Bacteria belonging to the genus Methylobacterium, popularly known as pink pigmented facultative methylotrophic (PPFM) bacteria, are well known for their distinct ability to use single-carbon compounds like methanol, formate and formaldehyde, and also a variety of multi-carbon substrates lacking carbon-carbon bonds. These bacterial groups are ubiquitously distributed, especially in phyllosphere and rhizosphere, and their occurrence have been reported in more than 100 species of plants so far. PPFMs have profound influence on soil fertility, crop growth and yield. The ability for phosphate acquisition, nitrogen fixation, iron chelation and phytohormone production indicate the possibility of developing them as promising biofertilizer candidates. In addition, many of them possess biocontrol activity against several phytopathogens. PPFMs induce several physiological changes in plants, making the plants more resistant to biotic and abiotic stress. They can therefore be promising alternatives to conventional chemical inputs in sustainable agricultural systems.
Methylobacterium, Methylotrophs, Plant Growth Promotion, PPFMs, Biocontrol
Phyllosphere harbours large, complex and dynamic communities of microorganisms, where bacteria constitute the dominant microbial inhabitants. It forms a significant microbial habitat that supports organisms of diverse nutritional and physiological requirements. The leaf surface area of terrestrial plants possibly be occupied by microbes is reported to be around 6.4 × 108 km2, that would harbour bacterial populations of about 1026 cells. Plants are potential reservoir of structurally and functionally diverse natural compounds ranging from quite simple esters to more complex molecules like carbohydrates, polyketides, flavanoids, lignans, terpenoids, alkaloids, and tannins.
Of the diverse natural products, methanol, a simple organic molecule, formed as a by-product of pectin demethylation during cell wall metabolism is released from plants via epidermal stomatal pores. Pectin, a major plant cell wall component, is structurally a heteropolysaccharide enriched with α-D-galacturonate residues, and a number of sugars such as α-L-arabinose, α-L-rhamnose and β-D-galactose in smaller amounts. Galacturonate methyl esters that are present in the cell wall helps in the transport of compounds through it during cell wall expansion. Demethylation of these methyl esters by methylesterase produces methanol as a by-product.1 Emission of methanol through stomata by the transpiration stream leads to the enrichment of plant surfaces with methanol.2,3
Several microorganisms have evolved the interesting characteristic to utilize mono carbon (C1) compounds like methanol and methane or complex carbon compounds lacking carbon-carbon bonds (dimethyl ether and dimethylamine) as the carbon source. These bacteria are commonly referred to as methylotrophs and the ability of an organism to utilize single carbon compounds as the exclusive energy source for its growth is known as methylotrophy.4 Among the methylotrophic organisms, facultative methylotrophic (FM) bacteria of the genus Methylobacterium and Methylorubrum have been widely studied and are generally known by the term pink pigmented facultative methylotrophic (PPFM) bacteria. These facultative methylotrophic bacteria with unique physiological characteristics are distributed ubiquitously in/on plants.
PPFMs have been studied widely for their plant growth promoting ability by a plethora of mechanisms. Important modes of action include secretion of plant growth stimulating compounds like indole-3-acetic acid (IAA), cytokinin, Gibberellic acid (GA), 1-aminocyclopropane-1-carboxylate deaminase (ACC) and increasing the availability of essential nutrients. These mechanisms alone or in combination positively influences the growth and development of the host plant. Moreover, biocontrol ability of PPFMs is considered another noticeable beneficial trait which lessen the detrimental effect of various phytopathogens. Thus, plant beneficial PPFMs can play a significant role in crop cultivation and many researches suggest the possibility of their application in agriculture as an excellent bioresource to reduce the detrimental impacts of chemical inputs. Keeping in view of all the beneficial attributes of PPFMs, this review aims to provide a concise summary of the findings from various relevant studies describing the potential of pink pigmented facultative methylotrophs as promising alternative to conventional hazardous chemical inputs for eco-friendly and sustainable crop production.
