ISSN: 0973-7510
E-ISSN: 2581-690X
Prior to the Severe Acute Respiratory Syndrome Coronavirus Disease 2 (SARS-CoV-2) pandemic, the rise in antimicrobial resistance was a major source of concern in public health. However, due to the novelty of SARS-CoV-2 infection during the pandemic, antibiotics were administered prior to laboratory testing for secondary gram-negative bacteria (SGNB) in order to avoid or reduce the occurrence of SGNB infection. The purpose of this study was to investigate the etiology, prevalence, and antimicrobial susceptibility pattern of gram-negative bacteria (GNB) isolated from SARS-CoV-2 positive patients. Respiratory and blood samples were collected from confirmed SARS-CoV-2 positive patients. They were subsequently cultured and bacterial isolates identified according to standard microbiological protocols. Antimicrobial susceptibility testing (AST) was performed and interpreted according to Clinical & Laboratory Standards Institute (CLSI) 2021 guidelines. A total of sixty-four non-repetitive GNB were isolated from respiratory samples and twenty-two GNB from blood samples. K. pneumoniae was the major cause of SGNB, followed by Acinetobacter species. K. pneumoniae had over 60% resistance to β-Lactam combination agents, cephalosporin, and the carbapenem group of antibiotics. In the current study, we observed that K. pneumoniae was the major cause of SGNB and had high resistance to the antimicrobial agents. Hence, it is important that the epidemiology and susceptibility patterns of circulating organisms causing SGNB infection are always monitored to inform clinical treatment and decrease the occurrence of antibiotic-resistant bacteria.
Antimicrobial Susceptibility Patterns, Gram Negative Bacteria, Multidrug-resistance, SARS-CoV-2
The past two decades have recorded the emergence of severe viral infections, with SARS-CoV-2 being the most recent. SARS-CoV-2 started in Wuhan, China, and has since then quickly spread over the rest of the world.1 As of August 15, 2022, the global incidence of SARS-CoV-2 was over 587 million cases with over 6.4 million deaths.2 Its clinical presentation varies from the patient being asymptomatic to having severe pneumonia, causing respiratory failure and eventually death.3 The SARS-CoV-2 often affects ciliated cells in the alveolar epithelium and its barrier function due to viral-induced immune-mediated damage and dysregulated host-immune response, thus facilitating bacterial attachment and colonization.4
Secondary infections, which are caused either by bacteria, fungi, or viruses, are a major consequence of viral infection progression.5 They appear during or after viral infection and are associated with prolonged hospital stays, increased respiratory failure, and mortality, especially in critically ill patients.6,7 Generally, the overall incidence rate of secondary bacterial infection (SBI) in patients infected with SARS-CoV-2 varies substantially. Researchers reported an incidence rate of 14%–44% for SBI and 0.8%–19.8% for co-infection, respectively.7,8 The variation could be due to differences in geographical locations, sample sizes, and populations, among others. Zhou et al. report in their study that the ratio of secondary infections in SARS COV-2 is 1:7, with over 50% of the fatality rate caused by untreated or untreatable SGNB.9
The rapid rise in antimicrobial resistance, which was a major cause of public health concern before SARS-CoV-2, became much more of a worry earlier during the pandemic because of uncertainty regarding the new infectious disease, the surge in critically ill patients, and the lack of effective therapies for SARS-CoV-2, resulting in the excessive and unnecessary use of antibiotics, which were commonly administered before or at the time of hospital admission.10-12 As described by the World Health Organization (WHO) in 2017, GNB accounts for the vast majority of antibiotic-resistant organisms because of their ability to alter their outer membrane, thus making them more resistant to antibiotics than gram-positive bacteria (GPB). In addition, the critical priority pathogens on the WHO list were all GNB (i.e., carbapenem-resistant Acinetobacter baumannii, carbapenem-resistant Pseudomonas aeruginosa, and 3rd generation cephalosporin-resistant Enterobacteriaceae).13 As a result, they require close regional monitoring and the development of practical guidelines to limit access to unprescribed antibiotics and their overuse. This could help to reduce resistance among GNB.14-16 This study focuses on identifying the prevalence of SGNB and their antimicrobial susceptibility patterns. To the best of our knowledge, limited studies have focused on the prevalence and susceptibility pattern of SGNB infections in our region. Regional knowledge is important because it will play a significant role in predicting clinical outcomes and potentially aiding in the creation of strategies that could assist in lowering the severity and complications associated with the presence of secondary bacteria in the case of a future viral pandemic.
