ISSN: 0973-7510
E-ISSN: 2581-690X
In this study, one hundred and thirty isolates were isolated from water and sludge samples taken from hot springs located in different regions of Turkey. Among them, eleven isolates were chosen according to conventional (morphological, physiological and biochemical tests) and molecular methods (rep-PCR and 16S rRNA sequencing). These bacteria were then tested for their capability to produce valuable enzymes. As a result; species belonging to Bacillus, Anoxybacillus, Aeribacillus, Enterococcus, Exiguobacterium and Paenibacillus were identified. Test strains were found to have optimum reproductive potential at pH 5.0-9.0 and 15-65°C, usually at a concentration of 1.0-10.0% (w/v) NaCl. In addition, all thermo-tolerant bacteria were Gram, endospore (except E. profundum), catalase and oxidase (except E. faecium and E. profundum) positive, and rod-shaped (except E. faecium). It was observed that all isolates had a 99% similarity percentage as a result of 16S rRNA sequence analysis. All of the isolates were capable of producing industrially important enzymes moreover, eight of them could produce at least two of these enzymes. Test strains had high potential of industrial enzyme production, and the enzymes from these thermo-tolerant isolates will be widely used in biotechnological processes.
Thermo-tolerant, isolation, 16S rRNA, rep-PCR, industrial enzymes.
The extraordinary diversity of life on earth has attracted researchers’ attention from the past. Not only the diversity of living things, but also their distribution and function in different geographies is very attractive. However, an important part of the information on diversity is focused on the dissemination and protection of biodiversity on animals and plants. Most of the microbial diversity is still not discovered (>95%). Only 1% of the species diversity of prokaryotes in the world has been identified; therefore, little is known about prokaryotic diversity26,54,56.
For each microorganism, there is the minimum temperature at which the breeding is not possible, the optimum temperature at which the breeding is fastest and the maximum temperature values at which the breeding is not possible. These three temperatures, which are called cardinal temperatures, are typical for each organism and vary significantly between species34. Thermophilic bacteria which grow in hot springs, tropical soils, fertilizer heaps, manure forming fodder and garbage10 the temperature at which the can generally live optimally at 60°C. However, it is stated that very few thermophilic bacteria can live above 75°C22. In order to live and reproduce in extreme conditions, these organisms must adapt their metabolic and other cellular functions to these environments. The cell membrane of thermophilic is composed of saturated fatty acids. These fatty acids provide the cell with a hydrophobic environment and keep the cell tight and firm enough to survive at high temperatures. The cellular elements (cell membrane) and components (enzymes, proteins, nucleic acids, etc.) of thermophilic organisms are resistant to high temperatures. They are also resistant to denaturants such as extremely acidic and alkaline conditions and to proteolysis14,24. The DNA of thermophiles contains a DNA Gyrase, which provides the reversible in the DNA to create positive supercoils. This increases the melting point of the DNA to at least the organism’s maximum growth temperature. Thermophiles also tolerate high temperatures using increasing interactions such as electrostatic disulfide bridge and hydrophobic interactions using non-thermotolerant organisms24.
Natural geothermal areas are found in tectonic active zone regions where earth crust movements in the world. Turkey is one of the richest countries in the world in the aspect of geothermal sources. However, the number of scientific studies on the determination of microorganism flora in thermal sources in different geographic regions of our country and determination of species with potential to be used in industry is very limited41. Until now, conventional methods based on phenotypic characters have been used in the diagnosis of thermophilic microorganisms in scientific studies conducted both in the world and in our country. The identification of microorganisms by morphological, physiological and biochemical tests has been insufficient to determine the differences between the desired microorganisms. Moreover, the conventional methods need long-term experiments, over labor, experienced and trained researchers, and the results are open to alternative interpretations, other known disadvantages of these methods36. Therefore, in recent years, molecular techniques have been developed to be used in the diagnosis of microorganisms. Genetic profiles 16-23S rRNA-PCR, rep-PCR, ERIC-PCR, BOX-PCR and (GTG)5-PCR) are commonly used methods35.
Rai and Mukherjee45 defined enzymes as green chemicals, specific biocatalysts that provide the realization and control of all reactions occurring in the living organism under suitable conditions, most of which are of protein structure and naturally produced only by living things. microbial enzymes which have applications in food industry, food and beverage production, cleaning and processing of garments in the textile industry, paper and detergent industries, medicine and medical field diagnosis of diseases continue to change with each passing day especially with biotechnological approaches and can be included in different application areas. The worldwide production volume of industrial enzymes, most of which have extracellular properties, was $ 1 million in 1995; In 2000, approximately 60% of the industrial enzymes used in the world market were produced in Europe and 40% in the USA and Japan30. The need for non-toxic and more effective methods has enabled the production of enzymes to scale up and penetrate into different markets thanks to the rapidly growing biotechnology.
Thermophilic organisms have a very important place due to their biotechnological potential. Their most important biotechnological features are; to produce enzymes that can catalyze biochemical reactions at much higher temperatures than normal organisms23. These enzymes are thermophilic enzymes, and these enzymes are preferred in many industrial areas due to their pH changes and their stability to high temperatures. Thermostable enzymes play a role in the hydrolysis of protein, lipid and polysaccharide substrates in molecular biology7. Proteases from thermophilic enzymes are used in food industry, pharmacology, leather and detergent industry in a wide industrial area. enzymes such as lipase, esterase, amylase, cellulase and xylanase; used in detergent, sugar, textile and paper industries55.
Thermostable enzymes are more advantageous than other enzymes, and both the industrially important and the thermostable enzyme producer, the introduction of new source microorganisms into the literature, will be able to respond to the needs of industry today and to shed light on possible future studies. The aim of the study was isolation and identification of new and industrially valuable bacterial strains from different geothermal resources of Turkey, presentation of their novel potential to produce industrially important enzymes and multi-enzyme productions.
Purification of thermo-tolerant isolates
The water and sludge samples from Armutlu (Yalova), Germencik (Aydn), Yildizburnu (Izmir), Havza (Samsun), Guroymak (Bitlis) and Karakurt (Krsehir) hot springs were collected and the samples were transferred to Nutrient Agar (NA) medium (peptone from meat 5.0 gL-1; meat extract 3.0 gL-1; agar 12.0 gL-1) in order to incubate (Thermo Scientific Heratherm, the USA) at 55°C for 24 h. The growing colonies were spread on NA to obtain pure cultures and were eliminated by morphology differences in plates, as a first step. The pure, single and different colonies were stored in the Nutrient Broth (NB) with 15% glycerol content at -86°C for further studies3.