Characteristics of PPFMs
PPFMs are obligate aerobic Gram-negative rods, which can grow on single-carbon substrates especially methanol and methylamine and also on an array of multicarbon containing compounds.3,4 The average cell size of the bacterium is approximately 1.0 µm long by 0.5 µm wide. The major bacterial storage compound, poly-b-hydroxy butyrate (PHB) granules were identified in cells of Methylobacterium spp. using PHB granule staining.5 Most studies have shown that Methylobacterium are Gram negative; however, some reports observed them to be Gram variable.6 Many studies documented the ubiquitous presence of PPFM in soil, freshwater, lake sediments, leaf surface, nodules and dust. Bassalik7 described the first Methylobacterium strain isolated from earth worm casts and called it as Bacillus extorquens. Later, Kuono and Ozaki8 isolated 59 PPFM strains from many soil and water samples. Patt et al.9 isolated and reported the first PPFM strain with methane utilization ability. The ubiquitous nature of PPFM was first described by Green and Bousefeild.10 Considering the methanol utilization ability, PPFMs are usually isolated on Ammonium Mineral Salt (AMS) agar medium amended with methanol as the exclusive carbon source. They utilize methanol by oxidizing it to formaldehyde by means of pyrroloquinoline quinone (PQQ)-dependent methanol dehydrogenases (MDHs).11 There are two paralogous MDH enzymes present in PPFMs, viz. a Ca2+-dependent MxaFI and lanthanide (Ln3+)-dependent XoxF.12 Lanthanide (Ln3+)-dependent XoxF is highly conserved in Methylobacterium species than Ca2+-dependent MxaFI.13
PPFM as plant associated bacteria
Presence of Methylobacterium has been described on a wide array of sources like root of rice,14 leaves of rice,15,16 stem of rice,17 root nodule of crotalaria,18 South African legumes,19 stem nodule of Sesbania rostrata,20 soil,21-27 soyabean,28 vegetables,29 Cucurbita pepo,30 seeds of rice,31 brassica,32 Combretum erythrophyllum,33 potato,34 banana,35 palm oil tree,36 palm oil,37 siam squash and corn,38 and soyabean39 by various workers. All these evidences helped gaining insights into the distribution of PPFMs in nature especially on plants. Plant-Methylobacteria beneficial communication could be considered as a best model for symbiotic association between plants and microbes. Host plants release metabolic by-products including carbon source majorly in the form of methanol which are utilized by associated Methylobacteria. In turn, bacteria offer phytohormones needed for the growth and metabolism of the host. Such mutually beneficial relations generally indicate the chance of coevolution of symbiotic bacterial partner.40 Generally, Methylobacterium spp. are present throughout the plant, especially the leaf surfaces, stem, flowers and roots. Austin et al.41 first described the diversity of Methylobacterium in the phyllosphere region of Lollium perenne. Later, they have been reported as the most dominant phyllosphere population from more than seventy plant species tested and are mostly found as common prokaryotic epiphytes.42-45 However, they can colonize plants as endosymbionts46 and a few members can reside in intracellular spaces of meristematic cells.47 Endophytic and intercellular colonization of bacteria supports active interaction of bacterial symbionts with host plant. Colonization of Methylobacterium spp. in intercellular spaces of tomato were observed by Poonguzhali et al.48 Undoubtedly, all these previous reports establish the ubiquitous presence of PPFM bacteria in the nature. Almost all the information pertaining to colonization of Methylobacterium in plants imply their active cross talk with host plants. Besides, a few researchers have made attempts to understand and elucidate the biology of Methylobacterium spp. found in untapped habitats such as biological soil crusts,49 Lichen like Lepraria sp.,50 spores of the mycorrhizal fungus Glomus iranicum var. tenuihypharum,51 tungsten mine tailings52 and Philippine fermented food.53
The genetic diversity of these pink microbial community has been studied by different techniques such as Restriction fragment length polymorphism, Restriction analysis of polymorphic DNA and Amplified ribosomal DNA restriction analysis. These finger printing techniques could discriminate distinct microbial communities amongst PPFMs. For the first time, PPFMs inhabiting maize, cotton, and sunflower phyllosphere were detailed based on utilization profile of carbon source and RAPD data by Balachander et al.54 It revealed the presence of six diversified groups of PPFM, wherein four different groups were found harbouring the phyllosphere of sunflower and maize and only two groups on cotton phyllosphere.
Genetic diversity of PPFM assessed using molecular tools and differential carbon source utilization profile identified Methylobacterium populi, M. thiocyanatum, M. suomiense, M. aminovorans, and M. fujisawaense as the predominant colonizers in the phyllosphere of crop plants like cotton, sunflower, maize, soybean and mentha.55 Further, the genetic diversity of the heavy metal tolerant Methylobacterium spp. community in a mangrove forest has been unveiled with the help of in vitro assays.56 Kaur et al.57 assessed the diversity and heterogeneity of PPFM bacteria from the leaves of five kharif crops viz., rice, maize, millet, mung bean and urad bean through plant growth promotion screening and ARDRA profiling and noticed the prevalence of four distinct groups of PPFMs in these plants. Based on 16S rRNA phylogenetic gene sequencing, a high abundance of sequences closely related to Methylobacterium radiotolerans was reported on sugarcane plants.58
Assessment of the influence on the diversity and community changes of PPFM in transgenic Bt-cotton compared with the conventional cotton plants by means of a polyphasic approach that included differential carbon source utilization profiling and DNA based techniques showed that diversity richness of PPFMs in the phyllosphere, rhizoplane and internal tissues has not differed among Bt and non-Bt-cotton plants.59 Although much remains to be explored, the documented data pertaining to the genetic diversity of Methylobacterium might give a crucial insight to select potential plant associated bacteria with plant growth-promoting characteristics for utilization in sustainable agriculture.
Carotenoid Pigment Production: A major characteristic of PPFM
Carotenoids are a group of yellow, orange, or red-colored lipophilic isoprenoid molecules present in all kingdoms of life. These natural pigments play diverse functional roles such as capturing and processing of light, photoprotection, specific coloration across genera and species, and pollinator attraction in both photosynthetic and non-photosynthetic biological systems.60,61 Approximately, 1100 different carotenoids compounds have been identified so far from varied sources with diverse colours and they provide attractive colours to its source of origin.62 Bacteria belong to the genus Methylobacterium possess a characteristic pinkish colour due to the presence of non-diffusible and non-fluorescent carotenoids, mainly xanthophylls63,64 and hence named as pink-pigmented facultative methylotrophs.
