Plasmodium knowlesi is a parasite that can spread from animals to humans. Over the past 20 years, scientists have become more and more interested in this parasite. This parasitic zoonotic infection is common in Southeast Asia, and 6-9 % of symptomatic adult who get it have severe symptoms. In this review paper, we will discuss recent and past studies on P. knowlesi. Additionally, we will describe the epidemiology, clinical aspects, diagnosis, and therapy of this infectious disease. In addition to this, we concentrate on the difficulties associated with the prevention and management of this important parasitic zoonotic disease.
Plasmodium knowlesi, Methods, Treatment, Diagnosis, Malaria
Malaria is an acute, febrile sickness caused by protozoan parasites that belong to the genus Plasmodium. It is spread via the infected mosquito. According to the WHO, there were 241 million occurrences of malaria in 2020, out of which there were 627,000 fatalities, an alarming 77% of which were under the age of 5 years and ninety-six per cent of mortality was attributed to the African region. Around 150 different species of the Plasmodium parasite have been identified as being infectious to birds, mammals, and reptiles.1 Even though they are only able to infect certain types of hosts, they occasionally spread to people as well. Plasmodium knowlesi is one example of a newly discovered parasite. The Presbytis mela lophos (banded leaf monkey), Macaca nemestrina (pig-tailed macaque), Macaca arctoides (stump-tailed macaques) and Macaca fascicularis (long-tailed macaque) are all natural hosts for the Plasmodium knowlesi parasite.2-4
Prior to the description of significant cases of P. knowlesi infections in Kapit District, Malaysia, by Singh et al.,5 malarial infections caused by P. knowlesi in human beings were thought to be extremely uncommon. After that, Plasmodium knowlesi infection has been described in many parts of the Southeast Asia region. As a result, Plasmodium knowlesi is currently believed to be the fifth species that causes malaria in humans.6 P. knowlesi is the cause of an upsurge in zoonotic malaria cases in Malaysia, which rose from 1600 cases to more than 4000 cases between 2016 and 2018.7 Although the number of P. knowlesi cases decreased significantly in 2019 and 2020 (to 3213 and 2609 cases, respectively) there were six and five fatalities in those years respectively.7
In India, P. knowlesi malaria infections are not frequently described; nonetheless, a research conducted in the Andaman and Nicobar Islands demonstrated that 53 of the 445 samples analysed had P. knowlesi-specific gene sequences.8 As a result of this, it is abundantly obvious that the population outside of South-East Asia is also at risk of infection with P. knowlesi, specifically in India.
It is quite difficult to identify P. knowlesi by microscopy on peripheral blood smear due to its similarities with other species of plasmodium, especially with P. falciparum and P. malariae.9
As P. knowlesi can be incorrectly identified as P. malariae or P. falciparum by conventional microscopy method, cases of P. knowlesi may already be prevalent in South East Asian countries but have been overlooked. In light of this, the utilisation of modern molecular diagnostic assays in conjunction with routine microscopy is required in order for us to be able to identify instances of P. knowlesi. In this review paper, we will examine and detail many studies on P. knowlesi. Additionally, we will describe the epidemiology, clinical aspects, diagnosis, and therapy of this infectious disease. In addition to this, we concentrate on the difficulties associated with the prevention and management of this important parasitic zoonotic disease.
