ISSN: 0973-7510
E-ISSN: 2581-690X
https://dx.doi.org/10.22207/JPAM.13.1.21 | © The Author(s). 2019
Antibiotic resistance is an essential issue mostly in the intensive care units (ICUs). The Goal of this study was to investigate the widespread of multidrug resistance (MDR) gram-negative bacterial pathogens isolated from ICUs at King Khalid University Hospital (KKUH), Riyadh, KSA, and their ability to produce ESBL and MBL enzymes. All organisms were isolated from different ICUs at (KKUH) between June to December 2016. Identification and antimicrobial susceptibility were committed according to the laboratory policy. The bacterial Isolates flagged as ESBL or MBL by Vitek 2 were confirmed using E-test method recommended by CLSI. 70 isolates from different body sites comprising 25 (35.7 %) were P. aeruginosa, 23 (32.9 % ) were K. pneumoniae, 16 (22.9%) were E. coli, and 6 (8.6 %) were A. baumannii. Among the 23 isolates K. pneumonia and 16 of E. coli, 19 (82.6%) and 16 (100%) were detected as ESBL (+) by double-disk diffusion method according to guidelines of CLSI. On the contrary, ESBL was not detected in any isolates of P. aeruginosa or in A. baumannii. All P. aeruginosa and A. baumannii isolates were carbapenem resistant. MBL was found in all P. aeruginosa, A. baumannii and 4 (17.4 %) of K. pneumonia where E. coli strains did not appear any MBL action. The essential resistance mechanisms in the evaluated strains were ESBL and MBL. Molecular testing is recommended to confirm the phenotypic results and to detect the resistant genes.
MDR gram negative bacteria, P. aeruginosa, K. pneumoniae, E. coli, A. baumannii, ESBL, MBL.
The growth of MDR gram negative bacteria is an important healthcare challenge worldwide1. The infections by MDR gram negative pathogens often lead to enhanced mortality, prolonged hospital stays and unreasonable healthcare costs. In total, this evidence emphasize the significance for hospitals to start reacting in a proactive way rather than in a reactive one to withstand the increasing resistance rates2.
The increase at intervals percentages of antibiotic resistance can be a serious interest in every non-fermenting bacilli and isolates of the Enterobacteriaceae family. b-lactams are the base of medication for genuine contaminations. The Most effective of those are the carbapenems, which are recommended to be used for the treatment of infections brought about by strains of the emerging Enterobacteriaceae, principally Escherichia coli and Klebsiella pneumonia, and non-fermenters, Pseudomonas spp., and Acinetobacter spp3, which possess extended-spectrum-b-lactamase (ESBL).
b-lactamases are proteins made by specific bacteria that break down the b-lactam ring of b-lactam anti-infection agents (monobactams, penicillin’s, cephalosporins, carbapenems, and monobactams), is one amongst the foremost necessary mechanisms of microbial resistance to b-lactam antibiotics4. The decompounded b-lactam drugs result in a dormant product once the ring is broken5.
One of the foremost crucial resistant mechanisms in Gram-negative bacteria against b-lactam anti-infection agents is iatrogenic by the creation of b-lactamase catalysts. b-lactamase proteins could be characterized into four gatherings which are: A, B, C and D. Predictable with this order, expansive range b-lactamases are sorted among gathering A. The Gram-negative microscopic organisms have brisk enlarged protection from expansive range b-lactam antimicrobial agents. In excess of 200 kinds of expanded range b-lactamases (ESBLs) are discovered around the world, most having a place with the Enterobacteriaceae family6 .
Resistance in Gram-negative microscopic organisms is principally mediated through the yield of (ESBL), AmpC b – lactamases and Carbapenemases7.
Extended-spectrum b-lactamases (ESBLs) are bacterial enzymes that hydrolyze b-lactam anti-infection agents, such as, penicillins, cephalosporins and monobactams. They represent the foremost mechanism of resistance to second, third, and fourth generation cephalo-sporins, for example, cefuroxime, cefotaxime, ceftriaxone and ceftazidime8.
Carbapenemases are b- lactamases that exemplify Metallo-b-lactamases (MBL) and serine- b-lactamases (KPC, GES, OXA, etc). Conversely, throughout the past few years, Carbapenem resistance has been progressively recorded among Enterobacteriaceae and is predominantly recognized by the production of Ambler class B procured Metallo-b-lactamases (MBLs)9.