Generally, accessory light-harvesting carotenoid complexes are found with chlorophyll molecules in green plants. In photosynthetic organisms, these light capturing complexes protects the chlorophyll molecules from photooxidation by absorbing and transferring light energy to chlorophyll molecules. More importantly, its anti-oxidant or oxygen free radical quenching activity protect the cell in both phototrophic and non-phototrophic organisms, usually under biotic and abiotic stress conditions. The production of carotenoid pigments makes the producer bacteria tolerant to extreme light condition and radiation.65 Abiotic stresses cause accumulation of reactive oxygen species, that leads to oxidative injury to the organisms. To overcome the lethal effects of ROS, many organisms possess an evolved strategy viz., accumulation of carotenoid pigment by up-regulating carotenoid biosynthetic pathway.61 Carotenoid pigments produced by PPFMs as secondary metabolites serve as a reliable and useful chemotaxonomic identification marker of the genus.66 Biosynthesis of carotenoids makes them resistant to extreme light conditions, high or low temperatures and freezing conditions and also to UV and ionizing radiations.67
The colonies formed by Microbacterium arborescens-AGSB, a Gram-positive bacterium, collected from coastal sand dune vegetation, Ipomea pes-caprae were predominantly orange pigmented.68 This study revealed the light induced biosynthesis of carotenoids pigment in Microbacterium arborescens-AGSB helps them to survive under stress conditions. Detailed chromatographic and spectrophotometric analysis of carotenoids in Methylobacterium genus has shown that majority belong to oscilloxanthin,64 C40 carotenoid astaxanthin69 and lycopene type carotenoids.68 The ability to produce carotenoid pigments by PPFM can be made use of in different industries.70-73 Several attempts have been made to recover the maximum amount of carotenoid pigments from PPFM,74,75 and during recent year, the optimum conditions for the extraction when bacteria were grown in Ammonium mineral salt medium was reported to be at 25˚C, pH of 7.5, having 0.5% of methanol as carbon source.76
Plant Growth Promotion Mediated by PPFMs
PPFMs promote crop growth through multitude of mechanisms like indole-3-acetic acid production,77,78 cytokinin production,79 vitamin B12 production,80 siderophore production,81 ACC deaminase production,82 nitrogen fixation and nodule formation83 and phosphorus solubilization.84 Furthermore, significant biocontrol activity of Methylobacterium spp. against phytopathogens protects the plants from destructive pathogens and thereby improves the health of plants. All these characteristics emphasize the potential of the PPFMs in crop production. Strikingly, much progress has been made over the past few years in understanding the beneficial traits of PPFM. A concise information on plant growth promoting traits of Methylobacterium is given below (Table 1; Figure 1).
Table (1):
Plant growth promoting attributes of PPFM in various crops.
Crop plant |
Associated methylotrophs |
Source |
Activity |
References |
|---|---|---|---|---|
Rice |
Methylobacterium extorquens, Methylobacterium fujisawaense |
Phyllosphere |
IAA production |
91 |
Rice |
Methylobacterium sp. CBMB-20 |
Rhizosphere |
N2 fixation |
15 |
Soybean |
Pink-pigmented facultative methylotroph |
Phyllosphere |
IAA production |
93 |
Tomato |
Methylobacterium suomiense |
Rhizosphere |
Root colonization |
48 |
Red pepper |
Methylobacterium suomiense |
Rhizosphere |
Root colonization |
192 |
Groundnut |
Pink-pigmented facultative methylotroph |
Phyllosphere |
IAA production |
78 |
Sugarcane |
Methylobacterium sp. |
Stem, root, Rhizosphere |
IAA production, PGP activities |
193 |
Red pepper |
Methylobacterium oryzae |
Rhizosphere |
Biofertilizer |
194 |
Mung bean |
Methylobacterium organophilum |
Mud |
Biofertilizer |
133 |
Red pepper |
Methylobacterium sp. |
Phyllosphere |
IAA and cytokinin production |
195 |
Combretum erythrophyllum |
Methylobacterium radiotolerans MAMP 4754 |
Seed |
Synthesis of heavy metal resistant proteins plant growth-promoting compounds |
196 |
Banana |
Methylobacterium salsuginis TNMB03 |
Leaf |
Cytokinin production ACC deaminase activity Biochemical changes in plant |
35 |
Poplar |
Methylobacterium sp. CP3 |
Seed of Crotalaria pumila |
IAA production Phytate mineralization Zn tolerance |
197 |
Phytohormone Production
Soil and plant-associated bacterial groups can synthesize and excrete one or more phytohormone. Among the various plant growth hormones, Auxins are found to be the most essential phytohormone for normal plant growth and development and till date, there have been no reports on plants lacking auxin synthesizing ability. It has been well documented that microbially excreted auxins exert positive influence on plant growth. Though, plants synthesize various auxins, Indole-3-acetic acid (IAA) is the most profound auxin as it is directly involved in several crucial developmental processes. Hence, IAA producing bacteria can potentially involve in increasing the plant’s auxin pool.85