History
P. knowlesi was first seen and studied in detail at the Kolkata School of Tropical Medicine, India, by Napier and Campbell in the peripheral blood smear of the Cercopithecus pygerythrus and Macacus rhesus in 1932.10 A further detailed study on P. knowlesi was executed by Knowles and Das Gupta. They reported that rhesus macaque shows fulminating and life-threatening parasitemia compared to long-tailed macaque. In the rhesus macaque, all phases of schizogony were evident in the peripheral circulation, and the terminal phases of infection were intense, with parasite numbers up to 3.25 million parasites per cubic millimeter. There were no appreciable symptoms, and the infection cleared up spontaneously in the long-tailed macaque.11 The first known natural infection of humans with P. knowlesi occurred in 1965, when a traveller acquired the parasite in Southeast Asia.12 The Kapit Division of Malaysian Borneo was the location where the largest number of cases was discovered.5 There have been reports of infections in humans not just in Malaysia but also in other countries of the world, including Thailand13,14 the Philippines.15,16 Myanmar,17,18 Singapore,19,20 Vietnam,21 Indonesia,22 and Cambodia.23
Vectors
The Anopheles Leucosphyrus group of mosquitoes are believed to be responsible for the transmission of knowlesi malaria, which are typically located in the same areas as are home to pig-tailed and long-tailed macaques.24
An. balabacensis is the most effective vector for the knowlesi infection.25,26 By employing An. balabacensis, it has been shown that P. knowlesi can be transmitted from monkeys to humans, as well as from humans to other humans, and from humans to monkeys. An. latens was identified as the vector in Kapit, Division of Malasia.27 An. cracens has also been found to be a carrier of knowlesi malaria in Pahang, Peninsular Malaysia. It bites most often between 8 and 9 p.m. An. cracens is highly zoophilic and has been seen feeding on macaques in the treetops and people on the ground.28, 29 The malaria parasites Plasmodium falciparum, Plasmodium malariae, Plasmodium vivax, and Plasmodium ovale were not discovered to be present in any of the P. knowlesi vectors that were examined in a recent entomological study in Sarawak,27,30 or in any of the other regions of Malaysia.28 In Vietnam, it has been determined that An. dirus is the most effective vector for the transmission of P. knowlesi.21 In this region, An. dirus is also known to act as a vector for P. vivax.
Evolutionary and Demographic History and Molecular Epidemiology
The research that was carried out in Sarawak demonstrated that many of the P. knowlesi isolates collected from wild macaques and those isolated from people shared identical sequences of circumsporozoite protein gene (csp) and mitochondrial DNA (mtDNA). Comparing the DNA sequences of nuclear P. knowlesi genes taken from human hosts and macaques makes it difficult to say whether or not knowlesi malaria is a recently emerging zoonosis. The examination of P. knowlesi mitochondrial DNA (mtDNA) sequences, on the other hand, makes it feasible to expand our knowledge of the demographic history and the evolution of the species P. knowlesi. Based on these types of analysis, the time to the most recent common ancestor (TMRCA) of P. knowlesi was estimated to be between 98,000 to 478,000 years ago.31 This suggests that P. knowlesi is descended from a population of ancestral parasites. It is at least as old as, and possibly even older than, the human malaria parasites P. vivax and P. falciparum, for which the TMRCAs have been estimated to be anywhere between 50,000 to 330,000 years ago,32,33 and 53,000 to 265,000 years ago respectively.34,35
Clinical presentation
An infection caused by Plasmodium knowlesi can manifest clinically in a variety of different ways, ranging from asymptomatic illness to severe disease. The incubation period is typically between three and fourteen days; however, higher incubation times have been reported in some instances.36-38 In the three largest prospective studies, the most common presenting symptoms of knowlesi malaria were found fever (100%), shivering, headache (89–94%), myalgia (47–88%), nausea, vomiting (24–34%), cough (35–56%), and abdominal pain (23–52%).39-41 The fever spikes occur every 24 hours. There are no specific features or clinical presentation of P. knowlesi infections that help in the diagnosis from any other type of the malaria infections.42
Lab diagnosis of P. knowlesi infection
Microscopy
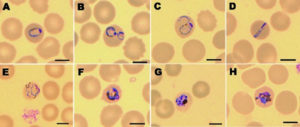
Due to the fact that P. knowlesi shares morphological similarities with other species of Plasmodium, the study of peripheral blood smears under a microscope is not the definitive method for identifying this Plasmodium.
The early trophozoites of P. knowlesi have the same morphological traits as P. falciparum, including double chromatin spots (Figure 1C), multiple infections per erythrocyte (Figure 1B), and no expansion of infected erythrocytes, making microscopical identification of P. knowlesi difficult (Figure 1A). Band-form trophozoites and the rest of P. knowlesi’s blood stages are similar to those of P. malariae (Figure 1D to 1H). When Knowles and Das Gupta introduced knowlesi malaria into three humans via blood passage in 1932, they first noticed the morphological similarities between P. knowlesi and P. malariae.