MBLs producing Gram-negative bacteria is an emerging threat worldwide ascribed to their resistance to any or all b-lactams except aztreonam10. The genes of MBL are either transferable plasmids or a piece of the bacterial chromosome11.
Aim of the work
To detect the antimicrobial resistance pattern, ESBL and MBLase producers among strains of gram-negative bacteria were secluded from King Khalid University Hospital ICUs.
Bacterial isolates
This retrospective study was done at King Khalid University Hospital, Riyadh, Saudi Arabia. The study included 70 successive, non-copied clinical isolates of ESBL K. pneumoniae, E .coli, MDR P. aeruginosa and A. baumannii that were identified from June to December 2016. These isolates were from various specimen types including, blood, wound, sputum, tracheal aspirate, eye, and groin, which were routinely cultured in the bacteriology laboratory.
Identification of the organisms and susceptibility testing
Identification and antimicrobial susceptibility of 70 isolates belonging to different species of gram negative microscopic organisms were carried out according to the laboratory policy by Vitek 2 (Biomerieux, Marcy l’Etoile). As indicated by the instructions of the Clinical Laboratory Standard Institute12 (CLSI), antibiotic susceptibilities were confirmed by Kirby-Bauer disk diffusion method. The selected strains of gram negative bacteria were tested for a group of antibiotics such as ceftazidime (30mg), ceftriaxone (30mg), ciprofloxacin (5mg), levo-floxacin (5mg), gentamicin (10mg), imipenem (10mg), meropenem (10mg), piperacillin- tazobactam (100/10mg), cefoxitin (30mg), cefuroxime (30mg), amoxicillin/ clavulanic acid (20/10mg), aztreonam (30mg) and amikacin (30mg), tigecycline (15mg), and colistin (10mg), (Oxoid, England).
Phenotypic detection of ESBls and MBLs
All strains were examined for ESBL production by CLSI phenotypic corroborative test by double-disk diffusion method12. ESBL generation was affirmed by E-test ESBL strips (AB Biodisk, Solana, Sweden).
All strains were investigated for Metallo-betalactamase production by utilizing the Etest MBL strip (AB Biodisk, Solana, Sweden), containing two-sided seven-dilution scope of imipenem(IP) and imipenem+ EDTA (IPI). The test was done with accordance to the manufacturer’s directions MIC ratio of ³8 for the two reagent sides, a phantom zone between IP/IP inhibitor and distortion of each ellipse was considered as a positive result13.
Statistical analysis
All data were stored in Microsoft Excel, Version 2016. Data management and statistical analyses were also performed in Excel. descriptive statistics of the data and variables are shown in the form of frequencies and percentages.
ETHICAL STATEMENT
The Institutional Review Board, college of medicine, King Khalid University Hospital, Riyadh, Saudi Arabia, has granted permission to conduct this study.
Bacterial isolates
The study included 70 isolates from different body sites comprising 25 (35.7%) were P. aeruginosa, 23 (32.9 %) were K. pneumoniae, 16 (22.9 % ) were E. coli, and 6( 8.6 %) were A. baumannii, are shown in table 1.
Table (1):
Frequencies and percentages of various Gram-negative bacteria isolated from ICUS ( n = 70)
Organisms |
Frequency (N=70) |
Percentage |
|---|---|---|
Pseudomonas aeruginosa |
25 |
35.7 % |
Klebsiellapneumoniae |
23 |
32.9% |
Escherichia coli |
16 |
22.9% |
Acinetobacterbaumannii |
6 |
8.6 % |
The samples were secluded from numerous clinical specimens such as tracheal aspirate(n=23; 33%), sputum (n= 13 ;19%),urine (n= 11 ;16%), blood (n=10 ; 14%), wound ( n=5 ;7% ), eye (n=3; 4%), body fluid (n= 2; 3%), genital, groin and tissue ( n=3;3%), mainly from ICUs (SICU, n=23 ; 33%, MICU, n=20; 29%, NICU, n=15; 21% , CCU, n= 6; 9%, PICU, n= 4;6% , CICU, n=2;3%. From different age groups, (>61-100 ,n/N: 24/70: 34%, >31-60, n/N: 16/70: 23%,<1, n/N: 15/70: 21%, >16-30, n/N: 11/70: 16%, >1-15, n/N: 4/70: 6 %. Forty isolates which represents (57%) were from female and thirty isolates (43%) from male. The demographic characteristics of the patient populations are shown in ( Fig. 1).