A large number of reports have described indole-3-acetic acid (IAA) production by methylotrophs. IAA is synthesized using different pathways by the producer organisms. Based on the intermediaries formed, these pathways are classified into indole-3-acetamide (IAM), tryptamine and indole-3-acetonitrile and indole-3-pyruvic acid (IPyA) pathway.86 Of these, most important IAA biosynthetic pathways operating in PPFM are the IAM and the IPyA pathways. IAA produced by methylotrophic bacteria via, IAM and IPyA pathways has significant effects on growth of plants.87 Ivanova et al.87 first reported the production of significant amount of indole acetic acid in culture supernatants of four different methylotrophic bacteria. IAA produced by Methylobacterium has been found to influence seed proliferation and seedling growth of various plants. 77, 88-90 Based on colorimetric assay the presence of indole compounds was observed in PPFM culture supernatants also.77 IAA synthesized by PPFM has influence on the root growth and development of various host plants also.91 Thangamani and Sundaram92 reported that among 16 Methylobacterium isolates tested three isolates indicated the accumulation of indole compounds in PPFM culture supernatants. These three isolates produced IAA amounting from 6 to 13.3 µg mL-1 in the presence of L-tryptophan. Radha93 and Jones94 have independently documented the production of IAA by different PPFM strains ranging from 9.04 to 28.15 μg mL-1 and 0.14 to 25.15 μg mL-1 of culture filtrate, respectively. Anitha78 isolated PPFMs from the phyllosphere of different crops by leaf impression method and screened eight isolates for their influence on seed germination and production of IAA. Using HPLC, amount of IAA produced by different isolates was estimated and a maximum of 2.32 μg mL-1 of IAA was produced even in the absence of tryptophan by the isolate obtained from ground nut leaf (PPFM-GN). An increase in plant IAA concentration with the inoculation of Methylobacterium isolate and subsequent plant growth promotion was reported by Lee et al.15
Besides Indole acetic acid, Cytokinins also influence physiological functions of plants. Root functions of plants can be altered by change in level of cytokinin concentration in plants. Potential involvement of plant-growth promoting rhizobacteria (PGPR) like Azospirillum, Azotobacter, Bacillus, Pseudomonas spp. and Rhizobium in cytokinin production in pure cultures has already been described.95-102 Methylotrophic strains present on phyllosphere have also been found to produce cytokinin, with many of them able to excrete it into the growing medium.103-105 Study by Madhaiyan et al.106 demonstrated that PPFM inoculation on true seeds of sugarcane increases the percentage and rate of germination. A higher PPFM population has been observed when a combination of seed imbibition, soil application and phyllosphere spray were given. An immunological assay reported a significant increase of cytokinin both in mature and young leaves of sugar cane plants. Foliar spray of methanol or PPFMs results in increased PPFM populations which in turn caused a higher concentration of bacterially produced cytokinin and increased cytokinin contributed to improvement in yield of cotton and sugarcane.107 Quantification of cytokinin produced by phyllosphere inhabiting Methylobacteria isolated from sugarcane, pigeon pea, mustard, potato and radish ranged between 1.09 to 9.89 µg mL-1 in the culture filtrate and treating wheat seeds with these cell-free culture filtrates registered a significant improvement in seed germination.79 With a similar approach, El-Gawad et al.108 described cytokinin production of PPFM bacteria obtained from cotton, datura, snap bean, castor oil and peanut plants with a maximum of 2.07 µg ml-1 of culture filtrate. An exhaustive cytokinin profiling of Methylobacterium strains was conducted by Palberg et al.109 recently. Analytical results obtained from High performance-liquid chromatography-tandem mass spectrometry (HPLC–MS/MS) could uncover the immense potential of Methylobacterium strains to produce most active cytokinin form, trans-Zeatin (tZ), ranged from 0.46 to 82.16 pmol mL−1 and which marked higher than tZ produced by any other plant growth promoting bacteria reported so far. Despite a number of exclusive studies on the cytokinin production potential of PPFMs, still there is lack of knowledge on exact role of Methylobacterium produced cytokinins in plants. Therefore, further elucidation is required to understand its precise mechanism in plants.
Gibberellic acid (GA3), a plant growth hormone, is a key growth regulator involved in cell division and tissue differentiation, net assimilation rate, dry matter accumulation, leaf expansion and elongation, regulation of transpiration rate, flowering and photosynthesis.110,111 Apart from these functions, GA plays a pivotal role in regulating plant development and growth under various abiotic stress conditions.112 Plant growth promoting effect of gibberellins produced by many plant-growth promoting bacteria (PGPB) were reported by several workers.113-116 For the first time, Rajan et al.117 reported gibberellic acid production in Methylobacterium obtained from vegetable crops and was found to be ranging from 10.9 μg mL-1 to 106.97 μg mL-1 of the culture filtrate. Studies by Radha93 and Jones94 also revealed the production of gibberellic acid by methylotrophs which ranged from 24.11 to 70.30 μg mL-1 and 53.2 to 273.2 μg mL-1, respectively. Further, Sheela et al.118 have successfully demonstrated the GA production potential of different Methylobacterium strains and estimated a maximum amount of 59.13 μg mL-1 of GA. PPFM strains isolated from chilli leaves, rhizosphere soil and roots samples produced GA from 4.77 µg mL-1 to 128.28 µg mL-1 of culture filtrate among different isolates as reported by Savitha et al.119
Detailed investigations on phytophormone synthesize and release by plant associated Methylobacterium community, generally encountered on leaf surfaces, may provide insight into their beneficial activities. It is also possible to improve the PPFM strains by developing hormone over-producers which may have direct influence on plant growth and development.