Peripheral blood films stained with Giemsa to demonstrate the various stages of P. knowlesi in RBCs are shown in Figure 1.43
Molecular identification
The accurate identification of malaria parasites has been made possible with the development of molecular detection technologies, which have consistently demonstrated to be more sensitive and specific than microscopy. The molecular detection methods for P. knowlesi, gene targets for PCR primers are summarized in Table. Primers Pmk8 and Pmkr9 were constructed to be used in a nested PCR assay for the identification of P. knowlesi. This assay was designed based on the small-subunit rRNA genes. In nested PCR assays, this gene was utilised extensively for the purpose of identifying Plasmodium vivax, Plasmodium falciparum, Plasmodium ovale, and Plasmodium malariae. The first round of PCR amplification was carried out with primers that were specific to the genus (primer rPLU6 in combination with either primer rPLU1 or rPLU5), and then a separate second round of PCR amplification was carried out with primers that were specific to the species. This method has high sensitivity and it is able to identify between one and six parasites per microliter of blood when a DNA template is created by a simple boiling procedure from blood spots collected on filter paper in the presence of a chelating agent.44
Table:
Molecular assays for Plasmodium knowlesi.
Gene target |
Type of assay |
Primer or probes |
Sequence (5’→3′) |
References |
|---|---|---|---|---|
SSU rRNA |
Nested PCR |
Pmk8 + Pmkr9 PkF1140 PkR1550 |
Pmk8:GTTAGCGAGAGCCACAAAAAAGCGAAT Pmk9: ACTCAAAGTAACAAAATCTTCCGTA PkF1140: GATTCATCTATTAAAAATTGCTTC PkR1550: CTTTTCTCTCCGGAGATTAGAACTC |
39 61 |
SSU rRNA |
Real-time PCR |
pk |
pk: CTCTCCGGAGATTAGAACTCTTAGATTGCT |
40 |
Apical membrane antigen 1 β Tubulin |
LAMP |
F3, B3, FIP, BIP, FLP, BLP |
F3: GCCAAGGATATTTATCTCCAC B3: CTTCTTCTTATGTTTGCCGT FIP:TTACAAACGTAAAAGTTGCAGGTACGATAAGGAGAGTATCAAATGTCCA BIP:CTGTGTAGAGAAGAGAGCAGAAATTCCGGATTTTCATAATCCTCC FLP: GGAAATGTGTTCAGGCTCAC BLP: CATAAAGGAAGAATTTAA |
42 |
However, these primers still yielded false positive results. Cross-reactivity exists between P. vivax and P. knowlesi’s18S ribosomal RNA (rRNA) target area.22,45,48,49 The sequences of the area that the Pmkr9 primer was designed to target were identical when the ssrRNA-S genes from P. knowlesi and P. vivax from GenBank were aligned. The P. vivax sequence, in contrast, only had a change at the 32 end and two mismatches in the first 19 bases in the area corresponding to the Pmk8 primer.45
Imwong developed two new primer sets, PkF1060-PkR1550 and PkF1040-PkR1550, as a solution to this issue. These primer sets consist of three primers suited for semi-nested PCR amplification of the ssrRNA-A fragment of the P. knowlesi rRNA gene expressed during the asexual stages (ssrRNA-A). This study demonstrate a specific identification of P. knowlesi without any overlap with other species.45,50
Real-time PCR assay targeting the SSU rRNA genes demonstrated by Divis et al. was showed excellent sensitivity and specificity. Real-time PCR techniques are more rapid and can provide quantitative data than nested PCR, but they are costly and necessitate a significant initial investment.46
Loop-mediated isothermal amplification (LAMP), discovered by Notomi et al., can amplify DNA with significant efficacy as its detection limit for DNA in the reaction mixture is as low as six copies within an hour at 65°C.51 In comparison to nested PCR, LAMP is highly sensitive, with detection limits as low as ten copies for the AMA-1 plasmid (10 copies). Lau YL et al demonstrated that all 13 P. knowlesi suspected samples by microscopy were positive under LAMP while Nested PCR using species-specific primers found P. knowlesi in 12 samples.47 And another study the 71 P. knowlesi blood samples, LAMP detected 69 microscopy-positive samples. LAMP exhibited higher sensitivity than nested PCR assay. The SYBR green I LAMP assay was 97.1% sensitive (95% CI 90.2–99.7%) and 100% specific (95% CI 83.2–100%).52 The real-time PCR techniques and other molecular detection assays, such loop-mediated isothermal amplification (LAMP) assays, are very expensive and will not completely replace microscopy in routine diagnostic labs in developed countries where resources are limited.
