of Staphylococcus spp. in meat and dairy products (CFU/gm).
| Samples | Bacterial isolates | CFU/gm | |
|---|---|---|---|
| Dairy Products | Turkish labneh | S. saprophyticus | 9*104 |
| Cow cheese | S. saprophyticus | 12*104 | |
| Goat labneh | S. lentus | 4*105 | |
| Nabulsy cheese | S. hominis | 11*104 | |
| Haloumi goat cheese | S. sciuri | 14*104 | |
| Akawy white cheese | S. saprophyticus | 12*104 | |
| Meat Products | Camel meat | S. vitulinus | 13*104 |
| S. saprophyticus | 12*104 | ||
| S.hominis | 9*104 | ||
| Beef mortadella | S. equorum | 10*104 | |
Table (4):
Distribution of CoNS isolates in food products and in which sample isolated.
| Samples | Bacterial isolates | No. (%) of isolates in 45 samples | % of isolates in 30 samples | |
|---|---|---|---|---|
| Dairy Products | Turkish labneh | S. saprophyticus | 2 (4.4) | 6.6 |
| Cow cheese | S. saprophyticus | 10 (22.2) | 33.3 | |
| Goat labneh | S. lentus | 2 (4.4) | 6.6 | |
| Nabulsy cheese | S. hominis | 4 (8.8) | 13.3 | |
| Haloumi goat cheese | S. sciuri | 4 (8.8) | 13.3 | |
| Akawy white cheese | S. saprophyticus | 2 (4.4) | 6.6 | |
| Meat Products | Camel meat | S. vitulinus | 1 (2.2) | 3.3 |
| S. saprophyticus | 2 (4.4) | 6.6 | ||
| S.hominis | 1 (2.2) | 6.6 | ||
| Beef mortadella | S. equorum | 2 (4.4) | 3.3 | |
The detection of mecA gene in the recovered CoNS
Agarose gel electrophoresis shows amplification of 1319 bp fragments of mecA gene of CoVS. Fourthly percent (40%) of the CoNS were positive and harboring the coagulase gene and amplification of 1319 bp fragments were observed (Fig. 2).
Fig. 2. Agarose gel electrolysis. The gel shows amplification of 1319 bp pair fragments of mecA gene. Lanes 1,2 (from left) shows coagulase negative, while lane 3 showing negative samples, and lane 8 shows 100bp ladder. Lanes 4,5,6,7,9,10,11 shows positive amplification of 1319 bp of Staphylococcus species harboring mecA gene.
CoPS and CoNS are both considered as commensal opportunistic infections of human and animal.13 These species are known to cause wound infection in human and mastitis and skin infection in animals.14 Also, they can cause food poisoning through secretion of enterotoxins.15 Sometime the acquisition of antimicrobial resistance might be occurred.16 This study was evaluating the prevalence of Staphylococcus spp. in retail markets of meat and dairies. Other species of Staphylococcus were found, but S. aureus was not. Different CoNS were isolated and identified by VITEK 2 system as S. vitulinus, S. saprophyticus, S. hominis, S. quorum, S. lentus, S. sciuri from dairy and meat products.17-19
In this study, a total of 30 samples (33.3%) CoNS were isolated from 90 tested samples of meat and dairy products. The proportion of CoNS in both products was 6.6% and 26.6%, respectively. The higher prevalence of CoNS isolates in diary samples was S. saprophyticus (58.33%) followed by S. hominis and S. sciuri 16.66% each, then the lower isolates of CoNS was S. lentus (8.88%). We have found that CoNS was significantly exist in meat and dairy products without existence of CoPS in the same products. That might provide a significant association between staphylococci and live stock.13 As reported in different study, the standard hygienic process in dairy farms usually conducted,20 but the high prevalence might be attributed to insufficient standard hygiene during the process or following the distribution of food products in retail markets. Moreover, the poor quality of milking and udder preparation might play an important role in contamination during the milking process.13,21
Our findings demonstrated that the prevalence rate of Staphylococcus spp. in meat product was 13.3% and the camel meat has the highest contamination rate of Staphylococcus spp. (8.9%) of all analyzed samples, including S. vitulinus, S. saprophyticus and S. hominis (Table 2). Similarly, the poor hygienic process during or post slaughtering.13,22 and transportation of meat product to different retails markets might be considered. In comparison to meat products, the dairy products have higher prevalence rate of of Staphylococcus spp. (53.33%). Our study supports the findings of other authors who isolated CoNS from milk. However, their report disclosed toxigenic strains of Staphylococci in goat milk.23 As reviewed, the isolates from 15-55% from dairy products produce enterotoxin.24
We have found that CoNS was significantly exist without existence of coagulase positive Staphylococci in meat and dairy products. Another study claimed that the dairy products that contains CoNS is tend to be safe in regards to health hazard associated with CoNS.25 Vasileiou et. al., have isolated and studied epidemiological characterizations of S. saprophyticus S. xylosus, S. carnosus and S. equorum from a wide range of foods products such as meat, cheese and milk.9 The existence numbers of these bacteria might attribute to inappropriate heat treatment and temperatures; and poor hygiene during food processing or after the food production due to contamination after pasteurization.26 Furthermore, S. saprophyticus was the highest (n= 16; 53.3%) in food products (Fig. 1). S. saprophyticus was the most predominant species of CoNS in dairy products with approximate count (11*104 cfu/gm). However, the highest growth of species was S. lentus with average count 4*105 cfu/gm (Table 3). However, S. vitulinus was the lowest prevalence (n=1; 3.3%) among these products (Fig. 1). The existence of Staphylococci species in food product seems to have their ability to colonize and cause a disease in different hosts.27