Antibiotic susceptibility testing
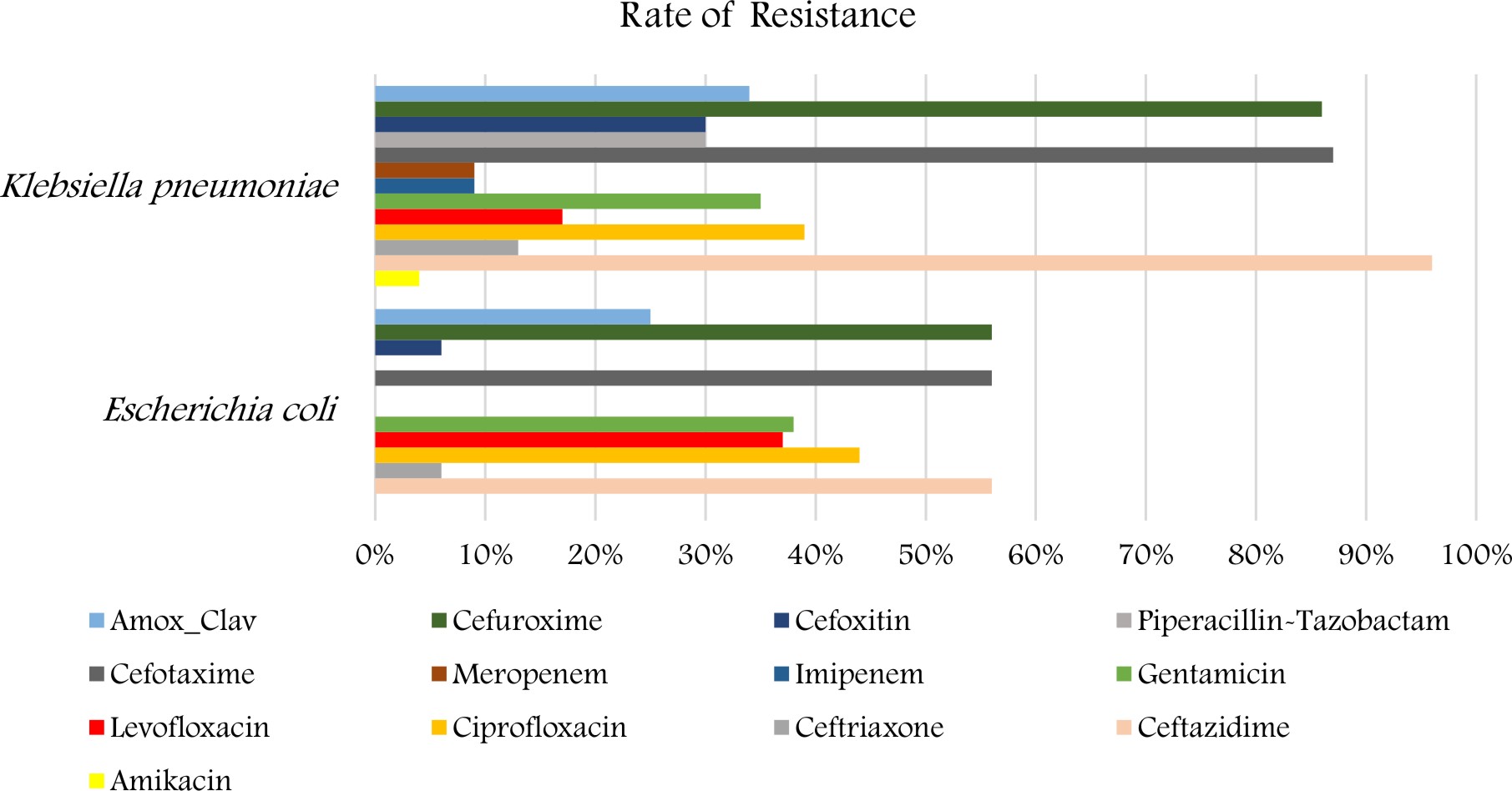
The antibiotic resistance pattern of K. pneumoniae and E. coli is shown in (Fig. 2). The isolates of K. pneumoniae demonstrated a high percentage of resistance to Ceftazidime 22 (96%), followed by cefotaxime and cefuroxime 20 (87%), ciprofloxacin 9 (39%), amoxicillin/clavulanic, gentamicin 8 (35 %). The strains revealed a low percentage of resistance to meropenem, I mipenem 2 (8.6% ) and to amikacin 1 (4%). Also for E.coli displayed a high percentage of resistance to Ceftazidime, cefotaxime and cefuroxime 9 (56%) also followed by Ciprofloxacin 7 (44%), levofloxacin and gentamicin 6 (38%), amoxicillin/clavulanic 4 (25%). The strains revealed a low percentage of resistance to Ceftriaxone 1(6%).