Siderophore Production
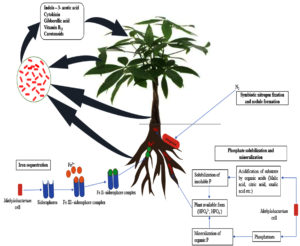
Siderophore mediated sequestration and transport of Fe3+ is an efficient strategy evolved in bacteria to meet iron requirements.120 Bacteria produces low-molecular-weight Fe chelating compounds, siderophores, that helps to sequester and transport the element into bacterial cells. Siderophore production by rhizospheric microorganisms is beneficial to plants as phytopathogens are out-competed by the producer strain under limited iron supply.121 This competitive advantage determines how bacteria can survive and provide benefit to the host plants.
Siderophores, either hydroxamate-type or catecholate-types are produced by plant growth promoting bacteria. Under iron limiting conditions, Methylobacterium spp. also produce siderophore which transport the Fe-siderophore complex by the use of specific proteins. Siderophores produced by them are anti-pathological factors which can also be considered as indicator of biocontrol efficiency.122 In vitro production of siderophores by the pathogen Xylella fastidiosa and PPFM Methylobacterium extorquens tested by Silva-stenico et al.123 found that the culture supernatants of X. fastidiosa did not contain both hydroxamate and catechol siderophores, whereas hydroxamate siderophores have been detected in the culture supernatants of M. extorquens. In citrus plants, Methylobacterium spp. inhabiting same ecological niche of Xylella fastidiosa subsp. pauca (Xfp) was found to be incapable of producing catechol-type siderophores but were also producing hydroxamate-type siderophores. Interestingly, in vitro growth of the pathogen was found to be increased in the presence of endophytic M. mesophilicum produced siderophores which occupy the same niche.124 Simionato et al.122 reported the production of siderophores by Methylobacterium mesophilicum (ARS 1/5 and ARS 1/6 strains). Celosia species is an iron rich plant with pink pigmented leaves and flowers. Gholizadeh and Kohnehrouz125 postulated the occurrence of both highly efficient iron-uptake bacteria and PPFM bacteria on the leaves of Celosia species. The prediction was experimented by probing with a cDNA fragment coding for Methylobacterial-type Fe siderophore receptor within Celosia leaf cDNA microflora. The results indicated the inhabitation of efficient iron scavenging bacteria, most probably PPFM, on the surface of plants with high iron content.
Positive results were obtained when Methylobacterium phyllosphaerae MB-5 and CBMB-27 were screened for synthesis of amino acid conjugated hydroxamate type of siderophore under laboratory conditions, whereas, both the strains not contained catecholate type of siderophores during iron limitation.126 According to Senthilkumar et al.,35 all the 28 endophytic Methylobacterium isolates obtained from south Indian banana cultivars were shown to have iron siderophore complexes production in CAS agar medium, indicated by an yellow halo zone formation around the bacterial colonies. The experimental results demonstrated that these endophytic Methylobacterium species ensure plant growth promoting nutrients/compounds to the host plants. Methylobacterium genomes contain different types of siderophores and TonB-dependent receptors (TBDRs) with diverse roles. Siderophore mutant Methylobacterium strain was unable to utilize methanol as carbon source, but they could solubilize insoluble anthanides oxide, suggesting their crucial role in methylotrophy. Besides, siderophores have important role in lanthanide uptake, oxidative and nitrosative stress tolerance, biofilm formation, and heavy metal sequestration.127
Reports suggest members of Methylobacterium as a potent source of siderophores which are successfully involved in iron scavenging and transport. Furthermore, siderophore production helps in root pathogen suppression by competitive exclusion, and promotes growth and development of the host plants.
Nitrogen fixation
Nitrogen, a vital mineral nutrient, is indispensable for development and growth of plants.128 It forms the building blocks of many structurally and physiologically relevant molecules like proteins, nucleic acids, chlorophyll and coenzymes. Moreover, it constitutes an important component of ATP, energy currency of the cell.129 There are numerous microorganisms, referred as diazotrophs, especially eubacteria, which can add substantial amount of nitrogen into soil by N2 fixation. Diazotrophic microorganisms provide fixed nitrogen in exchange of carbon source from plants.130 The beneficial effects of diazotrophic organisms have been well documented.