Rapid Diagnostic Method
Malaria rapid diagnostic tests (RDTs) are immunochromatographic tests that look for malaria antigens that are released from red blood cells that have been infected by parasites. RDTs need a blood sample from the patient, which is taken with a lancet. The sample and a buffer solution are then put in a test cassette, and the results are read. The tests do not need electricity or special equipment, and the results are available within 30 minutes. HRP-2, pLDH, and Plasmodium aldolase malaria antigens are detected in various types of RDT.53
There have been various different RDTs tested for knowlesi malaria in many studies. The outcomes of these tests have been mixed. Foster et al. discovered that when compared to microscopy, the sensitivity of P. knowlesi identification by the three RDTs (OptiMAL-IT, BinaxNOW® Malaria, and Paramax-3) was 71% (20/28; 95% CI = 54-88%) for fresh and 73% (30/41; 95% CI = 59-87%) for frozen knowlesi samples., 29% (8/28; 95% CI = 12-46%) of fresh and 24% (10/41; 95% CI = 11-37%) of frozen samples, 40% (10/25; 95% CI = 21-59%) with fresh and 32% 13/41; 95% CI = 17-46%) with frozen samples.54,55
Treatment
Artemether-lumefantrine and Artesunate- mefloquine is the preferred ACT for knowlesi malaria as listed by the Malaysian Ministry of Health lists.56 The knowlesi malaria was misdiagnosed as P. malariae due to its similarity on microscopy and treated with chloroquine, which was the first-line P. malariae treatment in Malaysia during the last decade. The majority of patients in the early case series of knowlesi malaria were also treated with chloroquine. Singh et al. reported 92 individuals with knowlesi malaria treated with chloroquine and primaquine with a median parasite clearance was 2.4 days in Sarawak, Malaysia. Ninety six people with uncomplicated knowlesi malaria received chloroquine in a hospital study and were successfully treated and all patients were PCR negative on days 7, 14, 21, and 28.57
Death is reported in unsuspected severe knowlesi malaria treated with chloroquine.58 For severe knowlesi malaria, parenteral artesunate is the treatment of choice, and once oral consumption is tolerated, 3-day course of an oral ACT is recommended, such as artemether-lumefantrine, in accordance with local recommendations and availability.59 Drug resistance has not been a problem due to the parasite’s zoonotic nature, and for uncomplicated illness,the first-line ACTs artemether-lumefantrine and artesunate-mefloquine have both been demonstrated to be very effective and for severe illness Intravenous artesunate must be started immediately.60
Control of Plasmodium knowlesi malaria
The knowlesi malaria transmission in humans involves multiple components, namely humans as accidental hosts, monkeys as natural hosts, and Anopheles with zoo-anthropophilic feeding behaviour. Humans are now very close to wildlife due to various activities, including logging industry, gathering jungle resources, subsistence agriculture, extension of home development to the border of forests, and eco-tourism. This makes the natural hosts (monkeys) and vectors (Anopheles), and humans close together and helps the knowlesi malaria transmission in humans.