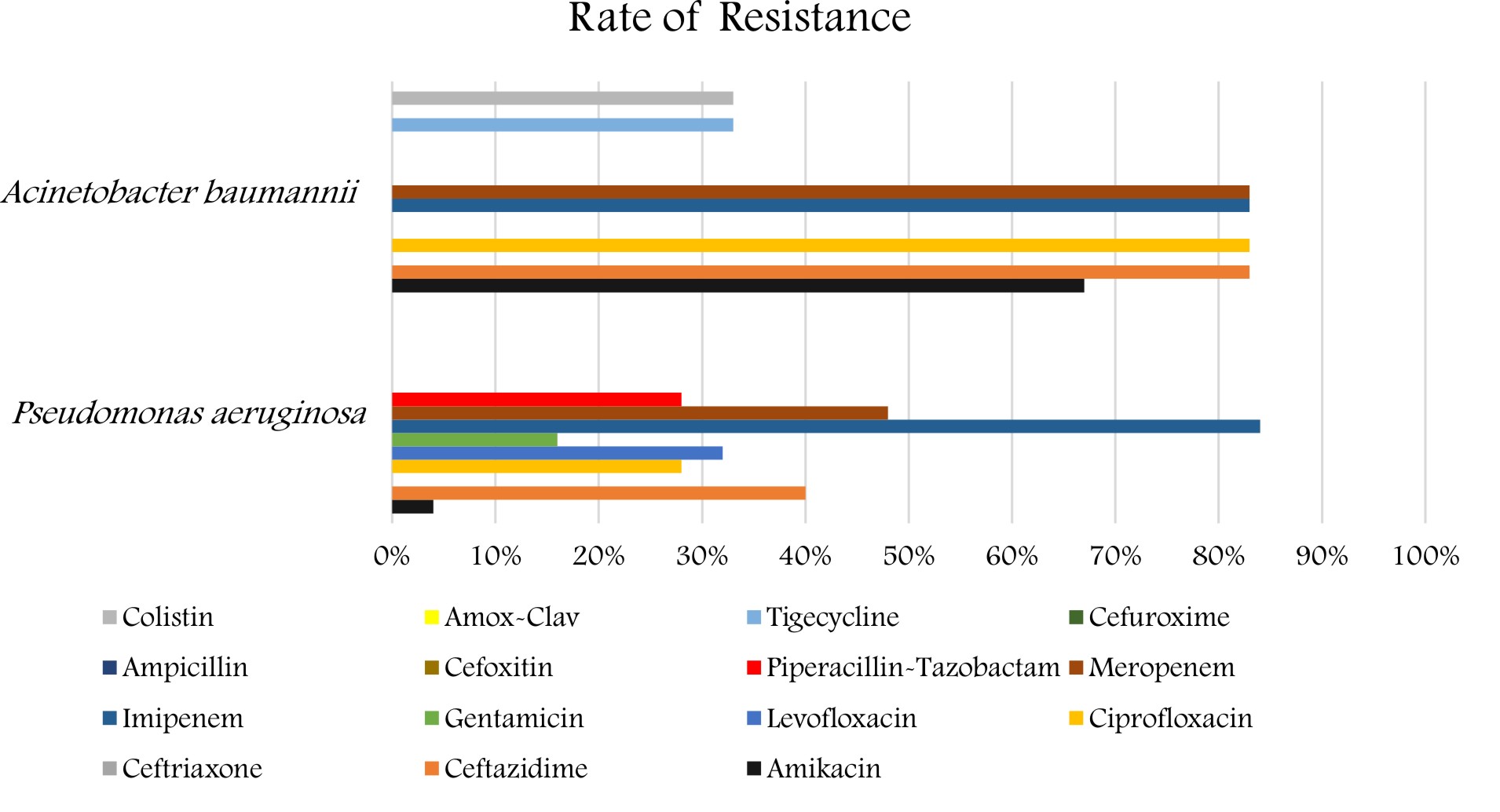
As for the P. aeruginosa and A. baumannii, the antibiotic resistance pattern is shown in (Figure 3). The isolates of the P. aeruginosa revealed a high percentage of resistance to imipenem and meropenem 21(84%) and 12 (48%), followed by ceftazidime 10(40%), levofloxacin 8 (32%), ciprofloxacin and pipe-racillin/tazobactam showed the same percentage of resistance 7(28%), followed by amikacin 1 (4%). The strains revealed a low percentage of resistance to gentamicin 4(16%). Also, the A. baumannii strains revealed a high percentage of resistance 5(83%) for imipenem, meropenem, ciprofloxacin, ceftazidime, and gentamicin, followed by amikacin 4(67%) followed by Tigecycline and colistin 2 (33%).
Phenotypic detection of ESBLS and MBLS
Among the 23 isolates K. pneumonia and 16 of E. coli ,19 (82.6%) and 16 ( 100%) were ESBL producers, sequentially. As opposed to this outcome, ESBL was not detected in any strains of P. aeruginosa or in A. baumannii.
All P. aeruginosa and A. baumannii strains were carbapenem-resistant. MBL was detected in all the specimens of P. aeruginosa, A. baumannii and 4 (17.4%) of K. pneumonia where E coli specimens did not demonstrate any MBL action (Table 2).
Table (2):
A number of ESBL and MBL producing bacteria detected by phenotypic tests.
Organism |
ESBL ( N = 35) |
MBL N = 35 |
|---|---|---|
Acinetobacterbaumannii |
0 |
6 |
Klebsiellapneumoniae |
19 |
4 |
Escherichia coli |
16 |
0 |
Pseudomonas aeruginosa |
0 |
25 |