Gene amplification, cloning and genomic fingerprinting of isolates
For DNA extraction of each isolate, a single colony from NA medium was selected and inoculated to NB at 55°C 150 rpm. After the incubation period, genomic DNA isolation was carried out according to Promega WizardR DNA purification kit and purified DNA was stored at +4°C until use47. Test isolates were subjected to rep-PCR with the special primers of (GTG)5 and BOX elements to obtain genomic fingerprinting4,6,20. To obtain the PCR products, 50 ng of purified DNA was used as the template in 30 ml reaction mixture. Twenty-seven microliters of the reaction cocktail were prepared as follows: Specific Gitschier Buffer 5 mL, dimethyl sulfoxide 2.5 mL (100%, 20X), dNTPs (10 mM) 1.25 mL, bovine serum albumin 1.25 mL (20 gL-1), primers (5 mM) 3.0 ml, Taq polymerase (250 U) 0.3 mL and water 13.7 mL. A negative control (no DNA) was included in each PCR assay. PCR reactions were performed with a 3×32-well ProFlexTM PCR System, using the following conditions for (GTG)5-PCR: an initial denaturation at 94°C for 7 min; 36 cycles consisting of 94°C for 1 min and annealing at 53°C for 1 min with (GTG)5 primer, extension at 65oC for 8 min; and a final polymerization at 65°C for 16 min before cooling at 4°C. For BOX-PCR; an initial denaturation at 95°C for 7 min; 36 cycles consisting of 94°C for 1 min and annealing at 53oC for 1 min with BOX primer, extension at 65°C for 8 min; and a final polymerization at 65°C for 16 min before cooling at 4°C.
The 16S rRNA of the isolates was amplified by Polymerase Chain Reaction (PCR) with the forward primer 27-F:(5′ AGAGTTTGATYMTGGCTCAG3′) and the reverse primer 1492-R:(5′ GGTTACCTTGTTACGACTT 3′)5. The amplified fragments were cloned into E. coli JM101 strain with a vector system (pGEM-T, Promega, the UK) and the clones were sequenced (Macrogen, Amsterdam, the Netherlands). The results of 16S rRNA gene sequencing were analyzed using the GenBank (http://blast.ncbi.nlm.nih.gov/blast.cgi) and EzTaxon (http://www.eztaxon.org/) servers9. Considering the results of the study, a phylogenetic tree was formed via the neighbor-joining method by using the software package MEGA 4.013.
The PCR products (27 mL) were mixed with 3 mL gel loading buffer (6X) and subjected to agarose (1.5% w/v) gel electrophoresis in Tris–Acetate–EDTA (TAE) buffer at 100 V for 110 min. After separation of the amplification products by the gel, the fragments were stained with ethidium bromide solution (2 mL Etbr/100 mL 1X TAE buffer). The amplified DNA product was monitored using Gel Documentation System.
Conventional identification
The test isolates were subjected to conventional tests. The pH (Mettler Toledo) and temperature range for bacterial growth were tested between pH 3.0–11.0 and 15–65°C. The NaCl requirement for growth was also tested in NB medium containing 1.0, 2.0, 4.0, 6.0, 8.0, 10.0 and 12.0% (w/v) NaCl. Cell and colony morphology (Leica ICC50 HD light microscope), Gram and endospore staining, motility and the presence of catalase and oxidase reactions were also investigated by the method of Prescott44 and Sari47.
Preliminary experiments of industrial enzymes
Amylase
Amylase assay was performed at 55°C for two days in a modified-medium which contained yeast extract (1 gL-1), starch (5 gL-1) and agar (15 gL-1)58. The amylase activity was screened by Lugol’s solution. Clear and large zones were evaluated as positive, or as amylase production; however, no zone formation was evaluated as negative, or as no amylase production.
Cellulase
A medium containing carboxymethyl-cellulose (10 gL-1), peptone (5 gL-1), yeast (5 gL-1), KH2PO4 (1 gL-1), MgSO4.7H2O (0.2 gL-1), NaCl (10 gL-1) and agar (15 gL-1) was used for cellulase assay. Incubation was carried out at 55°C for two days, and after the incubation, the plates were stained by Congo-Red Dye. The presence of clear zone was accepted as positive47.
Lipase
Test isolates were inoculated on tributyrin agar medium which contained 1% (w/v) tributyrin (glycerol tributyrate) and incubated at 55°C. Formation of hydrolysis zone around the culture on plate was controlled for two days, and the strains with transparent and the highest zone formation (lipolytic activity) were determined as lipase producers12.
Protease
Protease activity was conducted in medium which contained NB (8 gL-1), Skimmed Milk (10 gL-1) and agar (15 gL-1) at 55°C for two days. The plates were evaluated according to the zone formations, and the observation of a halo zone indicated positive protease activity42.
Xylanase
A solid medium with xylan (10 gL-1), NaNO3 (1.2 gL-1), KH2PO4 (3 gL-1), K2HPO4 (6 gL-1), CaCl2 (0.05 gL-1), MgSO4 (0.01 gL-1), ZnSO4 (0.001 gL-1) and agar (15 gL-1) was used to determine xylanase production. The plates were incubated at 55°C for four days and after the incubation, the plates were stained by Congo-Red Dye. The presence of clear zone was accepted as positive47.
Genomic fingerprinting of thermophiles
The first step of this study was the isolation of thermopilic bacteria from water and sludge samples of Armutlu (Yalova), Germencik (Aydn), Yildizburnu (Izmir), Havza (Samsun), Guroymak (Bitlis) and Karakurt (Krsehir) hot springs. One hundred and thirty bacterial isolates were obtained however this number was firstly reduced by eliminating according to colony differences observed on plate surface. As conventional analysis alone was not sufficient for the identification of bacteria, rep-PCR [(GTG)5-PCR and BOX-PCR] method was used which clearly showed the differences between microorganisms’ genomic fingerprinting. (GTG)5-PCR, gave the first information about isolates. It was observed that the isolates usually yielded up to sixteen polymorphic bands between 350-3500 bp and eight polymorphic bands between 700-2500 bp while EA 9 isolate gave no bands (Fig. 1). BOX-PCR, another genomic fingerprint method, was applied and it was determined that the test isolates were given two types of polymorphic bands, usually five bands between 300-1200 bp and two bands between 1200 – 4000 bp (Fig. 2). 16S rRNA region, which was evolutionarily conserved, was amplified using universal primers; to identify the isolates at species level. As a result of the analysis, eleven different species were detected, resulted in the fragments of 1500 bp. The sequences of cloned isolates were compared with the sequences of other bacteria in the GenBank and the nucleotides were analyzed using BLAST and EzTaxon (Table 1). The phylogenetic relationship between thermopilic bacteria was presented by phylogenetic tree (Fig. 3). As a result, the genus Bacillus with different species, Enterococcus and Exiguobacterium were determined. EA1 was similar to Aeribacillus pallidus, EA2 to Anoxybacillus geothermalis, EA3 to Anoxybacillus mongoliensis, EA5 to Bacillus albus, EA6 to Bacillus halodourans, EA7 to Bacillus licheniformis, EA8 to Bacillus paralicheniformis, EA9 to Enterococcus faecium, EA10 to Exiguobacterium profundum and EA11 to Paenibacillus dendritiformis species at a rate of 99%. Based on the knowledge that the similarity ratio of 16S rRNA sequence analysis among the strains of the same species should be more than 97%34, it was concluded that the bacterial strains were similar to test strains.
