ISSN: 0973-7510
E-ISSN: 2581-690X
Foodborne diseases and infection caused by associated pathogens is a public health concern. Majority of the investigations focus on common foodborne pathogens like Vibrio parahaemolyticus, Escherichia coli, Listeria monocytogenes, Shigella, Salmonella and Staphylococcus aureus. Limited knowledge has been accounted on Klebsiella pneumoniae. Presence of multidrug-resistant K. pneumoniae in the food supply is disturbing. Hence, this study assessed the presence of K. pneumoniae isolates from food samples (fresh vegetables and chicken), ascertained the presence of drug-resistant phenotypes, extended spectrum beta lactamase production, antibiotic resistance determinants, genes associated with virulence and their ability to form biofilm. Resistance towards ceftazidime and tetracycline was noted among all the isolates in the study, while they exhibited sensitivity to chloramphenicol and co-trimoxazole. All the isolates were potent ESBL producers carrying at least one ESBL encoding genes. Plasmid mediated quinolone resistance gene was detected in one isolate each from onion and chicken respectively. The isolates marked the absence of tetracycline and chloramphenicol resistance genes. Multiple virulence genes (ureA, khe, fimH, mrkD, wabG, uge and elt) were possessed by each of the isolates. K. pneumoniae from chicken and cucumber were moderate biofilm formers and those from tomato exhibited weak biofilm formation. Increased expression of the mrkA gene and reduction in the expression of the biofilm forming gene fimH gene was observed among the biofilm formers. One of the moderate and non-biofilm formers exhibited increased mrkD gene expression. The results from our study stipulate, that raw vegetables and meat serve as dormant source of drug-resistant and virulent K. pneumoniae.
Klebsiella pneumoniae, Multidrug-resistance, ESBL Production, Resistance Determinants, Virulence Factors, Biofilm Formation
Klebsiella pneumoniae is acknowledged as a momentous pathogen causing various infections in humans that include diarrhoea, pneumonia, septicaemia and liver abscesses. Besides, the clinical environment K. pneumoniae has been isolated from several foods including fish, meat, powdered infant food, raw vegetables and street foods.1 Extensive use of antibiotics in agriculture, veterinary and clinical practices has resulted the emergence of drug resistant K. pneumoniae strains.
Genetic elements viz plasmids, integrons and transposons further aid the spread of the resistance genes. In addition, persistence of these genes in the environment makes the control of drug-resistant pathogens strenuous. Currently, K. pneumoniae is resistant towards broad spectrum drugs like fluoroquinolones and aminoglycosides.2 Extended spectrum β-lactamases (ESBLs) offer broad spectrum of resistance towards penicillin’s, tetracyclines cephalosporins, sulfonamides, chloramphenicol and trimethoprim. K. pneumoniae are ready receptors of the R plasmids and found to have significant involvement in several nosocomial epidemic diseases. The plasmid mediated multi-drug resistant genes could possibly be transferred from pathogens of fish, animal and human origin to the susceptible strains of a diverse ecological niche.3 Therefore, the issue of resistance among the bacterial populations persists to be a critical public health concern.
Among all the other possible ESBL producers, K. pneumoniae is one of the dominant organisms. Although carbapenems are believed as the first line of drugs in the treatment of ESBL producing K. pneumoniae infections, the resistance has been steadily increasing.4 The ESBL producing K. pneumoniae is to be considered a critical affair due to limitations in the treatment choices and alarming rates of morbidity and mortality. Both humans and animals exposed to the ESBL producing strain by the consumption of contaminated water or food sources, could be at a greater risk of colonization and infection by the pathogen. In K. pneumoniae, ESBL genes belonging to the CTX-M, SHV and TEM families are familiar and clinically relevant, with CTX-M being the most predominant.
Formation of biofilms by food-related pathogens is a matter of consideration, in addition a predisposing factor for contamination of food sources. Microbes in a community secrete cement-like substances that serve as a glue, protecting them against host macrophages and antibiotics.5 Because, of which eradication of the biofilm producing pathogens from the indwelling surfaces. Pathogenic microorganisms like K. pneumoniae can attach, grow on food surfaces by forming biofilms. Formation of biofilms are one of the major virulence factors causing chronic infections.6 Inadequate hygiene in the food processing sectors facilitate bacterial adherence, biofilm formation and serve as reservoirs for various infectious agents.7 Thereby, food contamination continues to be a threat and burden on local and global markets.
Besides biofilms, fimbrial adhesins, production of siderophores, nature and synthesis of the polysaccharide capsule collectively contribute to the pathogenicity of K. pneumoniae. In addition, resistance to drugs, production of phospholipase and hemolysin are considered to be the potent virulence intensifiers of the pathogen. Conventionally, the food sector is considered to be a subtle affair that provokes fear to the food industry and also the common man, if contaminated.