There is a remarkable increase of multidrug-resistant (MDR) gram-negative bacteria in hospitals and significantly in the intensive care unit (ICU) setting. The clinical outcomes of infections brought about by MDR pathogens as yet dubious14. The resistance pattern is most likely noted in ICUs due to the unlimited use of antibiotics in these units compared to the other hospital departments15 and 70% of these infections were caused by GNB16.
In this study, most of bacterial pathogens were recovered from female patients. This may be due to higher antibiotic intake prior to hospitalization in women17. In contrast with Ibrahim’s findings (2018), who stated that the majority of bacterial pathogens were recovered from male patients 18. Moreover, most of the isolates were recovered from the respiratory system and this finding agrees with a previous surveillance studies in Riyadh indicated that the most of bacterial strains were from the respiratory tract19, 20. Our findings indicated that respiratory diseases were the most widely recognized in our ICUs.
In the current study, the most commonly isolated organism was P. aeruginosa followed by K. pneumoniae, E. coli, and A. baumannii.
In a study of Altun et al., (2013), who studied 78 specimens isolated from ICUs, Altun et al., (2013) found that A. baumannii was the most normally isolated organism pursued by Klebsiella spp., E. coli, and P. aeruginosa21.
According to Al Johani et al., (2010), who found that Acinetobacter baumannii was the most frequently organism, pursued by Pseudomonas aeruginosa, Escherichia coli, Klebsiella pnemoniae, Stenotrophomonas maltophilia, and Enterobacter22. Also, Ibrahim (2018) reported that the most commonlysecluded pathogens were Acinetobacter spp. (27.2%) followed by P. aeruginosa (23.8%) and K. pneumoniae (18.6%)18.