Our results demonstrated that 40% of the CoNS harboring the gene of mecA specific for methicillin resistance (Fig. 2). It suggested that the cause of methicillin resistance might be attributed to the mechanisms rather than expression of mecA.28 There are some findings of other authors who detected antibiotics resistance of CoNS which were isolated from various meat and dairy products.17-19 As stated in other studies that CoNS derived from camel milk was significantly resistance to around 70% of antibiotics such as erythromycin, colistin sulphate, penicillin G, oxacillin and vancomycin. On the other hand, CoPS of paired samples have showed resistance to only 3 antibiotics used such as penicillin G, oxacillin and vancomycin. Taken together, all isolates of CoNS from dairy products showed a full resistance to main antibiotics of penicillin G and oxacillin.3 Also, the CoNS might be easily developing to antibiotics resistant than S. aureus.29
Collectively, this study is considered as a key work to limit the prevalence of the CoNS and reduce the microbiological risk to the minimum. This will be approached through monitoring the animals and improving food safety as well.30 The existence of Satphylococci species in meat & dairy products might have the probability to serve as a transmission factors to spread more pathogenic and severe strains in our community.7 The contamination of retail meat and dairy products with Satphylococci species might be due to food production line and insufficient hygiene of food process that likely happened before marketing.
All samples isolated from meat and dairy products showed a significant existence of CoSN with around 33.3%. Moreover, not all the isolated CoNS from different food samples were resistant to methicillin as only 40% of which harboring mecA gene. As a conclusion, survey and diagnostic investigations are suggested as alternative solutions. The epidemiology of coagulase negative Staphylococci isolates from dairy and meat products is significant, and it should be incorporated in public health, food safety, and hygiene efforts.
Limitations
The number of collected samples is considered very low (90 samples of meat and diary products) to identify the the prevalence rate, as the negative samples of food product were high. Thus, more samples are required to be collected in the future and should be collected from different sources and different regions to make more comprehensive study. Furthermore, different antibiotics should be utilized to identify the resistance of antibiotics.
ACKNOWLEDGMENTS
The author would like to thank the Deanship of Scientific Research, and the College of Science Research Center at King Saud University for their support.
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
The datasets generated during the current study are available from the corresponding author on reasonable request.
- Arcuri EF, Angelo FF, Guimaraes MF, et al. Toxigenic status of Staphylococcus aureus isolated from bovine raw milk and Minas frescal cheese in Brazil. J Food Prot. 2010;73(12):2225-2231.
Crossref - Beyene T, Hayishe H, Gizaw F, et al. ‘Prevalence and antimicrobial resistance profile of Staphylococcus in dairy farms, abattoir and humans in Addis Ababa, Ethiopia. BMC Res Notes. 2017;10(1):171.
Crossref - Abulreesh H, Organji S. The Prevalence of Multidrug-resistant Staphylococci in Food and the Environment of Makkah, Saudi Arabia. Res J Microbiol. 2011;6(6):510- 523.
Crossref - Irlinger F. Safety assessment of dairy microorganisms: coagulase-negative Staphylococci. Int J Food Microbiol. 2008;126(3):302-310.
Crossref - Morgan M. Methicillin-resistant Staphylococcus aureus and animals: zoonosis or humanosis. J Antimicrob Chemother. 2008;62(6):1181-1187.
Crossref - Sawant AA, Gillespie BE, Oliver SP. Antimicrobial susceptibility of coagulase- negative Staphylococcus species isolated from bovine milk. Vet Microbiol. 2009;134(1-2):73-81.
Crossref - Shawish RR, Al-Humam NA. Contamination of beef products with Staphylococcal classical enterotoxins in Egypt and Saudi Arabia. GMS Hyg Infect Control. 2016;11:Doc08.
Crossref - Selvaggi M, D’Alessandro AG, Dario C. ‘Environmental and genetic factors affecting milk yield and quality in three Italian sheep breeds. J Dairy Res. 2017;84:(1)27-31.
Crossref - Vasileiou NGC, Chatzopoulos DC, Sarrou S, et al. Role of staphylococci in mastitis in sheep. J Dairy Res. 2019;86(3):254-266.