The reciprocity amidst the resistance mechanisms and presence of virulent traits in K. pneumoniae is not substantiated. However, it was previously established that drug-resistance comes with a fitness cost and decreased virulence. At present, reports have proposed increased virulence to the developing resistance. Additionally, virulence is noted to evolve naturally as a response to resistance acquired, or possibly shared among bacteria.8
In country like India, where antibiotics are used indiscriminately not only in the hospital settings, but also in the agriculture and food industries, drug-resistant pathogen like K. pneumoniae becomes prevalent and is a public health implication, often neglected.
Hence, the present study aimed to isolate and characterize K. pneumoniae from commonly used food sources. Antibiotic susceptibility pattern, ESBL production, antibiotic resistance genes and virulence factors of the isolates were also investigated. Ability of the isolated bacteria in forming biofilms and their associated gene expression were also studied.
Sample Collection
In total 30 samples derived from chicken (10), onion (10), tomato (5) and cucumber (5) collected from a local market in Mangaluru were used for the isolation of K. pneumoniae. Samples collection was done in pre-sterilized containers under aseptic conditions. The samples were stored in 4°C and were processed further.
Isolation of the K. pneumoniae and Phenotypic Identification
Collected samples (25 g) were weighed and enriched in 250 ml of heat sterilized nutrient broth (HiMedia, India) following an incubation of 37°C for 24 h. A loopful of the inoculum was streaked onto MacConkey agar (HiMedia, India) and further incubated at 37°C for 24 h. Pink mucoid colonies indicative of typical K. pneumoniae colonies were selected and re-grown on Luria Bertani (LB) broth (HiMedia, India). Gram-staining, motility, capsule staining, series of biochemical tests (Indole, methyl-red, Voges-Proskauer, citrate -utilization, catalase and oxidase) were performed for re-conformation of K. pneumoniae isolates. Phenotypically confirmed isolates were stored at -80°C in LB broth containing 30% glycerol.
Antibiotic Susceptibility Testing
Antibiotic susceptibility testing was carried out for all the confirmed isolates K. pneumoniae by Kirby Bauer’s disc diffusion following the guidelines of Clinical and Laboratory Standards Institute (CLSI).9 Antibiotics (HiMedia, India) included in the study were chloramphenicol (30 μg), cefotaxime (30 μg), ceftazidime (10 μg), cefuroxime (30 μg), ciprofloxacin (5 μg), nalidixic acid (30 μg), imipenem (10 μg), meropenem (10 μg), tetracycline (30 μg), co-trimoxazole (25 μg) and. Based on their zone of inhibition K. pneumoniae isolates were assigned as resistant, intermediate and susceptible. As a reference strain Escherichia coli ATCC 25922 was used.
Double Disk Diffusion to Assay Production of ESBL
Phenotypic confirmation of the ESBL production by potent ESBL producing K. pneumoniae from food samples was affirmed by the use of and by a combination of both ceftazidime (30 μg) and clavulanic acid (10 μg) placed 25mm away from each other.
Isolates of K. pneumoniae exhibiting ≥5 mm increase around combined clavulanate discs in comparison to the ceftazidime disc alone were confirmed to produce ESBL.
Molecular Confirmation
Isolates obtained were genotypically confirmed as genus Klebsiella and species K. pneumoniae by extraction of DNA from the isolates by cetyl trimethyl ammonium bromide (CTAB) as reported by Ausubel et al.10 and subjecting them to polymerase chain reaction. The gene gyrA was used as a target for the identification of isolates as genus Klebsiella, while the 16S rDNA -23S rDNA internal transcribed spacer (ITS) region was used for the species-specific confirmation. Details of the primers used is provided in Table S1.
Identification of Antibiotic Resistance Determinants and Genes Associated with Virulence
All the K. pneumoniae isolates were investigated for their respective antibiotic resistance determinants and virulence genes by PCR. Genes encoding for ESBL (blaTEM, blaCTX-M and, blaSHV). Tetracycline resistance (tetA, tetB and tetG), plasmid mediated quinolone resistance (qnrA, qnrB, qnrS), chloramphenicol resistance (catA1), sulphonamide resistance gene (sul1 and sul2) (Table S2).
Presence of nine different virulence genes viz ureA (urease), uge (epimerase), wabG (transferase), kfu (iron uptake system), khe (haemolysin), bfp (bundle forming pilli), elt (enterotoxin), fimH and mrkD (fimbrial adhesins) were investigated by PCR in the K. pneumoniae isolates (Table S3 and S4).