A one-year retrospective study was carried out between October 2020 and October 2021 at a 1500-bed tertiary care hospital in Tamil Nadu, India. SARS-CoV-2 positive patients were identified using Real Time RT-PCR, and blood as well as respiratory samples were collected by the clinician from all SARS-CoV-2 positive patients and sent to the microbiology laboratory to be tested for the presence of SBI. All collected blood samples were aseptically inoculated into the BacT/ALERT®3D microbial detection culture media bottles and incubated in the BacT/ALERT®3D microbial detection system until there was a positive signal, while respiratory samples were directly cultured on three culture media. They include blood, MacConkey, and chocolate agar, and were identified using biochemical tests at 37°C for 18-24 hours. Blood-positive samples were also cultured using the same media and incubated at 37°C for 18-24 hours. Inclusion criteria were SARS-CoV-2 positive patients of all age groups confirmed with SGNB, while exclusion criteria were SARS-CoV-2 positive patients with either GPB, contaminates, or colonizers. Demographic data such as the age and gender of patients was collected from the laboratory register.
SBI was defined as the presence of a true pathogen in either the blood or respiratory sample of a patient confirmed with SARS-CoV-2 using RT-PCR. Hence, results were corrected clinically in the laboratory and the presence of likely contaminants or colonizers was screened out. Only data from confirmed true pathogens was included in this study.
Antimicrobial Susceptibility Test (AST)
AST was performed by the disk diffusion method (using the Kirby-Bauer method) and the results were interpreted in accordance with the CLSI 2021 guidelines.17 Antibiotics tested for were amoxicillin-clavulanate, ceftazidime clavulanic acid, piperacillin-tazobactam, cefazolin, cefepime, cefotaxime, ceftriaxone, cefoxitin, cefuroxime, ceftazidime, ertapenem, meropenem, imipenem, gentamycin, tobramycin, amikacin, ciprofloxacin, and tetracycline.
Statistical Analysis
Data was entered into Microsoft Excel and statistical analysis was performed using IBM SPSS V25.
Ethics
Ethical permission was not required as the study was retrospective and data was collected from the microbiology register.
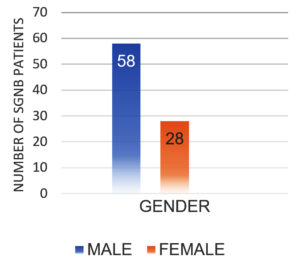
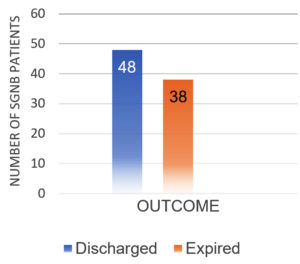
After excluding contaminants, the total number of SARS-CoV-2 positive patients diagnosed with SBI during the study period was 145. There were 86 (59.31%) GNB isolates and 59 (40.69%) GPB isolates. Table 1 shows the sample type distribution and prevalence of GNB organisms identified. Males had a higher prevalence of SGNB (67.44%) than females (32.55%) (Figure 1). 48 (55.81%) of SGNB patients recovered and were discharged, while 38 (44.18%) died (Figure 2).
Table (1):
Prevalence of SGNB Isolates among SARS-CoV-2 positive patients from different sample types.
| Isolates | Sample Types | |||
|---|---|---|---|---|
| Blood | Sputum | Tracheal Aspirate | Others | |
| K pneumoniae | 11 | 15 | 15 | 5 |
| Acinetobacter | 8 | 1 | 7 | – |
| P aeruginosa | 1 | 6 | 9 | 2 |
| E coli | 2 | – | 2 | 2 |
Antimicrobial Susceptibility Testing (AST)
Table 2 shows the antimicrobial susceptibility patterns of four predominant GNB organisms identified during our study. The organisms were K pneumoniae, followed by P. aeruginosa, Acinetobacter spp., and E. coli. A high rate of resistance was noticed in K. pneumoniae isolates, while P. aeruginosa was the most sensitive organism. Over 50% of Acinetobacter spp. were carbapenem-resistant, with over 80% of K. pneumoniae 3rd generation cephalosporin-resistant.