There has been remarkable research progress in the area of Methylobacterium mediated nitrogen fixation and the involvement of Methylobacterium mediated nitrogen acquisition by host plants. In the past few years, Rhizobia were classified under three different phylogenetic branches within the alpha-2 subclass of Proteobacteria. Of the three branches described, first branch contains the genera Rhizobium, Sinorhizobium, Mesorhizobium, and Allorhizobium with Agrobacterium, second branch with Bradyrhizobium with photosynthetic free-living Rhodopseudomonas, and third contains the genus Azorhizobium as well as the chemoautotroph Xanthobacter. Later, Sy et al.83 described Methylobacterium as a fourth rhizobial branch within alpha-subclass of Proteobacteria and introduced a new species of Rhizobium, Methylobacterium nodulans, isolated from Crotalaria legumes using nodA amplification assays. Though the wide spread occurrence of Methylobacterium in various crops has already been well known, their symbiotic relationship with host plants was unknown until recently. In this concern, Methylobacterium nodulans capable of inducing nodulation in plants was the first nodule forming Methylobacterium species identified. This study could reveal the relatedness of nodA gene present in M. nodulans with nodA gene of Bradyrhizobium and was believed to be acquired by horizontal gene transfer mechanisms. It was for the first time, Raja et al.131 reported that a phyllosphere colonizing, non-nodulating Methylotroph, Methylobacterium spp. MV10. possess functional nifH gene which was quite different from M. nodulans. A partial sequencing of genome of Methylobacterium extroquens helped identifying a few genes with close similarity to symbiosis-associated genes of Rhizobia and Agrobacterium.132
Lee et al.15 experimented the potentiality of diazotrophic rice methylotrophs like Methylobacterium sp. CBMB20, Enterobacter sp. CBMB30, Burkholderia sp. CBMB40 in improving the rice seedling growth. These isolates exerted a discernible influence on germination of seed and seedling vigour index, and biomass production of rice seedlings. Methylobacterium organophilum, isolated from hot spring mud, a thermophilic nitrogen-fixing species, fixed di-nitrogen efficiently even at elevated temperature.133 The characteristic feature of Methylobacterium to fix atmospheric nitrogen and colonize leaf tissues have been shown to improve tolerance of Jatropha (biodiesel crop) under low soil nutrient conditions. Exploitation of nitrogen fixing Methylobacterium was reported to have improved the productivity and green index of Jatropha biofuel.134 Recently, a preliminary assessment of three endophytic Methylobacterium sp. obtained from Palm oil revealed their ability to proliferate in nitrogen-free media, suggesting the nitrogen fixing ability of Methylobacterium.37
Despite considerable research on methylotroph mediated nitrogen fixation, there are still gaps in our understanding of its precise mechanism. When Methylobacterium mediated nitrogen fixing mechanisms are well understood, future researchers may be able to develop successful and promising strains to improve plant growth and development.
Phosphorus solubilization
Phosphorous is a limiting nutrient which is very essential for biological growth and development as it is involved with life sustaining metabolic processes of plants such as photosynthesis, signal transduction, energy transfer, macromolecular biosynthesis and respiratory process. Phosphate solubilization process is considered as equally important as nitrogen fixation process.135 Though P is quite abundant in soil, it remains mostly unavailable to plants. Generally, P in the soil exists in two different forms, either in inorganic (bound, fixed, or labile) or organic (bound) forms. However, plants can take up P only in soluble forms such as mono- and dibasic phosphate forms.136 Therefore, it is evident that the phosphate solubilization is an extremely important and necessary process. Supply of plant essential nutrient through any biological means, especially microorganisms, is a better and promising choice in sustainable agriculture. Microorganisms play a central role in solubilizing the unavailable inorganic, but insoluble P fraction of soil and make them available for uptake by the plants easily.
Microorganisms offer various means to mineralize phosphates like rock phosphate, tricalcium phosphate, aluminium phosphate and organic phosphorus which have complex-structural characteristics, present in soil to improve the availability of accessible forms of P, like orthophosphate to plants.137,138 The mechanisms underlying microbe mediated P solubilization has been well documented. Of various mechanisms, secretion of organic acids of low molecular weight is the primary mechanism of P solubilization by microorganisms. Amongst them, acids like carboxylic acid, formic acid, succinic acid, lactic acid, glycolytic acid, fumaric and propionic acid139 brings down the rhizosphere pH which in turn cause the release of bound phosphate forms.140 Another well studied phosphate solubilizing mechanism is the release of extracellular enzymes such as phytases, nonspecific acid phosphatases and C-P lyases phosphatases by phosphorous solubilizing bacteria that mineralize insoluble organic phosphate.141,142 Obviously, secretion of extracellular enzymes would help them to gain competitive advantage over deleterious pathogens.