Biofilm Formation Assay
96-well Microtiter Plate Method
Biofilm forming ability of K. pneumoniae food isolates was assessed by microtiter plate method. To brief, (10 μl) K. pneumoniae isolates grown overnight were allowed to form added biofilm by incubated them at 37°C for 24 h in wells containing LB broth (190 μl). Sterile distilled water
was used to wash and remove the cells from the well. Methanol was used to fix the cells followed by crystal violet (1%) staining and incubation for 15 min. Glacial acetic (30%) acid was used to solubilize the stain adhering the cells forming biofilm, followed by measuring their absorbance at 630 nm. True OD values were calculated by deducting the control value. The isolates were classified as follows (i) 4*O.D.c< O. D=strong biofilm former (ii) 2*O.D.c) <O. D≤ (4*O.D.c) moderate biofilm former (iii) O.D.c < O. D≤(2*O.D.c) weak biofilm former (iv) O D≤O.D.c non-biofilm former.11
Expression of Biofilm Related Genes
K. pneumoniae isolates were grown for 24 h at 37°C in a 6 well cell culture plate. As a negative control LB broth and as positive control K. pneumoniae ATCC 10031 were used following incubation, Trizol reagent (1 ml) was added to lyse the cells. Chloroform (200 µl) was added following centrifugation (12000xg at 4°C for 15 min). Isopropyl alcohol (0.5 ml) was used to precipitate the RNA from the aqueous phase. Ethanol (75%) was used to pellet the cells and was resuspended in nuclease-free water. Concentration and purity of the obtained RNA were examined by using a spectrophotometer (Biospectrometer, Eppendorf, Germany). The RNA samples obtained were stored (-80°C) for the synthesis of cDNA.
DNase Treatment and cDNA Synthesis
Thermo scientific kit was used to convert RNA to cDNA. To 0.5 g of the RNA sample reaction buffer (1 µl) with MgCl2 (10 x) and DNase (1 µl) was added. DEPC- treated water was used to make up the volume to 10 µl. Incubation step for 30 min at 37°C following deactivation at a temperature of 65° C for 10 min. For cDNA synthesis the RNA samples obtained were normalized to 500 nanograms. Prime Script one-step reverse cDNA synthesis kit (TaKaRa, Japan) was used to reverse transcribe the RNA samples. The cDNA samples were stored at -20°C for the expression studies.
Real-time PCR (q-PCR) Studies to Quantify Associated Biofilm Genes in K. pneumoniae
Relative gene expression (fimH, mrkA, and mrkD) of the biofilm forming genes in K. pneumoniae food samples were quantified. PRIMER 3 software was used to design primers for the biofilm-related genes in K. pneumoniae (Details of the primers used are given in Table S4). Relative expression of three target genes were standardize in comparison to the house keeping gene (rpoA). Standardization of primer concentration for all the biofilm genes was performed and used henceforth. The cycling conditions were as follows: initial denaturation of 95°C for 3 min, followed by 45 cycles of denaturation at 95°C for 10 sec, 55°C for 20 sec and extension of 72°C for 20 sec. Melt curve was analysed to exempt the amplification untargeted regions. Relative quantification of all the genes was calculated using the 2-ΔΔct formula.12
Isolation and Identification of K. pneumoniae
All the isolates formed lactose fermenting typical pink mucoid colonies on MacConkey agar. Gram-negative rods were observed, upon Gram staining. The isolates showed positive for capsular staining, Voges-Proskauer, citrate production and catalase test. Negative for indole, methyl-red, oxidase, motility test and spore staining.
Antibiotic Susceptibility Testing and ESBL Production
All the isolates tested for their antibiotic susceptibility were resistance towards ceftazidime and tetracycline. Among the 5 isolates resistance was observed towards cefotaxime (3), ciprofloxacin (2) and imipenem (1) respectively. Isolates exhibited intermediate susceptibility towards nalidixic acid (2), meropenem (2) and cefuroxime (3). All the isolates were sensitive to chloramphenicol and co-trimoxazole followed by imipenem (4). The antibiogram of the isolates is represented in Table (1).
All the K. pneumoniae isolates in the study were potent ESBL producers.
Table (1):
Isolation source, antibiogram and biofilm formation by K. pneumoniae food isolates.
| Isolates | Source of isolation | Biofilm forming ability | Antibiotic susceptibility Pattern | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAZ | CIP | C | COT | CXM | CTX | IPM | MRP | NA | TE | |||
| KP1 | Chicken | Moderate | R | R | S | S | I | S | S | S | I | R |
| S1B | Tomato | Weak | R | I | S | S | S | R | S | I | S | R |
| S2B | Tomato | Weak | R | I | S | S | S | R | S | S | S | R |
| S9 | Cucumber | Moderate | R | R | S | S | S | R | R | I | I | R |
| S13 | Onion | Non | R | S | S | S | S | S | S | S | S | R |
CAZ- ceftazidime, CTX- cefotaxime, CIP- ciprofloxacin, COT- co-trimoxazole, CXM- cefuroxime, C- chloramphenicol, IPM- imipenem, NA- nalidixic acid, MRP- meropenem, TE- tetracycline; R= Resistant, S= Sensitive, I=Intermediate
Genotypic Confirmation of K. pneumoniae
Genus confirmation of the isolates as Klebsiella was done by targeting the gene gyrA, while 16S rDNA -23S rDNA internal transcribed spacer (ITS) region of K. pneumoniae was targeted to confirm the isolates as K. pneumoniae. All the isolates included in the study were confirmed as K. pneumoniae.