Table (2):
Antimicrobial susceptibilities of SGNBI Isolated from SARS-CoV-2 positive patients.
| Organism names and numbers (%) | |||||
|---|---|---|---|---|---|
| Antibiotics | Interpretation (S, I, R) | K. pneumoniae (n=46) | Acinetobacter spp. (n=16) | E. coli (n=6) | P. aeruginosa (n=18) |
| Penicillin | |||||
| Ampicillin | S | 0 (0) | – | – | 1 (17) |
| I | 0 (0) | – | – | 0 (0) | |
| R | 46 (100) | – | – | 5 (83) | |
| Beta-lactam combination agents | |||||
| Amoxicillin- clavulanate | S | 0 (0) | – | 1 (17) | – |
| I | 2 (4) | – | 0 (0) | – | |
| R | 44 (96) | – | 5 (83) | – | |
| Ceftazidime clavulanic acid | S | 16 (35) | – | 5 (83) | – |
| I | 0 (0) | – | 0 (0) | – | |
| R | 30 (65) | – | 1 (17) | – | |
| Piperacillin-tazobactam | S | 17 (37) | 12 (75) | 4 (67) | 18 (100) |
| I | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| R | 29 (63) | 4 (25) | 2 (33) | 0 (0) | |
| Cephalosporins | |||||
| Cefazolin | S | 7 (15) | – | 1 (17) | – |
| I | 0 (0) | – | 0 (0) | – | |
| R | 39 (85) | – | 5 (83) | – | |
| Cefepime | S | 10 (22) | 11 (69) | 2 (33) | 18 (100) |
| I | 2 (4) | 1 (11) | 1 (17) | 0 (0) | |
| R | 34 (74) | 4 (25) | 3 (50) | 0 (0) | |
| Cefotaxime | S | 8 (17) | 7 (44) | 1 (17) | – |
| I | 0 (0) | 3 (19) | 0 (0) | – | |
| R | 38 (83) | 6 (38) | 5 (83) | – | |
| Ceftriaxone | S | 9 (20) | 9 (56) | 2 (33) | – |
| I | 0 (0) | 0 (0) | 0 (0) | – | |
| R | 37 (80) | 7 (44) | 4 (67) | – | |
| Cefoxitin | S | 11 (24) | – | 4 (67) | – |
| I | 2 (4) | – | 0 (0) | – | |
| R | 33 (72) | – | 2 (33) | – | |
| Cefuroxime | S | 8 (17) | – | 1 (17) | – |
| I | 0 (0) | – | 0 (0) | – | |
| R | 38 (83) | – | 1 (17) | – | |
| Ceftazidime | S | 8(15) | 11 (61) | 2 (33) | 17 (94) |
| I | 2 (4) | 0 (0) | 0 (0) | – | |
| R | 37 (81) | 5 (39) | 4 (67) | 1 (6) | |
| Carbapenems | |||||
| Ertapenem | S | 15 (33) | – | 4 (67) | |
| I | 2 (4) | – | 1 (17) | ||
| R | 29 (63) | – | 1 (17) | ||
| Meropenem | S | 17 (37) | 11 (69) | 16 (89) | |
| I | 0 (0) | 0 (0) | 0 (0) | ||
| R | 29 (63) | 5 (31) | 2 (11) | ||
| Imipenem | S | 17 (37) | 9 (56) | 4 (67) | 16 (89) |
| I | 1 (2) | 0 (0) | 1 (17) | 0 (0) | |
| R | 28 (61) | 7 (44) | 1 (17) | 2 (11) | |
| Aminoglycosides | |||||
| Gentamycin | S | 17 (37) | – | ||
| I | 0 (0) | – | |||
| R | 29 (63) | – | |||
| Tobramycin | S | – | 16 (89) | ||
| I | – | 0 (0) | |||
| R | – | 2 (11) | |||
| Amikacin | S | 20 (43) | 11 (69) | 4 (67) | – |
| I | 0 (0) | 0 (0) | 0 (0) | – | |
| R | 26 (57) | 5 (31) | 2 (33) | – | |
| Tetracyclines | |||||
| Tetracycline | S | 19(41) | 10 (63) | 2 (33) | – |
| I | 0 (0) | 1 (6) | 0 (0) | – | |
| R | 27 (59) | 5 (31) | 4 (67) | – | |
| Quinolones | |||||
| Ciprofloxacin | S | 2 (4) | 12 (75) | 0 (0) | 14 (78) |
| I | 11 (24) | 0 (0) | 3 (50) | 0 (0) | |
| R | 33 (72) | 4 (25) | 3 (50) | 4 (22) | |
S-sensitive, I-Intermediate, R-Resistance
Infection pathogenesis is a multifaceted process that involves several components, such as the state of the host immune system, the bacterial isolates, and antimicrobial susceptibility pattern, among other factors.18 Studies on viral infections over the past decades have shown that the presence of SBI during or after a viral infection further deteriorates patients’ health, especially when a patient is critically ill.4,5 Hence, this study was done to study the major GNB organisms causing SBI and their susceptibility patterns to antibiotics.