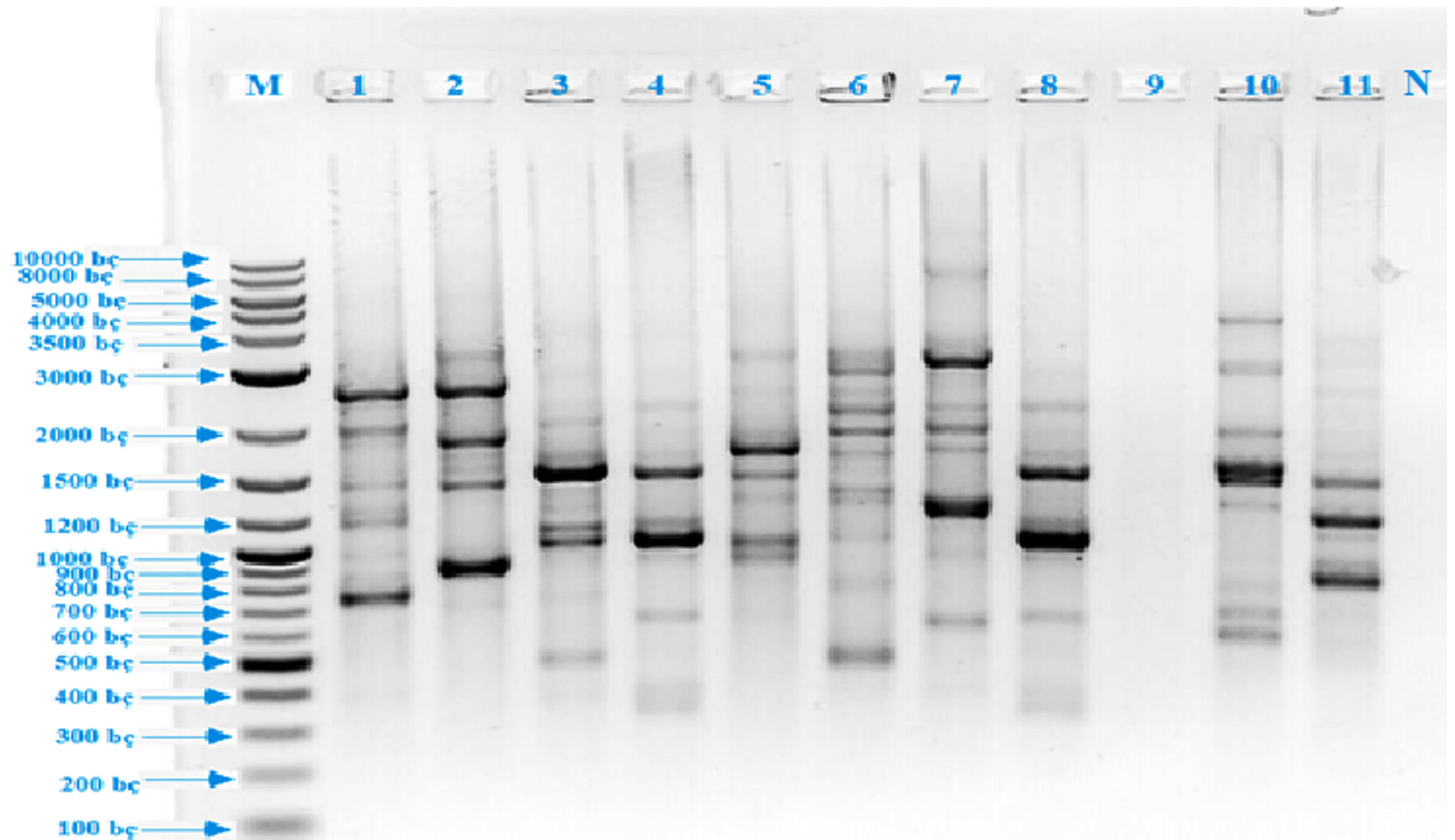 Fig. 1. (GTG)5 PCR band profiles of thermophilic isolates (M: DNA Marker, 1: EA 10, 2: EA 6, 3: EA 5, 4: EA 3, 5: EA 9, 6: EA 11, 7: EA 1, 8: EA 7, 9: EA 4, 10: EA 8, 11: EA 2, 12: Negative control)
Fig. 1. (GTG)5 PCR band profiles of thermophilic isolates (M: DNA Marker, 1: EA 10, 2: EA 6, 3: EA 5, 4: EA 3, 5: EA 9, 6: EA 11, 7: EA 1, 8: EA 7, 9: EA 4, 10: EA 8, 11: EA 2, 12: Negative control) Table (1):
Molecular identifications of the isolates
Code |
Name |
Similarity ratio (%) |
GenBank number |
|---|---|---|---|
EA1 |
Aeribacillus pallidus |
99.9 |
MH411153 |
EA2 |
Anoxybacillus geothermalis |
99.7 |
MH411161 |
EA3 |
Anoxybacillus mongoliensis |
99.3 |
MH411164 |
EA4 |
Anoxybacillus rupiensis |
99.5 |
MH411201 |
EA5 |
Bacillus albus |
99.8 |
MH411233 |
EA6 |
Bacillus halodurans |
99.7 |
MH411241 |
EA7 |
Bacillus licheniformis |
98.9 |
MH411598 |
EA8 |
Bacillus paralicheniformis |
99.8 |
MH411597 |
EA9 |
Enterococcus faecium |
99.9 |
MH411686 |
EA10 |
Exiguobacterium profundom |
99.9 |
MH411710 |
EA11 |
Paenibacillus dendritiformis |
99.7 |
MH411711 |
rep-PCR [(GTG)5-PCR and BOX-PCR], which was molecular method used for this purpose, was a highly effective method used to reveal the diversity in the ecosystem, the phylogenetic relationship between the strains and to distinguish genetically close micro-organisms at species and sub-species level4. BOX-PCR and (GTG)5-PCR methods were found to be very successful in demonstrating the genomic differences between strains in parallel with literature data3,4,6,58.