Human knowlesi malaria transmission increases with monkey-human overlap. Breaking this transmission circuit should prevent knowlesi malaria in humans. Knowlesi malaria vectors must be identified for vector control. Several Anopheles Leucosphyrus mosquitoes have been linked to knowlesi malaria.26,28,61 Vector control should include landscape and urban planning. Human dwellings and natural forests should be separated by a “buffer zone” free of primates (humans and simians) and knowlesi malaria vectors.61,62 To break the malaria transmission cycle from monkeys to humans via mosquitoes, in locations where Anopheles balabacensis vectors P. knowlesi, the “buffer zone” between human activity and monkey habitats should be at least 1 km.63
Our current understanding of the pathophysiology, clinical course, therapy, and epidemiology of knowlesi malaria is based on a small number of case series and limited prospective and retrospective studies. The data obtained suggest that knowlesi malaria is predominantly a zoonotic disease, the primary reservoir hosts of P. knowlesi are Macaca fascicularis and Macaca nemestrina. Although P. knowlesi infections can be fatal, they can be treated with antimalarials. P. knowlesi can be difficult to accurately diagnose using microscopy because its various stages, such as early trophozoite, are similar to those of P. falciparum, and its mature blood stages are comparable to those of P. malariae. This makes it difficult to distinguish between the two parasites. In addition, the sensitivity of the rapid diagnostic tests that are now commercially available is low for P. knowlesi. The most accurate methods available for dectecting the P. knowlesi are P. knowlesi-specific PCR tests, which can be performed by nested PCR, real-time PCR, or PCR combined with sequencing. Healthcare providers and other medical professionals should be aware that P. knowlesi infections can sometimes be fatal, and knowlesi malaria should be monitored continuously.
ACKNOWLEDGMENTS
The author would like to acknowledge the Department of Microbiology, Jawaharlal Nehru Medical College, AMU, Aligarh, India, for providing the necessary facilities for the work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies on human participants or animals performed by any of the authors.
- Garnham PCC. 1966. Malaria parasites and other haemosporidia. Blackwell Scientific Publications, Oxford, United Kingdom. https://www.worldcat.org/title/malaria-parasites-and-other-haemosporidia/oclc/598598941
- Eyles DE, Laing AB, Dobrovolny CG. The malaria parasites of the pig-tailed macaque, Macaca nemestrina nemestrina (Linnaeus), in Malaya. Indian Journal of Malariology. 1962;16(3):285-98. https://www.cabdirect.org/cabdirect/abstract/19632901595
- Eyles DE, Laing AB, Warren M, et al. Malaria parasites of the Malayan leaf monkeys of the genus Presbytis. Med J Malaya. 1962;17:85-86. https://nla.gov.au/nla.obj-743595832/view
- Fungfuang W, Udom C, Tongthainan D, Kadir KA, Singh B. Malaria parasites in macaques in Thailand: stump-tailed macaques (Macacaarctoides) are new natural hosts for Plasmodium knowlesi, Plasmodium inui, Plasmodium coatneyi and Plasmodium fieldi. Malar J. 2020;19(1):350.
Crossref - Singh B, Kim Sung L, Matusop A, et al. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363(9414):1017-1024.
Crossref - White NJ. Plasmodium knowlesi: the fifth human malaria parasite. Clin Infect Dis. 2008;46(2):172-173.
Crossref - World Malaria Report 2021. 2021. https://www.who.int/publications/i/item/9789240040496.
- Tyagi RK, Das MK, Singh SS, Sharma YD. Discordance in drug resistance-associated mutationpatterns in marker genes of Plasmodium falciparum and Plasmodium knowlesi during coinfections. J Antimicrob Chemother. 2013;68(5):1081-1088.
Crossref - Lee KS, Cox-Singh J, Singh B. Morphological features and differential counts of Plasmodium knowlesi parasites in naturally acquired human infections. Malar J. 2009;8:73.
Crossref - Napier LE, Campbell HGM. Observations on a Plasmodium infection which causes haemoglobinuria in certain species of monkey. Ind Med Gaz. 1932;67(5);246:249.
- Knowles RM, Das Gupta B. A study of monkey malaria and its experimental transmission to man. Ind Med Gaz. 1932;67(6):301-320.