Crossref - Faria CI, Vaz-Moreira E, Serapicos OC, Nunes, Manaia CM. Antibiotic resistance in coagulase negative staphylococci isolated from wastewater and drinking water. Sci Total Environ. 2009;407(12):3876-3882.
Crossref - Boamah VE, Agyare C, Odoi H, Adu F, Gbedema SY, Dalsgaard A. Prevalence and antibiotic resistance of coagulase-negative Staphylococci isolated from poultry farms in three regions of Ghana. Infection and Drug Resist. 2017;10:175-183.
Crossref - Moussa I, Shibl AM. Molecular characterization of methicillin-resistant Staphylococcus aureus recovered from outpatient clinics in Riyadh, Saudi Arabia. Saudi Med J. 2009;30:611-617.
- Von Eiff C, Peters G, Heilmann C. Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect. Dis. 2002; 2:677–685.
Crossref - Becker K, Heilmann C, Peters G. Coagulase-negative staphylococci. Clin Microbiol Rev. 2014;27(4):870-926.
Crossref - Hennekinne J-A, De Buyser M-L, Dragacci S. Staphylococcus aureus and its food poisoning toxins: characterization and outbreak investigation. FEMS Microbiol Rev. 2012;36(4):815-836.
Crossref - Morgenstern M, Erichsen C, Hackl S, et al. Antibiotic resistance of commensal Staphylococcus aureus and coagulase-negative staphylococci in an international cohort of surgeons: a prospective point-prevalence study. PLoS ONE. 2016;11(2):e0148437.
Crossref - Silva NCC, Guimaraes FF, Manzi MdeP, et al. Characterization of methicillin-resistant coagulase- negative staphylococci in milk from cows with mastitis in Brazil. Antonie Van Leeuwenhoek. 2014;106(2):227-233.
Crossref - Yurdakul NE, Erginkaya Z, Unal E. Antibiotic Resistance of Enterococci, Coagulase Negative Staphylococci and Staphylococcus aureus Isolated from Chicken Meat. Czech J Food Sci. 2013;31(1):14-19.
Crossref - Cicconi-Hogan KM, Belomestnykh N, Gamroth M, Ruegg RL, Tikofsky L, Schukken YH. Short communication: Prevalence of methicillin resistance in coagulase- negative staphylococci and Staphylococcus aureus isolated from bulk milk on organic and conventional dairy farms in the United States. J Dairy Sci. 2014;97(5):2959-2964.
Crossref - Getaneh AM, Gebremedhin EZ. Meta-analysis of the prevalence of mastitis and associated risk factors in dairy cattle in Ethiopia. Trop Anim Health Prod. 2017;49(4):697-705.
Crossref - Anderson K, Pritchard D. An update on Staphylococcus aureus mastitis. 4th edition. San Francisco: Benjamin/Cummings Publishing Company; 2008:38.
- Haileselassie M, Taddele H, Adhana K, Kalayou S. Food safety knowledge and practices of abattoir and butchery shops and the microbial profile of meat in Mekelle City, Ethiopia. Asian Pac J Trop Biomed. 2013;3(5):407-412.
Crossref - Sindhu N, Sharma A, Jain VK. Coagulase Gene Based Molecular Detection of Staphylococcus aureus Directly from Mastitic Milk Samples of Murrah Buffalo. Buffalo Bulletin. 2010;29(1):62-69.
- Smith K. Food borne pathogenic microorganisms and natural toxins. Food and Drug Administration. Center Food Safety Appl Nutr. 2007;10:119-150.
- Even S, Leroy S, Charlier C, et al. Low occurrence of safety hazards in coagulase negative Staphylococci isolated from fermented foodstuffs. Int J Food Microbiol. 2010;139(1-2):87-95.
Crossref - Rosengren A, Fabricius A, Guss B, Sylven S, Lindqvist R. Occurrence of foodborne pathogens and characterization of Staphylococcus aureus in cheese produced on farm-dairies. Inter J Food Microbiol. 2010;144(2):263-269.
Crossref - Fitzgerald JR. Livestock-associated Staphylococcus aureus: origin, evolution and public health threat. Trends Microbiol. 2012;20(4):192-198.
Crossref - Fowoyo PT, Ogunbanwo ST. Antimicrobial resistance in coagulase-negative staphylococci from Nigerian traditional fermented foods. Ann Clin Microbiol Antimicrob. 2017;16(4).
Crossref - Taponen S, Pyorala S. Coagulase-negative staphylococci as cause of bovine mastitis-Not so different from Staphylococcus aureus. Vet Microbiol. 2009;134(1-2):29-36.
Crossref - Basanisi MG, La BellaG, Nobili G, Franconieri I, La Salandra.G. Genotyping of methicillin-resistant Staphylococcus aureus (MRSA) isolated from milk and dairy products in South Italy. Food Microbiol. 2017;62:141-146.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.