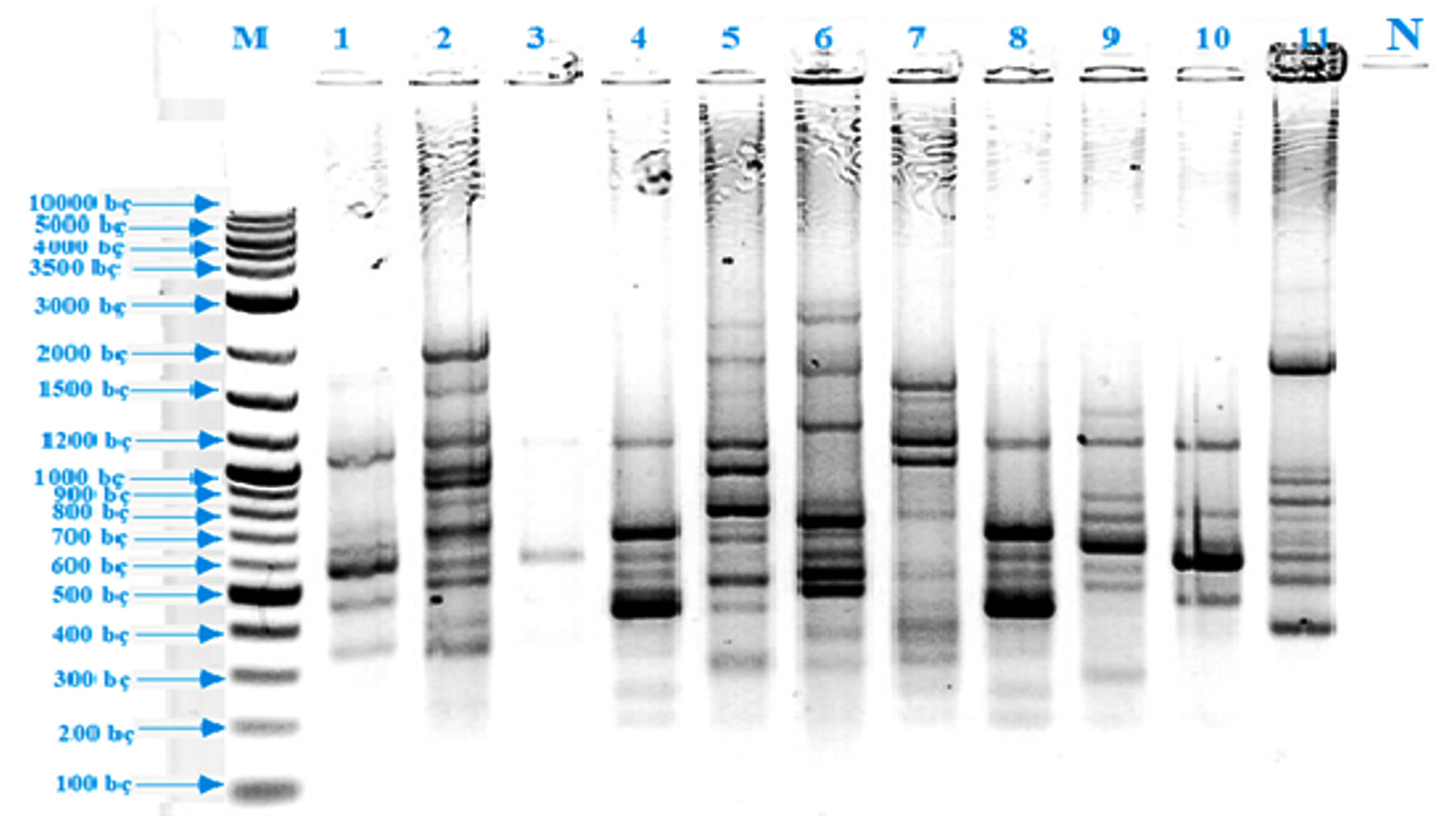 Fig. 2. BOX-PCR band profiles of thermophilic isolates (M: DNA Marker, 1: EA 2, 2: EA 8, 3: EA 4, 4: EA 7, 5: EA 1, 6: EA 11, 7: EA 9, 8: EA 3, 9: EA 5, 10: EA 6, 11: EA 10, 12: Negative control)
Fig. 2. BOX-PCR band profiles of thermophilic isolates (M: DNA Marker, 1: EA 2, 2: EA 8, 3: EA 4, 4: EA 7, 5: EA 1, 6: EA 11, 7: EA 9, 8: EA 3, 9: EA 5, 10: EA 6, 11: EA 10, 12: Negative control) Morphology, physiology and biochemical characteristics of thermophiles
Among eleven different bacterial isolates, all of isolates were Gram positive and motile, endospore producing (except EA10), catalase and oxidase positive (except EA9 and EA10) and bacilli-shaped (EA9 was coccus-shaped) (Table 2). To determine the salt concentration for growth of the isolates, 1.0, 2.0, 4.0, 6.0, 8.0, 10.0 and 12% (w/v) NaCl were used (Table 2). As a result of spectrophotometric measurements made at OD600, the test strains showed development in 1.0-10.0% salt concentration values; EA1 and EA4 was in 1.0-2.0%, EA3, EA7, EA8 and EA11 were in 2.0-4.0%, EA9 was in 2.0-6.0%, EA2 and EA6 were 2.0-8.0% and EA5 and EA10 were 2.0-10.0% range. In order to determine the pH ranges of test strains; microbial growth in pH 3.0, 5.0, 7.0, 9.0 and 11.0 were monitored; EA4 and EA11 were detected to develop in pH 6.0-7.0; EA1, EA3, EA5, EA6, EA7 and EA8 were in pH 7.0-9.0; EA2, EA9 and EA10 were in pH 5.0-9.0. The isolates were incubated at 15°C, 25°C, 35°C, 45°C, 55°C and 65°C to determine the temperatures at which they developed and the temperature values of the test strains were determined. Although the temperature ranges of thermophiles varied, EA9 and EA10 were determined that they could develop at 15-55°C, EA5 and EA6 were at 25-55°C, EA3 and EA7 were at 35-65°C, others were 35-55°C.
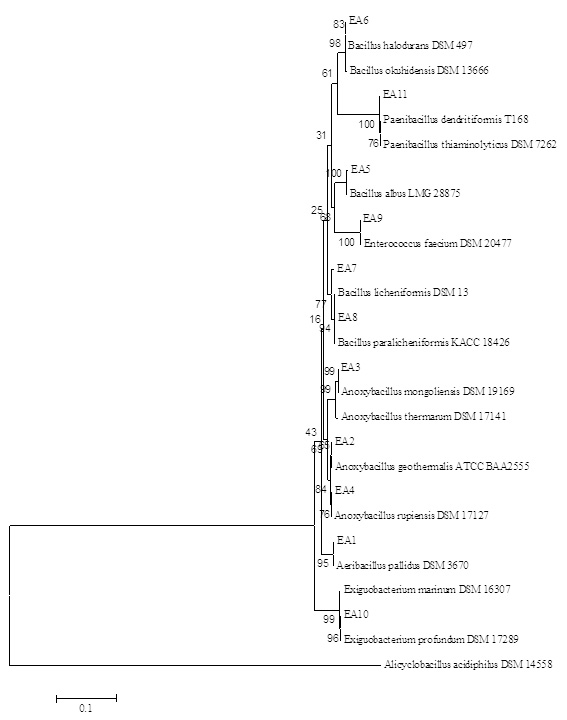 Fig. 3. Neighbor-Joining phylogenetic tree of thermophilic bacteria isolated from different hot springs of Turkey based on 16S rRNA gene analysis results. Alicyclobacillus acidophilus used as an external group
Fig. 3. Neighbor-Joining phylogenetic tree of thermophilic bacteria isolated from different hot springs of Turkey based on 16S rRNA gene analysis results. Alicyclobacillus acidophilus used as an external groupAs a result of the morphological analysis of the isolates obtained in parallel with the literature data, all test strains were Gram and endospore (except E. profundum) positive, motile and had bacilli cell morphology except E. faecium (coccus). Growth ranges for pH, temperature and salt concentration were 5-9 pH, 15-65°C and 1-10%(w/v). Finally, all the isolates were catalase and oxidase positive (except E. faecium and E. profundum). Yadav57 carried out the isolation and identification of 150 thermophilic bacteria from hot springs in Nepal. The 16S rRNA fragments of the isolates were amplified with a size of about 1.5 kbs (1500 bp) and the microorganisms were belonging to the genus Anoxybacillus, Aeribacillus, and Bacillus at a ratio of ³95. The bacilli isolates were gram positive, endospore forming and their optimum growth temperature were between 55-65°C. However, in this study, the isolates were thermo-tolerant due to large temperate range, 15-65oC. Aanniz1 isolated and identified 240 bacteria from different sources in Morocco. All isolates were Gram and endospore positive. When they examined the BOX-PCR and 16S rRNA sequence of the isolates, the dominant species was Bacillus (97.5%); 119 of them were B. licheniformis and 6 of the species belonging to Aerobacillus. Norashirene38 obtained six thermophilic isolates from Malaysia. The optimum growth temperatures of these isolates were 55°C and pH was 7.5, and they were aerobic, catalase and oxidase positive. Poli43 carried out the isolation of thermophilic bacteria by taking water and mud samples from the geothermal areas in Italy. These test strains were aerobic, endospore and Gram positive, motile, rod shaped and their optimum growth was at 65°C and pH 7.2. The salt concentrations in which the test strains could develop were tested at 1-10%(w/v), and the results were generally parallel to the literature data. Tambekar52 revealed the presence of the strains that could reproduce in 8% (w/v) salt concentration, in parallel with our data. However, B. halodurans (EA6) was found to be able to withstand up to 8% (w/v) salt concentration in contrast to the literature51.