- Chin W, Contacos PG, Coatney GR, Kimball HR. A Naturally Acquited Quotidian-Type Malaria In Man Transferable To Monkeys. Science. 1965;149(3686):865.
Crossref - Jongwutiwes S, Buppan P, Kosuvin R, et al. Plasmodium knowlesi malaria in humans and macaques, Thailand. Emerg Infect Dis. 2011;17:1799 -1806.
Crossref - Sermwittayawong N, Singh B, Nishibuchi M, Sawangjaroen VN, Vuddhakul V. Human Plasmodium knowlesi infection in Ranong province, southwestern border of Thailand. Malar J. 11:36.
Crossref - Luchavez J, Espino F, Curameng P, et al. Human infections with Plasmodium knowlesi, the Philippines. Emerg. Infect. Dis. 2008;14:811-813.
Crossref - Kuo MC, Chiang TY, Chan CW, Tsai WS, Ji DD. A case report of simian malaria, Plasmodium knowlesi, in a Taiwanese traveler from Palawan Island, the Philippines. Taiwan Epidemiol Bull. 2009;25:178-91. https://scholar.google.com/scholar?cluster=16190211041226804054&hl=en&as_sdt=0,5
- Zhu HM, Li J, Zheng H. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 2006;24(1):70-71. https://pubmed.ncbi.nlm.nih.gov/16866152/
- Jiang N, Chang Q, Sun X, et al. Coinfections with Plasmodium knowlesi and other malaria parasites, Myanmar. Emerg Infect Dis. 2010;16:1476-1478.
Crossref - Ong CW, Lee SY, Koh WH, et al. Monkey malaria in humans: a diagnostic dilemma with conflicting laboratory data. Am J Trop Med Hyg. 2009;80(6):927-928.
Crossref - Jeslyn WP, Huat TC, Vernon L, et al. Molecular epidemiological investigation of Plasmodium knowlesi in humans and macaques in Singapore. Vector Borne Zoonotic Dis. 2011;11(2):131-135.
Crossref - Marchand RP, Culleton R, Maeno Y, et al. Coinfections of Plasmodium knowlesi, P. falciparum, and P. vivax among humans and Anopheles dirus mosquitoes, southern Vietnam. Emerg. Infect. Dis. 2011;17:1232- 1239.
Crossref - Sulistyaningsih E, Fitri LE, Löscher T, Berens-Riha N. Diagnostic difficulties with Plasmodium knowlesi infection in humans. Emerg Infect Dis. 2010;16(6):1033-1034.
Crossref - Khim N, Siv S, Kim S, et al. Plasmodium knowlesi infection in humans, Cambodia, 2007-2010. Emerg Infect Dis. 2011;17:1900 -1902.
Crossref - Sallum MA, Peyton EL, Wilkerson RC. Six new species of the Anopheles leucosphyrus group, reinterpretation of An. elegans and vector implications. Med Vet Entomol. 2005;19(2):158:199.
Crossref - Collins WE, Contacos PG, Skinner JC, Guinn EG. Studies on the transmission of simian malaria. IV. Further studies on the transmission of Plasmodium knowlesi by Anopheles balabacensis balabacensis mosquitoes. J Parasitol. 1971;57(5):961-966.
Crossref - Collins WE, Contacos PG, Guinn EG. Studies on the transmission of simian malarias. II. Transmission of the H strain of Plasmodium knowlesi by Anopheles balabacensis balabacensis. J. Parasitol. 1967;53(4): 841- 844.
Crossref - Tan CH, Vythilingam I, Matusop A, Chan ST, Singh B. Bionomics of Anopheles latens in Kapit, Sarawak, Malaysian Borneo in relation to the transmission of zoonotic simian malaria parasite Plasmodium knowlesi. Malar J. 2008;7:52.
Crossref - Vythilingam I, Noorazian YM, Huat TC, et al. Plasmodium knowlesi in humans, macaques and mosquitoes in peninsular Malaysia. Parasit. Vectors. 2008;1:26.