Detection of Antibiotic Resistant Determinants
All the ESBL producers in the study carried either one of the ESBL encoding genes. None of the isolates harboured tetracycline resistance genes nor the chloramphenicol resistance genes. One isolate each from chicken (KP1) and onion (S13) harboured the plasmid mediated quinolone resistance (qnrS) and sulphonamide resistance genes (sul1) respectively. Details of the antibiotic resistance genes harboured by the isolates is depicted in Table 2.
Table (2):
K. pnuemoniae isolates harbouring ESBL encoding and antibiotic resistance determinants.
Isolates |
blaSHV |
blaCTX-M |
blaTEM |
tetB |
tetG |
qnrA |
qnrB |
qnrS |
catA1 |
sul1 |
sul2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
KP1 |
+ |
– |
– |
– |
– |
– |
– |
+ |
– |
+ |
– |
S1B |
+ |
+ |
– |
– |
– |
– |
– |
– |
– |
– |
– |
S2B |
+ |
+ |
+ |
– |
– |
– |
– |
– |
– |
– |
– |
S9 |
+ |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
S13 |
– |
+ |
– |
– |
– |
– |
– |
– |
– |
+ |
– |
Determination of Virulence Genes
All the K. pneumoniae isolates in the study carried the genes ureA, khe, fimH and mrkD. None of the isolates harboured the bfp gene. The genes wabG, uge and elt were detected in four of the isolates. One of the isolates from tomato (S1B) carried the kfu gene. Four isolates (KP1, S1B, S2B and S9) found to harbour multiple virulence genes in them. Virulence genes carried by each of the isolates is represented in Table 3.
Table (3):
Details of K. pnuemoniae isolates carrying virulence genes.
Isolates |
ureA |
uge |
wabG |
kfu |
khe |
bfp |
elt |
fimH |
mrkD |
|---|---|---|---|---|---|---|---|---|---|
KP1 |
+ |
+ |
+ |
– |
+ |
– |
+ |
+ |
+ |
S1B |
+ |
+ |
+ |
+ |
+ |
– |
+ |
+ |
+ |
S2B |
+ |
+ |
+ |
– |
+ |
– |
+ |
+ |
+ |
S9 |
+ |
+ |
+ |
– |
+ |
– |
+ |
+ |
+ |
S13 |
– |
– |
– |
– |
+ |
– |
– |
+ |
+ |
Ability of the K. pneumoniae from Food Sources to Form Biofilms
Among the five isolates of K. pneumoniae from food sources, four of the isolates could form biofilms (KP1, S9, S1B, S2B). Of which two isolates could form moderate (KP1, S9) and weak (S1B, S2B) biofilm formers respectively. K. pneumoniae isolated from onion (S13) did not show any biofilm formation. Isolation source and their biofilm forming ability has been represented in Table 1.
Co-relation among Biofilm Forming Ability and Antibiotic Resistance of K. pneumoniae Food Isolates
In our study, the moderate biofilm formers (KP1, S9) were resistant to ciprofloxacin and tetracycline, while they exhibited sensitivity towards co-trimoxazole and cefuroxime respectively. On the other hand, weak biofilm formers (S1B, S2B) were found resistant to tetracycline, cefotaxime and ceftazidime, and sensitive to imipenem, nalidixic acid, co-trimoxazole and chloramphenicol.
Strikingly imipenem, co-trimoxazole, meropenem, nalidixic acid, ciprofloxacin, chloramphenicol, ceftazidime and cefuroxime were effective against the non-biofilm former (S13) in the study.
Quantification of Associated Biofilm Genes in K. pneumoniae
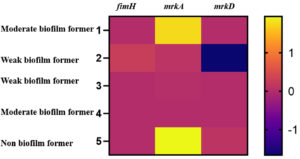
Relative gene expression of fimH (type I) and mrkA and mrkD (type III) fimbrial adhesins in the K. pneumoniae biofilm forming food isolates were analysed (Figure). Weak biofilm formers showed marginal level expression of all the three biofilm related genes. The gene mrkD which facilitates dense biofilm formation in K. pneumoniae exhibited enhanced expression in the non and one of the moderate biofilm formers. The other moderate biofilm former in the study exhibited reduced expression of all the three fimbrial genes (fimH, mrkA and mrkD).
Figure. Expression levels of biofilm associated genes in the K. pneumoniae isolated from food sources.
Levels of biofilm gene expression among the food isolates of K. pneumoniae is depicted by fold-changes transformed to log2 in comparison to the control. The fold-change (log2) value and the colour scale are indicated towards the right of the heat map.
Klebsiella pneumoniae is an opportunistic pathogen encountered from diverse microbiological niches, for instance, food, soil, skin, intestine and faeces of mammals.13 The bacterium has been substantiated to cause pneumonia, bacteraemia and urinary tract infections.