Table (2):
Physicochemical requirements and conventional identifications of the isolates
Isolate code |
Isolate name |
Morphology |
Gram |
pH |
Temperature (oC) |
Salt (%) |
Oxidase |
Catalase |
Endospore |
Motility |
|---|---|---|---|---|---|---|---|---|---|---|
EA1 |
Aeribacillus pallidus |
Bacilli |
+ |
7.0-9.0 |
35-55 |
1.0-2.0 |
+ |
+ |
+ |
+ |
EA2 |
Anoxybacillus geothermalis |
Bacilli |
+ |
5.0-9.0 |
35-55 |
2.0-8.0 |
+ |
+ |
+ |
+ |
EA3 |
Anoxybacillus mongoliensis |
Bacilli |
+ |
7.0-9.0 |
35-65 |
2.0-4.0 |
+ |
+ |
+ |
+ |
EA4 |
Anoxybacillus rupiensis |
Bacilli |
+ |
6.0-7.0 |
35-55 |
1.0-2.0 |
+ |
+ |
+ |
+ |
EA5 |
Bacillus albus |
Bacilli |
+ |
7.0-9.0 |
25-55 |
2.0-10.0 |
+ |
+ |
+ |
+ |
EA6 |
Bacillus halodurans |
Bacilli |
+ |
7.0-9.0 |
25-55 |
2.0-8.0 |
+ |
+ |
+ |
+ |
EA7 |
Bacillus licheniformis |
Bacilli |
+ |
7.0-9.0 |
35-65 |
2.0-4.0 |
+ |
+ |
+ |
+ |
EA8 |
Bacillus paralicheniformis |
Bacilli |
+ |
7.0-9.0 |
35-55 |
2.0-4.0 |
+ |
+ |
+ |
+ |
EA9 |
Enterococcus faecium |
Coccus |
+ |
5.0-9.0 |
15-55 |
2.0-6.0 |
– |
+ |
* |
+ |
EA10 |
Exiguobacterium profundom |
Bacilli |
+ |
5.0-9.0 |
15-55 |
2.0-10.0 |
– |
+ |
– |
+ |
EA11 |
Paenibacillus dendritiformis |
Bacilli |
+ |
6.0-7.0 |
35-55 |
2.0-4.0 |
+ |
+ |
+ |
+ |
*; endospore test was not applied.
It was noticeable that the growth of E. faecium, a lactic acid bacterium at 15-55°C, considering that mesophilic bacteria generally did not show growth above 45°C. However, Orr39 found some members of the microorganisms contained in Enterococci were thermo-tolerant. Svec50 reported that, (GTG)5-PCR method could be used effectively in the identification of the genus Enterecoccus at species and sub-species level. Abdel-Rahman2 identified E. faecium with 99% similarity ratio and as unable to utilize oxygen as an electron acceptor. They also concluded that; E. faecium could grow at 45-50°C and pH 9.6. Al-Mariri8 collected white cheese samples and identified thermophilic streptococci. As a result; E. faecium isolates were gram positive, non-sporulation and could grow at 4-6.5%(w/v) salt concentration while the salt concentration for this study was 2-6%(w/v). In another study; E. faecium was identified as catalase positive21 however Abdel-Rahman2 and Jensen28 identified as catalase negative. According to Schleifer and Kilpper-Bהlz48, some strains of E. faecium were motile. Crapart15 isolated lactic acid-producing thermo-tolerant bacterium, E. profundum, from hydrothermal vent and the growth conditions were parallel to our study with 12-49°C, 5.5-9.5 pH and 0-11%(w/v) salt concentration. This bacterium was characterized as motile, Gram and catalase positive, oxidase negative while Kasana and Pandey29 also concluded as non-sporulation.
Enzyme production capacities of thermophiles
Enzyme production abilities of these eleven isolates were tested in enzyme specific solid media. As a result; all of them were producers of at least one enzyme which were industrially valuable (Table 3). EA2, EA3, EA4, EA5, EA7 and EA8 were amylase producers. The only cellulase producer isolate was EA6. Except EA6 and EA11, the rest of the isolates were positive in terms of lipase production. EA6 and EA10 gave negative results in protease growth media however, the other isolates were protease producers. EA2, EA3, EA7 and EA8 were xylanase producers.
Table (3):
Industrial enzyme production profiles of the isolates and their first investments in thermophilic regions
Isolate code |
Isolate name |
A |
C |
L |
P |
X |
Global |
Turkey |
Reference(s) |
|---|---|---|---|---|---|---|---|---|---|
EA1 |
Aeribacillus pallidus |
– |
– |
+ |
+ |
– |
P |
P |
|
EA2 |
Anoxybacillus geothermalis |
+ |
– |
+* |
+ |
+* |
P |
* |
|
EA3 |
Anoxybacillus mongoliensis |
+ |
– |
+* |
+ |
+* |
P |
* |
|
EA4 |
Anoxybacillus rupiensis |
+ |
– |
+* |
+ |
– |
P |
P |
|
EA5 |
Bacillus albus** |
+* |
– |
+* |
+* |
– |
* |
* |
|
EA6 |
Bacillus halodurans |
– |
+* |
– |
– |
– |
P |
* |
|
EA7 |
Bacillus licheniformis |
+ |
– |
+ |
+ |
+ |
P |
P |
|
EA8 |
Bacillus paralicheniformis |
+* |
– |
+* |
+* |
+* |
* |
* |
|
EA9 |
Enterococcus faecium |
– |
– |
+* |
+* |
– |
P |
* |
|
EA10 |
Exiguobacterium profundom |
– |
– |
+* |
– |
– |
P |
* |
|
EA11 |
Paenibacillus dendritiformis |
– |
– |
– |
+* |
– |
* |
* |
(+):producer, (-): not producer; *: No study about related enzyme production ability/ No presence of bacteria at thermophilic regions; P:presence of the related bacteria in thermophilic regions;**: https://www.ncbi.nlm.nih.gov/genome/genomes/71466, Abbreviations: A-Amylase; C-Cellulase; L-Lipase; P-Protease; X-Xylanase
pallidus, which was isolated from desert soil by Aanniz1, could grow at 30-80°C, 0.5-10%(w/v) salt concentration and produce amylase and protease. Koc31 determined A. pallidus C196 and D642 as potential lipase producers due to use of olive oil as substrate. The isolation from different locations of Turkey resulted in identification of A. pallidus as amylase producer12. With optimum growth conditions between 40-65°C, 0-3% (w/v) NaCl and 5.0-9.5 pH A. geothermalis could hydrolyze casein and starch due to protease and amylase activity, respectively 19 while A. geothermalis in this study could also able to produce lipase and xylanase. Namsaraev37 determined A. mongoliensis, whose optimum growth was at 35-75°C, 0-5% (w/v) NaCl concentration and 5.5-10.8 pH, as a thermophilic protease and amylase producer due to the hydrolysis of starch, casein and gelatin. A. mongoliensis in this study was also identified as lipase and xylanase producer. Derekova16 was isolated and identified A. rupiensis, which could survive between 35-67°C, 5.5-8.5 pH, for the first time with ability to degrade xylan, starch and casein. Jardine27 was also proposed A. rupiensis as amylase, protease and celllulase producer. Yanmis58 isolated A. rupiensis from geothermal regions of Turkey and it was declared as amylase producer. However, lipase production of A. rupiensis was determined in this study.