Here in our study, the isolates of K. pneumoniae demonstrated a high percentage of resistance to Ceftazidime (96%), followed by cefotaxime and cefuroxime (87%), ciprofloxacin (39%), amoxicillin/clavulanic and gentamicin (35 %).Ibrahim (2018)reported that more than 60% of K. pneumoniae strains were resistant to cephalosporins, trimethoprim/sulfametho-xazole, aztreonam, piperacillin, and piperacillin/tazobactam. The resistance rates for amino-glycosides, carbapenems, and cipro-floxacin were between 40% and 55%. In another setting in Saudi Arabia, K. pneumoniae demonstrated the highest resistance rates for ceftriaxone (59.4%), aztreonam (58.3%), and ceftazidime (58.3%)18.
Results of the present study reported that E. coli specimens demonstrated a high percentage of resistance to Ceftazidime, cefotaxime, and cefuroxime (56%). Amer et al., (2017), stated that most of E. coli isolates exhibited ESBL activity, and resistance is significantly increased in all beta-lactams antibiotics especially ceftazidime (38% to 100%); while the previous surveillance study showed E. coli resistance to ceftazidime (24% to 54%)23.
In our study, P. aeruginosa showed a high percentage of resistance to imipenem and meropenem (84%) and (48%). Where Emeka et al., (2018), reported that resistance percentages for P. aeruginosa were as per the following: Augmentin (47.4%), Cefuroxime (52.6%), Ceftriaxone (100%)24.
In the present investigation, A. baumannii isolates showed a high percentage of resistance(83%) for imipenem and meropenem which is similar to the finding of Ibrahim (2018), who reported that high resistance rates for carbapenems such as imipenem (97.5%) and meropenem (96.2%)18. The high resistant rates pose a major concern in our ICU as carbapenems are the medication of decision for Acinetobacter infections. Our findings suggest the existence of carbapenemase-producing strains, which has also been reported in other studies in Saudi Arabia and Gulf Cooperation Council States25,26. Therefore, determining the carbapenem-resistance mechanisms could lead to improvements in the outcomes of infections brought about by these microscopic organisms25.
In the present investigation, ESBL was detected in 35 of 39 (90%) isolates (19 of K. pneumonia and 16 of E. coli). ESBL was distinguished by Vitek 2 compact system.
In the investigation of Altun et al.21. ESBL was affirmed in 19 of 27 (70%) of the bacterial specimens (12 E. coli and 19 K. pneumonia), ESBL was distinguished by Vitek 2 compact system. In the investigation of Baykal et al., ESBL was affirmed by combined disc method in 26 E. coli and 70 K. pneumoniae strains that were examined27.
In the current investigation, MBL was affirmed in all the bacterial specimens of P. aeruginosa and A. baumannii 31 of 35 (86%). P. aeruginosa displayed resistance to anti-infection agents as a consequence of a permeability barrier of the cell surface, multidrug efflux pumps, and yielding of b-lactamases (AmpC b- lactamase, ESBLs, and MBLS)28. MBL generation in P. aeruginosa in the current investigation (71% ) is comparable with Altun et al.21(57% ) and Tankhiwale (16.87%)29.
Carbapenemase- generation Acinetobacter strains are rising worldwide due to frequent carbapene mutilization and the choice of resistant bacteria under antibiotic pressure 30-32.
In our study, it was found that 6 of 6 (100%) A. baumannii strains were MBL producers by E test. Where Altun et al.(2013) stated that 31 of 44 (70%) A. baumannii strains were detected as MBL producers21.
To sum up, Gram-negative bacterial resistance is still a major issue in KKUH ICUs, Riyadh ESBL and MBL were the fundamental resistance patterns of the strains assessed in this investigation. The most commonly secluded organisms were P. aeruginosa pursued by K. pneumonia, E. coli, and A. baumannii. ESBL was confirmed in 35 of 39 (90%) isolates (19 of K. pneumonia and 16 of E. coli. And MBL was affirmed in all the specimens of P. aeruginosa and A .baumannii 31 of 35 (86%). Imipenem, Meropenem, and Amikacinwere the most effective agents for K. pneumonia and E. coli. Colistin was the drug of choice for most cases of A. baumannii. P. aeruginosa, Amikacin and Colistin were the most effective agents.
This research project was funded by the Deanship of Scientific Research, Princess Nourah Bint Abdulrahman University, through the Research Group Funding Program, grant No 37 – ص – 177.
The authors declares that there is no conflict of interest.