Crossref - Jiram AI, Vythilingam I, NoorAzian YM, Yusof YM, Azahari AH, Fong MY. Entomologic investigation of Plasmodium knowlesi vectors in Kuala Lipis, Pahang, Malaysia. Malar J. 2012;11:213.
Crossref - Vythilingam I, Tan CH, Asmad M, Chan ST, Lee KS, Singh B. Natural transmission of Plasmodium knowlesi to humans by Anopheles latens in Sarawak, Malaysia. Trans. R. Soc. Trop. Med. Hyg. 2006, 100:1087-1088.
Crossref - Lee KS, Divis PC, Zakaria SK, et al. Plasmodium knowlesi: reservoir hosts and tracking the emergence in humans and macaques. PLoS Pathog. 2011;7:1002015.
Crossref - Joy DA, Feng X, Mu J, et al. Early origin and recent expansion of Plasmodium falciparum. Science. 2003;300(5617):318-321.
Crossref - Krief S, Escalante AA, Pacheco MA, et al. On the diversity of malaria parasites in African apes and the origin of Plasmodium falciparum from Bonobos. PLoS Pathog. 2010;6(2):1000765.
Crossref - Escalante AA, Cornejo OE, Freeland DE, et al. A monkey’s tale: the origin of Plasmodium vivax as a human malaria parasite. Proc Natl Acad Sci USA. 2005;102(6):1980:1985.
Crossref - Mu J, Joy DA, Duan J, et al. Host switch leads to emergence of Plasmodium vivax malaria in humans. Mol Biol Evol. 2005;22(8):1686-1693.
Crossref - Berry A, IriartX, Wilhelm N, et al. Imported Plasmodium knowlesi malaria in a French tourist returning from Thailand. Am J Trop Med Hyg. 2011;84(4):535-538.
Crossref - Antinori S, Galimberti L, Milazzo L, Corbellino M. Plasmodium knowlesi: the emerging zoonotic malaria parasite. Acta Trop. 2013;125(2):191-201.
Crossref - TanizakiR,Ujiie M, Kato Y, et al. First case of Plasmodium knowlesi infection in a Japanese traveller returning from Malaysia. Malar J. 2013;12:128-131.
Crossref - Barber BE, William T, Grigg MJ, et al. A prospective comparative study of knowlesi, falciparum, and vivax malaria in Sabah, Malaysia: high proportion with severe disease from Plasmodium knowlesi and Plasmodium vivax but no mortality with early referral and artesunate therapy. Clin Infect Dis. 2013;56(3):383-397.
Crossref - Daneshvar C, Davis TM, Cox-Singh J, et al. Clinical and laboratory features of human Plasmodium knowlesi infection. Clin Infect Dis. 2009;49(6):852-860.
Crossref - Grigg MJ, William T, Barber BE, et al. Age-related clinical spectrum of Plasmodium knowlesi malaria and predictors of severity. Clin Infect Dis. 2018;67(3):350-359.
Crossref - Kantele A, Jokiranta TS: Review of cases with the emerging fifth human malaria parasite, Plasmodium knowlesi. Clin Infect Dis. 2011;52(11):1356-1362.
Crossref - Van Hellemond JJ, Rutten M, Koelewijn R, et al. Human Plasmodium knowlesi infection detected by rapid diagnostic tests for malaria. Emerg Infect Dis. 2009;15(9):1478-80.
Crossref - Lee WC, Chin PW, Lau YL, et al. Hyperparasitaemic human Plasmodium knowlesi infection with atypical morphology in peninsular Malaysia. Malar J. 2013;12:88.
Crossref - Imwong M, Tanomsing N, Pukrittayakamee S, Day NPJ, White NJ, Snounou G. Spurious amplification of a Plasmodium vivax small subunit RNA gene by use of primers currently used to detect P. knowlesi. J Clin Microbiol. 2009;47(12):4173-4175.
Crossref - Divis PC, Shokoples S.E, Singh B, et al.: A TaqMan real-time PCR assay for the detection and quantitation of Plasmodium knowlesi. Malar J. 2010;9:344.