albus was deposited in NCBI under the project numbers of PRJNA509543, PRJNA326285 and PRJNA516150. Liu33 identified the marine isolate as B. albus sp. nov., type strain N35-10-2T (=MCCC 1A02146T =KCTC 33710T =LMG 28875T). However, there is no publication about thermophilic source of the microorganism.
The lipolytic activity of B. halodurans was proved by many of the studies17,46,53 and Vargas53 determined the optimum ranges at 25-55°C and 2.5-10% (w/v) NaCl. As a potential producer, amylase25 and xylanase32 were other enzymes that B. halodurans produced. However, B. haloduransin this study was identified as cellulase producer. Aanniz1 isolated B. licheniformis, which could survive at 30-75°C, 0.5-10% (w/v) salt content and 7-8.2 pH, had the potential to produce amylase, protease and cellulase. Archana and Satyanarayana11 isolated cellulase-free xylanase from thermostable B. licheniformis. Baltaci12 classified two of the isolates as B. licheniformis and they were also amylase, lipase, protease and cellulase producers. B. licheniformis, from thermal regions of Turkey58, could also able to produce cellulase and amylase.
Although B. paralicheniformis was isolated from fermented foods18,40, there was no record of thermophilic B. paralicheniformis and its enzyme production. B. paralicheniformis in this study could able to produce amylase, lipase, protease and xylanase. E. faecium was firstly isolated by Orr39 from hospital samples as thermo-tolerant however there was no report about enzyme production of thermo-tolerant E. faecium. However, lipase and protease production ability of E. faecium was determined in this study. Moderately thermophilic E. profundum was defined by Crapart15 but its industrial enzyme abilities never examined while lipase production of E. profundum discovered in this study. P. dendritiformis, which was included in the genus Bacillus, was first discovered in 1990s and reclassified as a separate genus49. However, there is no record about P. dendritiformis from thermophilic sources while protease production was determined in this study.
In addition, the isolation locations of organisms were important during the isolation and identification of thermo-tolerant organisms (Table 3). In this study, the presence of B. albus, B. paralicheniformis and P. dendritiformis in thermophilic regions was introduced for the first time. The presence of A. geothermalis, A. mongoliensis, B. albus, B. halodurans, B. paralicheniformis, E. faecium, E. profundum and P. dendritiformis in the thermal sources of Turkey, has been detected for the first time. When amylase-producing isolates were screened, it was observed that most of the isolates were producers of amylase. However, among them, B. albus and B. paralicheniformis were firstly identified as thermo-tolerant amylase producers. Although thermophilic bacteria are frequently identified as cellulase producers, among the isolates in this study, only B. halodurans could produce cellulase and this was declared in this study. A. pallidus, A. geothermalis, A. mongoliensis, A. rupiensis B. albus, B. licheniformis, B. paralicheniformis, E. faecium and E. profundum were thermo-tolerant bacteria with lipolytic activity. However, the lipase production abilities of the isolates, except A. pallidus and B. licheniformis, were not determined before, until this study. While all isolates, except B. halodurans and E. profundum, were able to produce protease, the proteolytic activities of thermo-tolerant B. albus, B. paralicheniformis, E. faecium and P. dendritiformis were determined for the first time. A. geothermalis, A. mongoliensis, B. licheniformis and B. paralicheniformis were able to produce xylanase; however, A. geothermalis, A. mongoliensis and B. paralicheniformis were firstly determined as thermo-tolerant xylanase producers
When the multienzyme production capabilities of the isolates were examined, it was observed that all the isolates, except B. halodurans, E. profundum and P. dendritiformis, were able to produce more than one enzyme. In a study, in order to determine the strains which have the potential to produce industrially important enzymes, Yadav57 observed that 135 of 150 thermophilic isolates have the potential to produce at least one extracellular hydrolytic enzyme.
As a conclusion, this research contained remarkable points in terms of presenting different thermo-tolerant species in the World and Turkey, microbial flora of thermal regions in Turkey and unknown abilities of these thermo-tolerant species to produce industrially valuable enzymes. The first isolation of B. albus, B. paralicheniformis and P. dendritiformis from thermal regions in the World was carried out. A. geothermalis, A. mongoliensis, B. albus, B. halodurans, B. paralicheniformis, E. faecium, E. profundum and P. dendritiformis were firstly isolated from hot springs of Turkey. A. geothermalis, A. mongoliensis, B. albus, B. halodurans, B. paralicheniformis, E. faecium, E. profundum and P. dendritiformis were identified as producers of some of industrial enzymes, for the first time. Eight different species with the potential to produce multi-enzyme were identified. Since thermostable enzymes play an important role in industrial and biotechnological processes, the enzyme/multi-enzyme production potentials of the isolates identified in this study has been one of the another notable results.
Acknowledgements
None.
Conflict Of Interest
The authors declare that there is no conflict of interest.
Authors’ Contribution
AS and GB designed and carried out the study, GB and OH participated in design to draft the manuscript, TM and AA dealed with financial supports and funding. All the authors read and approved the final manuscript.
Funding
The authors acknowledge the support of Ataturk University, Turkey for this work. This work was supported by the Research Development Centre of Ataturk University (Grant number was FAD-2018-6352).
Data Availabity
All datasets generated or analyzed during this study are included in the manuscript
Ethics Statement
All text, data, figures/tables or other illustrations presented in the manuscript are completely original and does not contain or include material taken from other copyrighted sources. This article does not contain any studies about human or animal objects.
- Aanniz T., Ouadghiri M., Melloul M., Swings J., Elfahime E., Ibijbijen J., Ismaili M. and Amar M., Thermophilic bacteria in Moroccan hot springs, salt marshes and desert soils. Brazilian Journal of Microbiology, 2015; 46: 443-453.
Crossref - Abdel-Rahman M.A., Tashiro Y., Zendo T., Sakai K. and Sonomoto K., Enterococcus faecium QU 50: a novel thermophilic lactic acid bacterium for high-yield l-lactic acid production from xylose. FEMS Microbiology Letters, 2015; 362: 1-7.