Through the years, resistance towards broad spectrum drugs like the third and fourth generation cephalosporins and fluoroquinolones have been gradually acquired by K. pneumoniae prevalent in both community and hospital settings.14 Carbapenems, being the last line of resort has failed in treating the pathogen and is a matter of utmost concern.
The Centre for Disease Control and Prevention (CDC) stated millions of deaths occurring worldwide annually, is due to the foodborne pathogens. In addition, indiscriminate use of antibiotics in treating the foodborne diseases has increased the resistance offered by the pathogens, towards the antibiotics, eventually leading to ineffectiveness of the drugs.
The present study reports K. pneumoniae from raw vegetables and poultry sources resistant to multiple antibiotics, harbouring ESBL and virulence associated genes, capable of forming biofilms.
The food isolates of K. pneumoniae in our study exhibited higher rates of resistance towards third generation cephalosporins, ceftazidime and cefotaxime. Resistance to third-generation cephalosporins among K. pneumoniae isolates from food have also been reported in other studies.15,16
Cephalosporins have a major role in the hospital settings as therapeutics to Klebsiella related infections. In comparison to other beta-lactam antibiotics, cephalosporins exhibit less sensitivity to inhibition by beta-lactamase enzymes, possessing a greater spectrum of activity.16 World health organization, has entitled the third generation cephalosporins into “watch” category of antibiotics, i.e., drugs crucial in treating bacterial infections in humans and thus must be given precedence as objective of antibiotic resistance investigation and stewardship bulletin.17
K. pneumoniae food isolates exhibiting tetracycline resistance were reported in some of the studies.15,18 In our study there was a discrepancy between the resistance pattern of tetracycline phenotypically and genotypically. Though all the isolates exhibited phenotypic resistance to tetracycline, the tested corresponding genes were not present. This could be due to several reasons, (i) In this study we could test for the presence of tetB and tetG, while the isolates could harbour other variants of tetracycline resistance determinants (ii) resistance towards tetracycline among these isolates could involve other mechanisms like enzymatic inactivation, efflux pumps, mutations in the plasmids etc (iii) resistance to a particular antibiotic phenotypically is due to several genetic mechanisms, reasons of which remain unknown.19 Over and above, tet genes have been proved to persist in the environments, even in the absence of selection pressure established by the constant use of tetracycline.20
Finer understanding of the mechanisms of tetracycline resistance conferred by the pathogens, could direct the design of modified next generation tetracyclines that could cautiously evade inactivation and maintain the efficacy against pathogens.
The jeopardization of antibiotic resistance could magnify by lack of awareness, self-medication and abusage of antibiotics. Confoundingly, all the isolates in the study were susceptible to chloramphenicol and co-trimoxazole, which is in line with the few studies involving K. pneumoniae from raw vegetables, meat and poultry dishes.13,21 Contrary to our findings few authors report complete resistance towards chloramphenicol and co-trimoxazole by the K. pneumoniae isolates from enteral diets.22,23 According to the findings of the current study, chloramphenicol and co-trimoxazole could be the drugs effective on K. pneumoniae isolates from food samples.
ESBLs are rapidly evolving plasmid borne enzymes that can catalyse monobactams and cephalosporins, thereby contributing to a major threat. It was exceptional to note that all the isolates were potent ESBL producers and carried at least one beta lactamase encoding gene (Table 2). Potent ESBL producing K. pneumoniae isolates from food sources were also reported by few other studies.15,24
Dietary intake could be considered one of the possible routes for the introduction of antibiotic resistance genes in bacteria into human gut. Furthermore, such pathogens can transfer the antibiotic resistance determinants to the other strains.
Different food sources including the meat products could be a potential route through which the MDR and ESBL-producing strains of the Klebsiella species could be transmitted. The clinical importance of K. pneumoniae as a pathogen is prevalent, while its dominance as a public health hazard in food requires attention.
Presence of virulence factors viz siderophores, endotoxins and iron scavenging systems in the pathogen have collectively seem to have enhanced its pathogenicity. Assessment of virulence factors in our study suggested ureA, khe, fimH, mrkD, wab, uge, elt and kfu were some of the genes harboured among the K. pneumoniae isolates from food.
All the K. pneumoniae food isolates carried multiple virulence genes (Table 3). Other reports also demonstrate multiple virulence genes in K. pneumoniae food isolates.25,26 These genes aid the bacterium evade the immune system causing diversified infections in animals and humans. One of the isolates in our study from tomato (S1B) harboured the gene kfu. The gene is identified to be involved with capsule formation, hypermucoviscosity virulent phenotype, tissue infections. The gene kfu is recognized as a pivotal gene in up take from the host cell.27
Though the isolates in the current study were sensitive to few antibiotics, they were found to harbour virulence genes in them (Table 1 & 3). In order to improvise treatment and prevention of Klebsiella related infections, an epidemiological relationship amidst the food and clinical isolates must be scrutinized.