Crossref - Lau YL, Fong MY, Mahmud R, et al. Specific, sensitive and rapid detection of human plasmodium knowlesi infection by loop-mediated isothermal amplification (LAMP) in blood samples. Malar J. 2011;10:197.
Crossref - Lubis IND, Wijaya H, Lubis M, et al. Contribution of Plasmodium knowlesi to multispecies human malaria infections in North Sumatera, Indonesia. J Infect Dis. 2017;215(7):1148-1155.
Crossref - Anderios F, Naing DKS, Lin Z, Emran NA. Single nucleotide polymorphism of SSU rRNA gene among Plasmodium knowlesi isolates of Sabah. Int J Collab Res Intern Med Public Heal. 2012;4(6):1007-1016.
- Suhandi DA, Suwandi JF. Identification of Plasmodium knowlesi using polymerase chain reaction (PCR) method. Medula. 2017;7(5):177-182. https://juke.kedokteran.unila.ac.id/index.php/medula/article/viewFile/2004/pdf
- Notomi T, Okayama H, Masubuchi H, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):63.
Crossref - Lai MY, Ooi CH, Lau YL. Validation of SYBR green I based closed tube loop mediated isothermal amplification (LAMP) assay for diagnosis of knowlesi malaria. Malar J. 2021;20(1):166.
Crossref - Moody A. Rapid diagnostic tests for malaria parasites. Clin Microbiol Rev. 2002;15(1):66-78.
Crossref - Foster D, Cox-Singh J, Mohamad DS, Krishna S, Chin PP, Singh B. Evaluation of three rapid diagnostic tests for the detection of human infections with Plasmodium knowlesi. Malar J. 2014;13:60.
Crossref - Barber BE, William T, Grigg MJ, Piera K, Yeo TW, Anstey NM. Evaluation of the sensitivity of a pLDH-based and an aldolase-based rapid diagnostic test for diagnosis of uncomplicated and severe malaria caused by PCR-confirmed Plasmodium knowlesi, Plasmodium falciparum, and Plasmodium vivax. J ClinMicrobiol. 2013;51(4):1118-1123.
Crossref - Ministry Of Health Malaysia. Management Guidelines of Malaria in Malaysia. Vector Borne Disease Sector, Disease Control Division, Ministry of Health, Malaysia. 2014. https://www.moh.gov.my/index.php/file_manager/dl_item/ 554756755a584a69615852686269394859584a70637942515957356b64
- Daneshvar C, Davis TM, Cox-Singh J, et al. Clinical and parasitological response to oral chloroquine and primaquine in uncomplicated human Plasmodium knowlesi infections. Malar J. 2010;9:238.
Crossref - Rajahram GS, Barber BE, William T, Menon J, Anstey NM, Yeo TW. Deaths due to Plasmodium knowlesi malaria in Sabah, Malaysia: association with reporting as P. malariae and delayed parenteral artesunate. Malaria J. 2012, Malaysia: association with reporting as P. malariae and delayed parenteral artesunate. Malar J. 2012;11:284.
Crossref - World Health Organization. 2022. https://www.who.int/publications/i/item/guidelines-for-malaria.
- Barber BE, Grigg MJ, Cooper DJ, et al. Clinical management of Plasmodium knowlesi malaria. Adv Parasitol. 2021;113:45-76.
Crossref - Chua TH, Manin BO, Vythilingam I, Fornace K, Drakeley CJ. Effect of different habitat types on abundance and biting times of Anopheles balabacensis Baisas (Diptera: Culicidae) in Kudat district of Sabah, Malaysia. Parasit Vectors. 2019;12:364.
Crossref - Hawkes FM, Manin BO, Cooper A, et al. Vector compositions change across forested to deforested ecotones in emerging areas of zoonotic malaria transmission in Malaysia. Sci Rep. 2019;9(1):13312.
Crossref - Lee WC, Cheong FW, Amir A, et al. Plasmodium knowlesi: the game changer for malaria eradication. Malar J. 2022;21(1):140.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.