Crossref - Adiguzel A., Molecular characterization of thermophilic bacteria isolated from water samples taken from various thermal plants. PhD, Atat rk University, Graduate School at Natural and Applied Sciences, Erzurum, Turkey, 2006; 130.
- Adiguzel G., Bozoglu C., Yanmis D., Gormez A., Gulluce M. and Adiguzel A., Phenotyping and genotyping characterization of Salmonella strains isolated from retail beef in Erzurum, Turkey. Journal of Pure and Applied Microbiology, 2012; 6: 1581-1589.
- Adiguzel G., Faiz O., Sisecioglu M., Sari B., Baltaci O., Akbulut S., Genc B and Adiguzel A., A novel endo-b-1,4-xylanase from Pediococcus acidilactici GC25; purification, characterization and application in clarification of fruit juices. International Journal of Biological Macromolecules, 2019; 129: 571-578.
Crossref - Adiguzel G., Gulluce M., Bozoglu C., Yanmis D., Gormez A., Atasever M. and Adiguzel A., Molecular characterization of Escherichia coli O157: H7 from retail beef in Erzurum, Turkey. Journal of Pure and Applied Microbiology, 2012; 6: 1033-1041.
- Aguilar A., Extremophile research in the European Union: from fundamental aspects to industrial expectations. FEMS Microbiology Reviews, 1996; 18: 89-92.
Crossref - Al-Mariri A., Younes A.A. and Sharabi N-E., Characterization of thermophilic streptococci isolated from rustic white cheese. The Journal of General and Applied Microbiology, 2013; 59: 97-103.
Crossref - Altschul S.F. and Gish W., [27] Local alignment statistics, in Methods in Enzymology. Academic Press, 1996; 460-480.
Crossref - Aragno M. and Kristjansson J., Aerobic, chemolithoautotrophic, thermophilic bacteria. Thermophilic Bacteria, 1992; 77-103.
- Archana A. and Satyanarayana T., Xylanase production by thermophilic Bacillus licheniformis A99 in solid-state fermentation. Enzyme and Microbial Technology, 1997; 21: 12-17.
Crossref - Baltaci M.O., Genc B., Arslan S., Adiguzel G. and Adiguzel A., Isolation and Characterization of Thermophilic Bacteria from Geothermal Areas in Turkey and Preliminary Research on Biotechnologically Important Enzyme Production. Geomicrobiol. J., 2017; 34: 53-62.
Crossref - Benson G, Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999; 27: 573.
Crossref - Bjarni A. and Magn”?s MKn, Properties of Extremophilic Enzymes and Their Importance in Food Science and Technology, in Handbook of Food Enzymology. CRC Press, 2002.
- Crapart S., Fardeau M-L., Cayol J-L., Thomas P., Sery C., Ollivier B. and Combet-Blanc Y., Exiguo-bacterium profundum sp. nov., a moderately thermophilic, lactic acid-producing bacterium isolated from a deep-sea hydro-thermal vent. International Journal of Systematic and Evolutionary Microbiology, 2007; 57: 287-292.
Crossref - Derekova A., Sjרholm C., Mandeva R. and Kambourova M., Anoxybacillus rupiensis sp. nov., a novel thermophilic bacterium isolated from Rupi basin (Bulgaria). Extremophiles, 2007; 11: 577-583.
Crossref - Dua A. and Gupta R., Functional characterization of hormone sensitive-like lipase from Bacillus halodurans: synthesis and recovery of pNP-laurate with high yields. Extremophiles, 2017; 21: 871-889.
Crossref - Dunlap C.A., Kwon S-W., Rooney A.P. and Kim S-J., Bacillus paralicheniformis sp. nov., isolated from fermented soybean paste. International Journal of Systematic and Evolutionary Microbiology. 2015; 65: 3487-3492.
Crossref - Filippidou S., Jaussi M., Junier T., Wunderlin T., Jeanneret N., Palmieri F., Palmieri I., Roussel-Delif L., Vieth-Hillebrand A and Vetter A, Anoxybacillus-geothermalis sp. nov., a facultatively anaerobic, endospore-forming bacterium isolated from mineral deposits in a geothermal station. International Journal of Systematic and Evolutionary Microbiology, 2016; 66: 2944-2951.
Crossref - Gevers D., Huys G. and Swings J., Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiology Letters, 2001; 205: 31-36.
Crossref - Ghattargi V.C., Nimonkar Y.S., Burse S.A., Davray D., Kumbhare S.V., Shetty S.A., Gaikwad M.A., Suryavanshi M.V., Doijad S.P. and Utage B., Genomic and physiological analyses of an indigenous strain, Enterococcus faecium 17OM39. Functional & Integrative Genomics, 2018; 18: 385-399.
Crossref - Guagliardi A., Martino M., Iaccarino I., Rosa M.D., Rossi M. and Bartolucci S., Purification and characterization of the alcohol dehydrogenase from a novel strain of Bacillus stearother-mophilus growing at 70°C. The International Journal of Biochemistry & Cell Biology, 1996; 28: 239-246.
Crossref - Gugliandolo C., Maugeri T.L., Caccamo D. and Stackebrandt E., Bacillus aeolius sp. nov. a novel thermophilic, halophilic marine Bacillus species from Eolian Islands (Italy). Syst. Appl. Microbiol., 2003; 26: 172-176.
Crossref - Haki G. and Rakshit S., Developments in industrially important thermostable enzymes: a review. Bioresource Technology, 2003; 89: 17-34.
Crossref - Hashim S.O., Delgado O.D., Martםnez M.A., Kaul R-H., Mulaa F.J. and Mattiasson B., Alkaline active maltohexaose-forming a-amylase from Bacillus halodurans LBK 34. Enzyme and Microbial Technology, 2005; 36: 139-146.
Crossref - Hugenholtz P. and Pace N.R., Identifying microbial diversity in the natural environment: a molecular phylogenetic approach. Trends in biotechnology, 1996; 14: 190-197.
Crossref - Jardine J., Stoychev S., Mavumengwana V. and Ubomba-Jaswa E., Screening of potential bioremediation enzymes from hot spring bacteria using conventional plate assays and liquid chromatography-Tandem mass spectrometry (Lc-Ms/Ms). Journal of Environmental Management. 2018; 223: 787-796.
Crossref - Jensen M., Ardצ Y. and Vogensen F.K., Isolation of cultivable thermophilic lactic acid bacteria from cheeses made with mesophilic starter and molecular comparison with dairy related Lactobacillus helveticus strains. Letters in Applied Microbiology. 2009; 49: 396-402.
Crossref - Kasana R.C. and Pandey C., Exiguobacterium: an overview of a versatile genus with potential in industry and agriculture. Critical Reviews in Biotechnology, 2018; 38: 141-156.
Crossref - Kirk O., Borchert T.V. and Fuglsang C.C., Industrial enzyme applications. Current Opinion in Biotechnology, 2002; 13: 345-351.
Crossref - Koc M., Cokmus C. and Cihan A.C., The genotypic diversity and lipase production of some thermophilic bacilli from different genera. Brazilian Journal of Microbiology, 2015; 46: 1065-1076.