Foremost step in the establishment of infections by pathogens like K. pneumoniae is, their competence in forming biofilms on the surface of the host. Biofilm formation significantly scales down the activity of the antimicrobials making it less effective on the bacterium. Few distinctive genes involved in the formation of biofilms are type I fimbriae (fimA and fimH), type III fimbriae (mrkA and mrkD), allantoin (allS), capsular polysaccharide (CPS) (k2A, wzyk2, treC, wabG, cpsD, wcaG, and wzc), aerobactin (iutA), colonic acid (wcaJ) and quorum sensing (QS) (luxS).28 The type 1 fimbriae are significant in the establishment of urinary tract infections in bacteria like E. coli and Klebsiella.29 Binding of the pathogen to the epithelial and endothelial cells of respiratory tract and urinary tracts is mediated by the type III fimbriae.30
In our study, the moderate biofilm formers were from chicken and cucumber respectively. Weak biofilm formers were found to be from tomato and the non-biofilm former from onion.
The gene mrkA showed increased expression among the biofilm formers. It has been investigated that the expression of the type 1 fimbriae is regulated by the fim-switch regulatory mechanism that contains the promoter for the major fimbrial subunit fimA. Nullification in the expression of the type I fimbriae is compensated by the upregulation of the type III fimbriae and vice versa, indicating a strong cross-regulation of the two fimbrial gene clusters.31 Complementary to the above hypothesis it was also suggested that the type I and type III fimbriae are able to compensate for each other and both promote biofilm formation.32 Mutational studies have demonstrated that the type III fimbriae have seem to promote biofilms in K. pneumoniae.33 Whereas, type 1 fimbriae have been reported to aid formation of biofilm in E. coli. However, in the present study, there was no impact found on the formation of biofilms as a result of reduced expression of the fimbrial genes. Per contra, there is a contradictory in the conclusion with respect to the genes involved in the biofilm formation. Further studies are necessary to augment on the association between the biofilm formation and associated genes.
The investigations from the study favours the fact that K. pneumoniae is considered a commensal of the upper respiratory and the gastro-intestinal tract in both mice and humans. However, the ability of the food pathogen to produce ESBL, presence of multiple virulence genes aids the pathogen to evade and infect the GI tract is the most panicking. Multiple virulence genes harboured by the isolates might provide an explanation for infections caused by K. pneumoniae and a major threat for the microbiological security. Furthermore, the drug-resistant phenotypes of the K. pneumoniae from food samples make the treatment regimens firm.
Thereby, the present study aimed to generate knowledge on the prevalence, resistance genes, virulence and biofilm forming ability as a potential factor of vulnerability in the ESBL producing K. pneumoniae from raw vegetables and chicken available for human consumption.
Current study detects the presence of various virulence genes in the food isolates of K. pneumoniae conferring resistance to antibiotics. It is worrisome to observe drug-resistant and potentially virulent K. pneumoniae in foods that could be consumed raw (onion, cucumber and tomato). K. pneumoniae from chicken and vegetables exhibiting resistance to antibiotics is a state of concern, as choices exposed to their antibiotic treatment stands straitened, should these isolate cause infections in humans. On this account, our study elucidates the significance in controlling the dissemination of antibiotic resistance determinants by judiciously prescribing antibiotics in the clinical settings, avoiding haphazard recommendation of antibacterial for treatments. Upgrading the hygiene and sanitation standards with appropriate food handling protocols and food safety practices would contribute towards the same.
Additional file: Additional Table S1-S4.
ACKNOWLEDGMENTS
The authors would like to thank Nitte (Deemed to be University), Nitte University for Science Education and Research for the infrastructure and facilities provided.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This study was supported by Nitte (Deemed to be University), Mangaluru, India through Grant no. N/RG/NUFR2/NUCSER/2020/05.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript and/or in the supplementary files.
ETHICS STATEMENT
Not applicable.
- Kim HS, Chon JW, Kim YJ, Kim DH, Kim MS, Seo KH. Prevalence and characterization of extended-spectrum-β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in ready-to-eat vegetables. Int J Food Microbiol. 2015;207:83-86.
Crossref - Hasan ME, Shahriar A, Shams F, Nath AK, Emran TB. Correlation between biofilm formation and antimicrobial susceptibility pattern toward extended spectrum β-lactamase (ESBL)-and non-ESBL-producing uropathogenic bacteria. J Basic Clin Physiol Pharmacol. 2020;32(2):1-12.
Crossref - Sivaraman GK, Sudha S, Muneeb KH, Shome B, Holmes M, Cole J. Molecular assessment of antimicrobial resistance and virulence in multi drug resistant ESBL-producing Escherichia coli and Klebsiella pneumoniae from food fishes, Assam, India. Microb Pathog. 2020;149:104581.
Crossref - Indrajith S, Mukhopadhyay AK, Chowdhury G, et al. Molecular insights of carbapenem resistance Klebsiella pneumoniae isolates with focus on multidrug resistance from clinical samples. J Infect Public Healt. 2021;14(1):131-138.