The current study recovered 145 GNB from SARS-CoV-2 positive patients. Among them, 86 (59.31%) were GNB isolates and 59 (40.69%) were GPB isolates. Of the 86 GNB isolates, 64 were from respiratory samples and 22 were from blood samples.
K. pneumoniae (53.48%), P. aeruginosa (20.93%), Acinetobacter (18.60%), and Escherichia coli (6.97%) were the most commonly isolated SGNB in SARS-CoV-2 positive patients. A similar study by Vijay et al. also identified K. pneumoniae as the most common SGNB recovered from SARS-CoV-2 positive patients. In contrast, Sharifipour et al. found Acinetobacter to be the most common SGNB in their studies.19,20 SGNB was higher in males than females. This could be due to the fact that men express angiotensin-converting enzyme 2(ACE 2) at a higher level than females and that, in terms of immunity, reproductive-age women are more prone to autoimmune disorders than infectious diseases. Men’s lifestyles, such as heavy smoking and drinking, render them more susceptible to infection.21 studies by Ramiez et al. and Bwire correlate with our findings.21,22
Among the 86 patients studied, 48 (55.81%) were discharged, while 38 (44.18%) expired. An overall higher death rate was observed among elderly, critically ill patients with comorbidities admitted to the intensive care unit. Our findings are in agreement with the studies of Boorgula et al., who also observed a higher mortality rate in critically ill patients.12
In terms of antibiotic susceptibility pattern, we found K. pneumoniae to be the most resistant strain, while P. aeruginosa was the most sensitive strain. K. pneumoniae was resistant to more than one antibiotic agent, which makes them multi-drug resistant organisms.23 Studies by Jyoti et al.24 show that MDR impedes disease control by increasing the likelihood of resistance organisms spreading, resulting in lower efficacy of therapy and prolonging the patient’s infection. Antimicrobial resistance is linked to an increase in the mortality rate and high medical expenses, as well as a reduction in antimicrobial agent effectiveness.24 Acinetobacter was identified as the most prevalent SGNB in SARS-CoV-2 positive patients by Sharifipour et al.20 and Moradi et al.5 with high resistance to all antibiotics used in treatment except colistin. Multiple other studies also show unique variation in susceptibility patterns that could be caused by a variety of factors, such as differences in geographical location and sample size.5,20 The WHO and the US Centers for Disease Control (US-CDC) have identified MDR K. pneumoniae as a major threat to human health.13 Infections caused by K. pneumoniae are more threatening in neonates, the elderly, and those who are immunocompromised.5,20
Antimicrobial resistance has a significant influence on therapy and the outcome of viral infection. In our study, K. pneumoniae had high resistance to the antimicrobial agents used. Hence, it is important that the epidemiology and susceptibility pattern of circulating organisms causing SBI are always monitored to inform clinical treatment and decrease the occurrence of antibiotic-resistant bacteria.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
The datasets generated during and analyzed during the current study are available from the corresponding author on reasonable request.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee, SRM Medical College and Research Center, India (2196/IEC/2020).
- Manohar P, Loh B, Nachimuthu R, Hua X, Welburn SC, Leptihn S. Secondary bacterial infections in patients with viral pneumonia. Front Med. 2020;7:420.
Crossref - WHO. Coronavirus (COVID-19) dashboard. World Health Organization. https://covid19.who.int/. Accessed 17 August 2022.
- Li X, Geng M, Peng Y, Meng L, Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J Pharm Anal. 2020;10(2):102-108.
Crossref - Manohar P, Loh B, Athira S, et al. Secondary bacterial infections during pulmonary viral disease: Phage therapeutics as alternatives to antibiotics? Front Microbiol. 2020;11:1434.