Crossref - Lin X-Q., Han S-Y., Zhang N, Hu H., Zheng S-P., Ye Y-R. and Lin Y., Bleach boosting effect of xylanase A from Bacillus halodurans C-125 in ECF bleaching of wheat straw pulp. Enzyme and Microbial Technology, 2013; 52: 91-98.
Crossref - Liu Y., Du J., Lai Q., Zeng R., Ye D., Xu J. and Shao Z., Proposal of nine novel species of the Bacillus cereus group. International Journal of Systematic and Evolutionary Microbiology, 2017; 67: 2499-2508.
Crossref - Madigan M.T., Martinko J.M. and Brock T.D., Brock biology of microorganisms. Pearson Prentice HallUpper Saddle River, NJ., 2006.
- Manceau C. and Horvais A., Assessment of genetic diversity among strains of Pseudomonas syringae by PCR-restriction fragment length polymorphism analysis of rRNA operons with special emphasis on P. syringae pv. tomato. Applied and Environmental Microbiology, 1997; 63: 498-505.
- Miller SA, T. R. Joaquim Diognostik techniques for plant pathogens, Biotechnology in Plant Disease Control, 1993.
- Namsaraev Z., Babasanova O., Dunaevsky Y., Akimov V., Barkhutova D., Gorlenko V. and Namsaraev B., Anoxybacillus mongoliensis sp. nov., a novel thermophilic proteinase producing bacterium isolated from alkaline hot spring, central Mongolia. Microbiology, 2010; 79: 491-499.
Crossref - Norashirene M., Zakiah Y., Nurdiana S., Hilwani I.N., Khairiyah M.S. and Arif M.M., Identification of Cellulose-Hydrolytic Thermophiles Isolated from Sg. Klah Hot Spring Based On 16S rDNA Gene Sequence. World Academy of Science, Engineering and Technology, International Journal of Biological, Biomolecular, Agricultural, Food and Biotechnological Engineering, 2014; 8: 1041-1044.
- Orr K., Holliday M., Jones A., Robson I. and Perry J., Survival of enterococci during hospital laundry processing. Journal of Hospital Infection, 2002; 50: 133-139.
Crossref - Othoum G., Bougouffa S., Razali R., Bokhari A., Alamoudi S., Antunes A., Gao X., Hoehndorf R., Arold S.T. and Gojobori T., In silico exploration of Red Sea Bacillus genomes for natural product biosynthetic gene clusters. BMC Genomics, 2018; 19: 382.
Crossref - Ozel N. General Outlook on Geothermal Resources in GAP Region and Thermal Springs of Guclukonak (SIRNAK) District Geothermal Energy. Available from: www.emo.org.tr/ekler/ecbba75f70a1293_ek.doc, 2000.
- Panda M.K., Sahu M.K. and Tayung K., Isolation and characterization of a thermophilic Bacillus sp. with protease activity isolated from hot spring of Tarabalo, Odisha, India. Iranian Journal of Microbiology, 2013; 5: 159.
- Poli A., Romano I., Cordella P., Orlando P., Nicolaus B. and Berrini C.C., Anoxybacillus thermarum sp. nov., a novel thermophilic bacterium isolated from thermal mud in Euganean hot springs, Abano Terme, Italy. Extremophiles, 2009; 13: 867-874.
Crossref - Prescott L.M., Harley J.P. and Klein D.A., Laboratory exercises in microbiology. McGraw-Hill Companies, 2002.
- Rai S.K. and Mukherjee A.K., Statistical optimization of production, purification and industrial application of a laundry detergent and organic solvent-stable subtilisin-like serine protease (Alzwiprase) from Bacillus subtilis DM-04. Biochemical Engineering Journal, 2010; 48: 173-180.
Crossref - Ramchuran S.O., Vargas V.A., Hatti-Kaul R. and Karlsson E.N., Production of a lipolytic enzyme originating from Bacillus halodurans LBB2 in the methylotrophic yeast Pichia pastoris. Applied Microbiology and Biotechnology, 2006; 71: 463-472.
Crossref - Sari B., Faiz O., Genc B., Sisecioglu M., Adiguzel A. and Adiguzel.G, New xylanolytic enzyme from Geobacillus galactosidasius BS61 from a geothermal resource in Turkey. International Journal of Biological Macromolecules, 2018; 119: 1017-1026.
Crossref - Schleifer K.H. and Kilpper-Bהlz R., Transfer of Streptococcus faecalis and Streptococcus faecium to the genus Enterococcus nom. rev. as Enterococcus faecalis comb. nov. and Enterococcus faecium comb. nov. International Journal of Systematic and Evolutionary Microbiology, 1984; 34: 31-34.
Crossref - Sirota-Madi A., Olender T., Helman Y., Brainis I., Finkelshtein A., Roth D., Hagai E., Leshkowitz D., Brodsky L., Galatenko V., Nikolaev V., Gutnick D.L., Lancet D. and Ben-Jacob E., Genome sequence of the pattern-forming social bacterium Paenibacillus dendritiformis C454 chiral morphotype. Journal of Bacteriology, 2012; 194: 2127-2128.
Crossref - Svec P., Vancanneyt M., Seman M., Snauwaert C., Lefebvre K., Sedlacek I and Swings J, Evaluation of (GTG) 5-PCR for identification of Enterococcus spp. FEMS Microbiology Letters, 2005; 247: 59-63.
Crossref - Takami H., Akiba T. and Horikoshi K., Production of extremely thermostable alkaline protease from Bacillus sp. no. AH-101. Applied Microbiology and Biotechnology, 1989; 30: 120-124.
Crossref - Tambekar D., Tambekar S., Jadhav A. and Babhulkar B., Isolation and partial characterization of protease from Bacillus halodurans (AJ302709) from alkaline Lonar lake. International Journal of Pharmaceutical Sciences and Research, 2016; 7: 4546.
- Vargas V.A., Delgado O.D., Hatti-Kaul R. and Mattiasson B., Lipase-producing microorganisms from a Kenyan alkaline soda lake. Biotechnology letters, 2004; 26: 81-86.
Crossref - Ward D.M., Weller R. and Bateson M.M., 16S rRNA sequences reveal numerous uncultured micro-organisms in a natural community. Nature, 1990; 345: 63.
Crossref - Wery N., Cambon-Bonavita M-A., Lesongeur F. and Barbier G., Diversity of anaerobic heterotrophic thermophiles isolated from deep-sea hydrothermal vents of the Mid-Atlantic Ridge. FEMS Microbiology Ecology, 2002; 41: 105-114.
Crossref - Whitman W.B., Coleman D.C. and Wiebe W.J., Prokaryotes: the unseen majority. Proceedings of the National Academy of Sciences, 1998; 95: 6578-6583.
Crossref - Yadav P., Korpole S., Prasad G.S., Sahni G., Maharjan J., Sreerama L. and Bhattarai T., Morphological, enzymatic screening, and phylogenetic analysis of thermophilic bacilli isolated from five hot springs of Myagdi, Nepal. Journal of Applied Biology & Biotechnology Vol., 2018; 6: 1-8.
Crossref - Yanmis D., Baltaci M.O., Gulluce M. and Adiguzel A., Identification of thermophilic strains from geothermal areas in Turkey by using conventional and molecular techniques. Res. J. Biotechnol., 2015; 10: 39-45.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