Hitherto, elaborate studies are lacking on phosphate solubilization by PPFM. A detailed study to understand Methylobacterium-mediated phosphorous solubilization under in vitro conditions was done by Jayashree et al.143 This study recorded P-solubilization index of thirteen PPFM isolates grown for 7 days on NBRIP-BPB plates which ranged from 1.1 to 2.7. A thorough examination of these isolates under in vitro conditions could identify Methylobacterium as potential phosphate solubilizer with diverse mechanisms to solubilize organic phosphate. Later, Kumari144 reported P solubilization index ranging from 1.28 to 1.85 of four different facultative methylotrophic strains that solubilized tricalcium phosphate in Pikovskaya’s agar medium. Agafonova et al.84 reported phosphate solubilization activity as a newly revealed characteristic feature of methylotrophs and explicated their phosphate-solubilizing activity in association with different plants. A first report on the presence of all the genes responsible for the synthesize of three phosphatase enzymes in Methylobacterium oryzae was published by Kwak et al.145 Recently, Rahim et al.36 demonstrated the phosphate solubilizing potential of nine newly isolated Methylobacterium sp. In Pikovskaya broth, the Methylobacterium isolate EPPD1 solubilized 4.12 mg/mL of inorganic phosphate whereas isolate ENPD2 solubilized an amount of 3.97 mg/ml of inorganic phosphate in Pikovskaya and 3.3 mg/ml in NBRIP broth.
Significant advances have been made in the area concerning to Methylobacterium mediated phosphate solubilization. Nevertheless, search for new potential phosphate solubilizing Methylobacteria should be augmented to develop promising candidates of PPFM for commercial inoculum development.
Modulation of Ethylene Levels in Plants
The gaseous hydrocarbon ethylene is a unique phytohormone with a number of biological roles related to seedling growth, ripening of fruits, germination of seed, abscission of leaves and petals, organ senescence and biotic and abiotic stress induced responses. Ethylene improves the plant growth and also hinder the developmental processes which depends up on the cell type and plant species. Generally, decrease in levels of ethylene in plants enhances root extension, but increased ethylene levels in plants, especially in fast growing roots, can hinder important developmental processes, mainly root elongation.146 Thus, beneficial role of this vaporous hormone is reported at very low concentrations. Elongation of shoot and root is normally inhibited by action of ethylene. High levels of ethylene is harmful which causes small lateral root proliferation.147 Moreover, stress regulating activity of ethylene has also been elucidated well.148 Various abiotic stress like salinity, drought, flooding of water, and presence of heavy metals dramatically increases endogenous level of 1-aminocyclopropane-1-carboxylate (ACC), the immediate precursor of ethylene in plants, which in turn cause higher concentration of ethylene.149 Accumulation of larger volumes of ethylene leads to further stresses in plants like reduced ability to nutrient uptake, water absorption etc.150
Degradation of the precursor molecules, ACC, would forbid the synthesize and accumulation of ethylene and thereby prevent the detrimental effects of the high ethylene levels.151 The enzyme ACC deaminase (EC 4.1.99.4), first characterized by Honma and Shimomura152 has been shown to degrade ACC to alpha-ketobutyrate and ammonium. Therefore, it was assumed that plant growth promoting bacteria with high locally induced ACC deaminase activity could be a powerful strategy for ameliorating plant stress. In this regard, it has been proved that ACC deaminase-containing PGPB could effectively be used to overcome the deleterious consequences caused by abiotic stresses. Achromobacter,153 Bacillus,154,155 Burkholderia,156,157 Ensifer,158 Mesorhizobium,159 Pseudomonas,155, 160,161 Streptomyces,162 and Variovorax,163 are some examples of well documented ACC deaminase-producing bacterial genera.
Only a few research publications on ACC deaminase activity in Methylobacterium spp. exist. The inhabiting activity of rice 1-aminocyclopropane-1-carboxylate deaminase (ACCD) on phyllosphere Methylobacteria has been detected and assessment of its functional regulatory role in determining ethylene level in rice and tomato seedlings showed that the enzyme activity notably lowers the ethylene level (60–80%) in the plants. A key finding made in this study was the homology of accD gene sequence of the rice phyllosphere Methylobacterium with Rhizobium leguminosarum (98% similarity).164 Based on the results of a gnotobiotic root elongation assay conducted on canola seedlings, Madhaiyan et al.82 reported the occurrence of ACC deaminase in Methylobacterium spp. ACC deaminase activity of M. fujisawaense caused substantial lowering of ethylene levels in canola seedlings. Madhaiyan et al.17 also found the presence of M. oryzae sp. nov., an aerobic ACC deaminase producing PPFM bacterium in stem tissues of rice. The isolate was reported to be closely related to M. fujisawaense, M. radiotolerans and M. mesophilicum. Methylobacterium oryzae strains CBMB20 and CBMB110 showed significant variation in their ability to utilize ACC and they produced 94.5 and 24.7 nmol α-ketobutyrate mg-1 of protein h-1, respectively. Seed treatment with theses strains increased the root length of pepper and tomato plants attributed to their ACC deaminase activity compared to control plants under gnotobiotic conditions.165 Treatment of tomato and red pepper plants with Methylobacterium reduced the ethylene emission compared to control plants under greenhouse conditions also.166 Recently, Senthilkumar et al.35 demonstrated the positive influence of endophytic Methylobacterium possessing ACC deaminase activity isolated from tissue culture banana plantlets of South Indian banana cultivars. Regulation of acdS gene encoding aminocyclopropane carboxylate deaminase (AcdS) by an AcdR homologous protein in epiphytic phytosymbiotic methylotroph Methylobacterium radiotolerans JCM2831 was proposed by Ekimova et al.167 The transcriptional regulatory protein encoded by an open reading frame activates the acdS gene expression when ACC or 2-aminoisobutyrate is present as an inducer. It can regulate the transcription initiation process even in the absence of inducer molecule, when present excessively. A better understanding of ACC deaminase enzyme activity in Methylobacterium is needed to extend their integration in sustainable crop production.