Crossref - Moradi N, Kazemi N, Ghaemi M, Mirzaei B. Frequency and antimicrobial resistance pattern of bacterial isolates from patients with COVID-19 in two hospitals of Zanjan. Iran J Microbiol. 2021;13(6):769-778.
Crossref - Stefanini I, De Renzi G, Foddai E, Cordani E, Mognetti B. Profile of bacterial infections in COVID-19 patients: Antimicrobial resistance in the time of SARS-COV-2. Biology. 2021;10(9):822.
Crossref - Westblade LF, Simon MS, Satlin MJ. Bacterial coinfections in coronavirus disease 2019. Trends Microbiol. 2021;29(10):930-941.
Crossref - Polly M, de Almeida BL, Lennon RP, Cortes MF, Costa SF, Guimarães T. Impact of the COVID-19 pandemic on the incidence of multidrug-resistant bacterial infections in an acute care hospital in Brazil. Am J Infect Control. 2022;50(1):32-38.
Crossref - Zhou P, Liu Z, Chen Y, Xiao Y, Huang X, Fan X-G. Bacterial and fungal infections in COVID-19 patients: A matter of concern. Infect Control Hosp Epidemiol 2020;41(9):1124-1125.
Crossref - Aslam B, Wang W, Arshad MI, et al. Antibiotic resistance: A rundown of a global crisis. Infect Drug Resist. 2018;11:1645-1658.
Crossref - Ukuhor HO. The interrelationships between antimicrobial resistance, covid-19, past, and future pandemics. J Infect Public Health. 2021;14(1):53-60.
Crossref - Boorgula SY, Yelamanchili S, Kottapalli P, Naga MD. An update on secondary bacterial and fungal infections and their antimicrobial resistance pattern (AMR) in COVID-19 confirmed patients at a tertiary care hospital. J Lab Physicians 2022.
Crossref - WHO publishes list of bacteria for which new antibiotics are urgently needed. World Health Organization. https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed. Accessed 17 March 2022.
- Lee CR, Cho IH, Jeong BC, Lee SH. Strategies to minimize antibiotic resistance. Int J Environ Res Public Health. 2013;10(9):4274-4305.
Crossref - Breijyeh Z, Jubeh B, Karaman R. Resistance of gram-negative bacteria to current antibacterial agents and approaches to resolve it. Molecules. 2020;25(6):1340.
Crossref - Zabawa TP, Pucci MJ, Parr TR, Lister T. Treatment of gram-negative bacterial infections by potentiation of antibiotics. Curr Opin Microbiol. 2016;33:7-12.
Crossref - CLSI publishes M100-performance standards for antimicrobial susceptibility testing, 31st edition. Clinical & Laboratory Standards Institute. 2021. https://clsi.org/about/press-releases/clsi-publishes-m100-performance-standards-for-antimicrobial-susceptibility-testing-31st-edition/. Accessed 17 March 2022.
- JW Peterson. Bacterial pathogenesis. National Center for Biotechnology Information. 1996. https://pubmed.ncbi.nlm.nih.gov/21413346/. Accessed 17 March 2022.
- Vijay S, Bansal N, Rao BK, et al. Secondary infections in hospitalized COVID-19 patients: Indian experience. Infect Drug Resist. 2021;14:1893-1903.
Crossref - Sharifipour E, Shams S, Esmkhani M, et al. Evaluation of bacterial co-infections of the respiratory tract in COVID-19 patients admitted to ICU. BMC Infect Dis. 2020;20(1):646.
Crossref - Ramirez-Soto MC, Arroyo-Hernandez H, Ortega-Caceres G. Sex differences in the incidence, mortality, and fatality of covid-19 in Peru. PLOS ONE. 2021;16(6):0253193.
Crossref - Bwire GM. Coronavirus: Why Men are More Vulnerable to Covid-19 Than Women? SN Compr Clin Med. 2020;2(7):874-876.
Crossref - Basak S, Singh P, Rajurkar M. Multidrug Resistant and Extensively Drug Resistant Bacteria: A Study. J Pathog. 2016;2016:4065603.
Crossref - Tanwar J, Das S, Fatima Z, Hameed S. Multidrug resistance: an emerging crisis. Interdiscip Perspect Infect Dis. 2014;2014:541340.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.