Crossref - Abebe GM. The role of bacterial biofilm in antibiotic resistance and food contamination. Int J Microbiol. 2020;2020:1705814.
Crossref - Mishra RAK, Muthukaliannan GK. Role of microalgal metabolites in controlling quorum-sensing-regulated biofilm. Arch Microbiol. 2022;204(3):1-3.
Crossref - Dhivya R, Rajakrishnapriya VC, Sruthi K, Chidanand DV, Sunil CK, Rawson A. Biofilm combating in the food industry: Overview, non-thermal approaches, and mechanisms. J Food Process Preserv. 2022:e16282.
Crossref - Bonneaud C, Tardy L, Giraudeau M, Hill GE, McGraw KJ, Wilson AJ. Evolution of both host resistance and tolerance to an emerging bacterial pathogen. Evol Lett. 2019;3(5):544-554.
Crossref - CLSI C. Performance standards for antimicrobial susceptibility testing. Clinical Lab Standards Institute.,2016;35(3):16-38.
- Ausubel FM, Brent R, Kingston RE, et al. Current Protocols in Molecular Biology. Molecular Reproduction and Development. 1989;1(2):146.
- Stepanovic S, Cirkovic I, Ranin L, Svabic-Vlahovic M. Biofilm formation by Salmonella spp. and Listeria monocytogenes on plastic surface. Lett Appl Microbiol. 2004;38(5):428-432.
Crossref - Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402-408.
https://doi.org/10.1006/meth.2001.1262 - Hartantyo SH, Chau ML, Koh TH, et al. Foodborne Klebsiella pneumoniae: virulence potential, antibiotic resistance, and risks to food safety. J Food Prot. 2020;83(7):1096-1103.
Crossref - Devi LS, Broor S, Rautela RS, Grover SS, Chakravarti A, Chattopadhya D. Increasing prevalence of Escherichia coli and Klebsiella pneumoniae producing CTX-M-type extended-spectrum beta-lactamase, carbapenemase, and NDM-1 in patients from a rural community with community acquired infections: A 3-year study. Int J Appl Basic Med. 2020;10(3):156.
Crossref - Sivaraman GK, Rajan V, Vijayan A, Elangovan R, Prendiville A, Bachmann TT. Antibiotic resistance profiles and molecular characteristics of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli and Klebsiella pneumoniae isolated from shrimp aquaculture farms in Kerala, India. Front Microbiol. 2021;12.
Crossref - Rao M, Laidlaw A, Li L, Young K, Tamber S. Isolation of third generation cephalosporin resistant Enterobacteriaceae from retail meats and detection of extended spectrum beta-lactamase activity. J Microbiol Methods. 2021;189:106314.
Crossref - World Health Organization. Global antimicrobial resistance and use surveillance system (GLASS) report: 2021.
- Bhardwaj K, Shenoy S, Baliga S, Unnikrishnan B, Baliga BS, Shetty VK. Research Note: Characterization of antibiotic resistant phenotypes and linked genes of Escherichia coli and Klebsiella pneumoniae from healthy broiler chickens, Karnataka, India. Poult Sci. 2021;100(6):101094.
Crossref - Hughes D, Andersson DI. Environmental and genetic modulation of the phenotypic expression of antibiotic resistance. FEMS Microbiol Rev. 2017;41(3):374-391.
Crossref - Wallace JS, Garner E, Pruden A, Aga DS. Occurrence and transformation of veterinary antibiotics and antibiotic resistance genes in dairy manure treated by advanced anaerobic digestion and conventional treatment methods. Environ Pollut.2018;236:764-772.
Crossref - Zhang S, Yang G, Ye Q, Wu Q, Zhang J, Huang Y. Phenotypic and genotypic characterization of Klebsiella pneumoniae isolated from retail foods in China. Front Microbiol. 2018;9:289.
Crossref - Sikarwar AS, Batra HV. Prevalence of antimicrobial drug resistance of Klebsiella pneumoniae in India. Int J Biosci Biochem Bioinforma. 2011;1(3):211.
Crossref - Pereira SC, Vanetti MC. Potential virulence of Klebsiella sp. isolates from enteral diets. Braz J Med Biol. Res.2015;48(9):782-789.
Crossref - Giri S, Kudva V, Shetty K, Shetty V. Prevalence and Characterization of Extended-Spectrum β-Lactamase-Producing Antibiotic-Resistant Escherichia coli and Klebsiella pneumoniae in Ready-to-Eat Street Foods. Antibiotics. 2021;10(7):850.
Crossref - Raheem AA, Mohammed GJ. Molecular Detection of Some Virulence Factors genes of Klebsiella pneumoniae Isolated from Food Samples. Ann Romanian Soc Cell Biol. 2021:11007-11019.
- Cheng J, Zhou M, Nobrega DB, et al. Virulence profiles of Klebsiella pneumoniae isolated from 2 large dairy farms in China. Int J Dairy Sci., 2021;104(8):9027-9036.