- Exner, M., Bhattacharya, S., Christiansen, B., Gebel, J., Goroncy-Bermes, P., Hartemann, P., et al. Antibiotic resistance: What is so special about multidrug-resistant gram-negative bacteria? GMS Hyg. Infect.Control, 2017;.12: Doc05. doi: 10.3205/dgkh000290
- Slama, Thomas G. “Gram-Negative Antibiotic Resistance: There Is A Price To Pay”. Critical Care12, 2002; 4: S4. Web.
- Yong D, Lee K, Yum JH, Shin HB, Rossolini GM, et al. Imipenem-EDTA disk method for differentiation of metallo-beta-lactamase-producing clinical isolates of Pseudomonas spp. and Acinetobacter spp. J. Clin. Microbiol., 2002; (40): 3798-3 801.
- Noyal M J C, Menezes G A, Harish B N, Sujatha S and Parija S C. Simple screening tests for detection of carbapenemases in clinical isolates of non-fermentative Gram-negative bacteria. Indian Journal Medical Research, 2017; (129): 707-712.
- Simoens S, De Corte N and Laekeman G. Clinical practice and costs of treating catheter-related infections with teicoplanin or vancomycin. Pharmacy Practice, 2006; 4(2): 68–73.
- Yazdi M, Nazemi A, Mirinargasi M, Jafarpour M and Sharifi S H. Genotypic versus Phenotypic methods to detect Extended-Spectrum Beta lactamases (ESBLs) in Uropathogenic Escherichia coli. Annuals of Biological Research, 2012 ; 3(5): 2454-2458.
- Schill, F., Abdulmawjood, A., Klein, G., and Reich, F. Prevalence and characterization of extended-spectrum beta-lactamase (ESBL) and AmpCbeta lactamase producing Enterobacteriaceae in fresh pork meat at processing level in germany. Int. J. Food Microbiol., 2017; (257): 58–66.
- Lavilla S, Gonzalez-Lopez J J, Miro E, Dominguez A, Llagostera M, Bartolome R M, et al. Dissemination of Extended-Spectrum b-lactamase-producing bacteria: the food-borne outbreak lesson. Journal Antimicrobial Chemotherapy, 2008; (61): 1244-1251
- Queenan AM, Bush K . Carbapenemases: the versatile beta-lactamases. Clin. Microbiol Rev., 2007; (20): 440–458.
- Cornaglia G, Akova M, Amicosante G, Canton R, Cauda R, Docquier J D, et al.Metallo b- lactamases as emerging resistance determinants in Gram- negative pathogens. International Journal Anti-microbial Agents, 2007; (29) : 380- 388.
- Varaiya A, Kulkarni N, Kulkarni M, Bhalekar P, Dogra J. Incidence of metallo beta lactamase producing Pseudomonas aeruginosa in ICU patients. Indian J. Med. Res., 2008; (127): 398-402
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. Wayne (PA), USA: M100-S25; 2015
- Walsh Tr, Bolmstrom A, Qwarnstrom A, Gales A. Evalution of a new E test for detecting metallo – beta lactamase in routine clinical testing. J. Clin. Microbiol., 2002; 40: 2755-2759.
- Paramythiotou.E and Routsi. C. Association between infections caused by multidrug-resistant gram-negative bacteria and mortality in critically ill patients. World J. Crit. Care Med., 2016; 5(2): 111-120.
- Radji M, Fauziah S, Aribinuko N. Antibiotic sensitivity pattern of bacterial pathogens in the intensive care unit of Fatmawati Hospital, Indonesia. Asian Pacific Journal of Tropical Biomedicine, 2011; 1(1): 39-42.
- Hospital-Acquired Infections Due to Gram-Negative Bacteria”. New England Journal of Medicine, 363. 15(2010): 1482-1484. Web.
- Rammaert, B., Goyet, S., Beautי, J., Hem, S., Te, V., Try, P. L., Mayaud, C., Borand, L., Buchy, P.,Guillard, B., &Vong, S. Klebsiellapneumoniae related community-acquired acute lower respi-ratory infections in Cambodia: Clinical charac-teristics and treatment. BMC Infectious Diseases, 2012; 12(1), 3. DOI: 10.1186/1471-2334-12-
- Ibrahim ME. High antimicrobial resistant rates among Gram-negative pathogens in intensive care units. Saudi Med. J., 2018; 39(10): 1035–1043
- Sahu M, Siddharth B, Choudhury A, Vishnubhatla S, Singh S, Menon R, et al. Incidence, microbiological profile of nosocomial infections, and their antibiotic resistance patterns in a high volume cardiac surgical intensive care unit. Ann Card Anaesth. 2016; 19:281.