Crossref - Aher T, Roy A, Kumar P. Molecular detection of virulence genes associated with pathogenicity of Klebsiella spp. isolated from the respiratory tract of apparently healthy as well as sick goats. Isr J Vet Med. 2012;67(4):249-252.
- Wu MC, Lin TL, Hsieh PF, Yang HC, Wang JT. Isolation of genes involved in biofilm formation of a Klebsiella pneumoniae strain causing pyogenic liver abscess. PloS One. 2011;6(8):e23500.
Crossref - El Fertas-Aissani R, Messai Y, Alouache S, Bakour R. Virulence profiles and antibiotic susceptibility patterns of Klebsiella pneumoniae strains isolated from different clinical specimens. Pathol Biol. 2013;61(5):209-216.
Crossref - Sarowska J, Futoma-Koloch B, Jama-Kmiecik A, et al. Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: recent reports. Gut Pathog. 2019;11(1):1-6.
Crossref - Murphy CN, Mortensen MS, Krogfelt KA, Clegg S. Role of Klebsiella pneumoniae type 1 and type 3 fimbriae in colonizing silicone tubes implanted into the bladders of mice as a model of catheter-associated urinary tract infections. Infect Immun. 2013;81(8):3009-3017.
Crossref - Stahlhut SG, Struve C, Krogfelt KA, Reisner A. Biofilm formation of Klebsiella pneumoniae on urethral catheters requires either type 1 or type 3 fimbriae. FEMS Microbiol Immunol. 2012;65(2):350-359.
Crossref - Schroll C, Barken KB, Krogfelt KA, Struve C. Role of type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. BMC Microbiol. 2010;10(1):1-10.
Crossref - Liu Y, Liu C, Zheng W, et al. PCR detection of Klebsiella pneumoniae in infant formula based on 16S-23S internal transcribed spacer. Int J Food Microbiol. 2008;125(3):230-235.
Crossref - Brisse S, Verhoef J. Phylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing and automated ribotyping. Int J Syst Evol. 2001;51(3):915-924.
Crossref - Yigit H, Queenan AM, Anderson GJ, et al. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45(4):1151-1161.
Crossref - Mahrouki S, Belhadj O, Chihi H, et al. Chromosomal blaCTX-M-15 associated with ISEcp1 in Proteus mirabilis and Morganella morganii isolated at the Military Hospital of Tunis, Tunisia. J Med Microbiol. 2012;61(9):1286-1289.
Crossref - Batchelor M, Hopkins K, Threlfall EJ, et al. blaCTX-M genes in clinical Salmonella isolates recovered from humans in England and Wales from 1992 to 2003. Antimicrob Agents Chemother. 2005;49(4):1319-1322.
Crossref - Zhao S, White DG, Ge B, et al. Identification and characterization of integron-mediated antibiotic resistance among Shiga toxin-producing Escherichia coli isolates. Appl Environ Microbiol. 2001;67(4):1558-1564.
Crossref - Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis. 2006;6(10):629-640.
Crossref - Ma M, Wang H, Yu Y, Zhang D, Liu S. Detection of antimicrobial resistance genes of pathogenic Salmonella from swine with DNA microarray. J Vet Diagn. 2007;19(2):161-167.
Crossref - Brisse S, Fevre C, Passet V, ry s;. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PloS one. 2009;4(3):e4982.
Crossref - Neuberger A, Oren I, Sprecher H. Clinical impact of a PCR assay for rapid identification of Klebsiella pneumoniae in blood cultures. J Clin Microbiol. 2008;46(1):377-379.
Crossref - Izquierdo L, Coderch N, Pique N, et al. The Klebsiella pneumoniae wabG Gene: Role in Biosynthesis of the Core Lipopolysaccharide and Virulence. J Bacteriol. 2003;185(24):7213-7221.
Crossref - Yu WL, Ko WC, Cheng KC, Lee CC, Lai CC, Chuang YC. Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn Microbiol Infect Dis. 2008;62(1):1-6.
Crossref - Duong VT, Tu LT, Tuyen HT, et al. Novel multiplex real-time PCR assays reveal a high prevalence of diarrhoeagenic Escherichia coli pathotypes in healthy and diarrhoeal children in the south of Vietnam. BMC Microbiol. 2020;20(1):192.
Crossref - Gunzburg ST, Tornieporth NG, Riley LW. Identification of enteropathogenic Escherichia coli by PCR-based detection of the bundle-forming pilus gene. J Clin Microbiol. 1995;33(5):1375-1377.
Crossref - Ashwath P, Deekshit VK, Rohit A, et al. Biofilm Formation and Associated Gene Expression in Multidrug-Resistant Klebsiella pneumoniae Isolated from Clinical Specimens. Curr Microbiol. 2022;79(3):1-10.
Crossref - Ritz M, Garenaux A, Berge M, Federighi M. Determination of rpoA as the most suitable internal control to study stress response in C. jejuni by RT-qPCR and application to oxidative stress. J Microbiol Methods. 2009;76(2):196-200.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.