- Moolchandani K, Sastry AS, Deepashree R, Sistla S, Harish BN, Mandal J. Antimicrobial resistance surveillance among intensive care units of a tertiary care hospital in South India. J. Clin Diagnostic Res., 2017; 11: DC01–DC07.
- Altun S, Tufan Z K, Server Yaנcl S, ײnde U , Bulut C, Knk S, and Demiroz A P. Extended Spectrum Beta-lactamases, AmpC and Metallo Beta-lactamases in Emerging Multi-drug Resistant Gram-negative Bacteria in Intensive Care Units. Open Access Scientific Reports, 2013; 2: 707 doi:10.4172/scientificreports.707
- Al Johani SM, Akhter J, Balkhy H, El-Saed A, Younan M, Memish Z. Prevalence of antimicrobial resistance among gram-negative isolates in an adult intensive care unit at a tertiary care center in Saudi Arabia. Ann Saudi Med., 2010; 30(5):364-9. doi: 10.4103/0256-4947.67073
- Amer RR, Alzomaili BT, AlTuwaijri RM, AlZahrani R S, AlHarbi SH, AlThubaitiA and Al Johani SM. Prevalence of Antimicrobial Resistance among Gram-Negative Isolates in an Adult Intensive Care Unit at a Tertiary Care Center in Saudi Arabia (2010-2014). Global Journal of Medical Research K. Interdisciplinary, 2017; 17(4): 1.0
- Emeka L.B, ulhadi Al-Sultan, A.A, Alrashed A.S, Alhaddad, M.S and Al-Barjas, A.K . Antimicrobial susceptibility pattern of Gram negative bacteria isolated from intensive care units in Al-Ahsa, Kingdom of Saudi Arabia. African Journal of Microbiology Research, 2018; 12(31), pp. 747-753.
- Memish ZA, Assiri A, Almasri M, Roshdy H, Hathout H, Kaase M, et al. Molecular characterization of carbapenemase production among gram-negative bacteria in Saudi Arabia. Microb. Drug Resist., 2015;21:307–314
- Zowawi HM, Sartor AL, Sidjabat HE, Balkhy HH, Walsh TR, Al Johani SM, et al. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii isolates in the Gulf Cooperation Council States: dominance of OXA-23-type producers. J. Clin. Microbiol., 2015; 53: 896–903.
- Baykal A, ַצpl N, ×im÷ek H, Esen B, G r D . The presence of extended spectrum beta-lactamase, KPC-type carbapenemase and plasmid-mediated AmpC b-lactamase in E.coli and K. pneumoniae strains isolated from blood cultures. Mikrobiyol Bul., 2012; 46: 159-169
- Pai H, Kang CI, Byeon JH, Lee KD, Park WB, et al. Epidemiology and clinical features of blood stream infections caused by AmpC-type-b- lactamase producing Klebsiella pneumoniae. Antimicrob Agents Chemother., 2004; 48: 3720-3278.
- Tankhiwale S. Beta-lactamases in P. aeruginosa: A threat to clinical therapeutics. Curr. Pediatr. Res., 2016; 20(1&2): 253-257.
- Esterly J, Richardson CL, Eltoukhy NS, Qi C, Scheetz MH. Genetic mechanisms of antimicrobial resistance of Acinetobacter baumannii., Ann Pharmacother, 2011.
- Taneja N, Singh G, Singh M, Sharma M . Emergence of tigecycline and colistin resistant Acinetobacter baumanii in patients with complicated urinary tract infections in north India. Indian J. Med. Res., 2011; 133: 681-684.
- Goel N, Wattal C, Oberoi JK, Raveendran R, Datta S, et al. Trend analysis of antimicrobial consumption and development of resistance in non-fermenters in a tertiary care hospital in Delhi, India. J. Antimicrob. Chemother., 2011; 66: 1625-1630.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.