PPFM as potential plant-growth promoters
Plethora of methylotrophic bacterial species are known to live in close intimacy with plants, both terrestrial and aquatic, by living along on roots, leaf surfaces, buds and other plant parts. Numerous benevolent attributes of PPFM have been explored and reported by various researchers (Table 1; Figure 1). Inoculation of either Methylobacterium or methanol spray has shown profound increase in dry matter production and plant height of cotton against uninoculated control.168 Discernable variation in photosynthetic activity was noticed in rice cultivar Co-47 treated with Methylobacterium due to increased chlorophyll content, stomatal number and maleic acid content.14 A remarkable improvement in germination was noted compared to uninoculated control when sugarcane true seeds were treated with PPFM strains. Strikingly, a combination different method of PPFM application such as seed treatment, soil drenching and phyllosphere spraying helped increasing the height of plants, specific leaf area, internodal numbers and cane yield significantly.106 Yet another interesting evidence was provided by Lee et al.15, wherein diazotrophic bacterial strains including Methylobacterium spp., not only improved seed germination but also biomass and seedling vigour index (SVI) of rice seedlings.
ACC deaminase producing Methylobacterium fujisawaense could reduce the synthesis of ACC and hence lowered the deleterious ethylene levels in seedlings of canola when grown under gnotobiotic conditions. Lowered ethylene level could induce canola seedlings root elongation.82 A gnotobiotic root elongation assay was carried out to test the efficacy of two Methylobacterium strains namely Methylobacterium sp. strain CBMB20 and CBMB110. Seeds of red pepper and tomato imbibed with Methylobacterium strains showed substantial increment in root length compared to uninoculated control and M. extorquens miaA– knockout mutant treated plants. Furthermore, accumulation of indole-3-acetic acid, trans-zeatin riboside and dihydrozeatin riboside was noted in the extracts of red pepper plants. In the same way, accumulation of trans-zeatin riboside and dihydrozeatin riboside was recorded in tomato plants extracts.85 Radhika et al.169 recorded highest maize cob yield when plants were sprayed with methylotrophic bacteria. In another investigation, high chlorophyll content was measured from soyabean plants which received both seed inoculation and foliar spray of Methylobacterium.170
Methylobacterim spp. are known to enhance rice plant growth in a multitude of ways. When effect of eight Methylobacterium isolates on seed germination was monitored, PPFM-SOY (isolated from soybean leaf) and GN (isolated from groundnut leaf), were shown to improve the germination of heat-treated seeds of paddy, maize and soybean. When the heated seed of soybean was treated with PPFM-SOY, 14.28 per cent increase in germination was obtained compared to untreated heated seeds. Same level of increase in germination was observed on treatment with PPFM-GN. When normal seeds were treated with PPFM-SOY and PPFM-GN, 23.21 and 7.14 per cent increase in germination was observed respectively. Treatment of heated maize seeds with PPFM-SOY and PPFM-GN resulted in an increase of 27.50% and 30.0% over control respectively. For paddy seeds also, 13.88% and 11.11% increase over control was recorded on treatment with PPFM-SOY and PPFM-GN respectively.89 Among selected methylotrophic bacteria tested in a pot culture study, PPFM50 significantly increased the shoot biomass, height, stem girth, leaf area, chlorophyll content, and tuber yield of Coleus forskohlii. Markedly, 216.10 per cent increase in tuber yield was observed against reference strain (216.10%) and uninoculated control (136.07%).171
In a two-year field experiment, application of PPFM alone was found improving growth attributes like leaf number per plant, chlorophyll content, and yield attributes like pod number per plant of snap bean. Moreover, total sugars, ascorbic acid, amino acids, and protein content of pods were also increased significantly.108 Effect of inoculation of Methylobacterium spp. possessing ACC deaminase (ACCD) and indole-3-acetic acid activity on tomato and red pepper seedling performed under gnotobiotic and greenhouse condition was found to be comparable to exogenous applications of synthetic IAA.172 Increased production of IAA by the foliar application of M. extorquens MP1 isolated from Peach (Prunus persica L.) phyllosphere and M. zatmanii MS4 from Strawberry (Fragaria ananassa L.) employing leaf imprint method augmented the growth of tomato plants compared to uninoculated control.173 Methylobacterium extorquens MM2 obtained from the phyllosphere of mustard leaf increased the production of IAA in plants which in turn enhanced the plant growth.66 Methylobacterium sp. 2A isolated from Solanum tuberosum L. cv. Desirée plants significantly increased biomass of potato plants along with root hair density. Besides, its strong biocontrol activity against fungal phytopathogens has been evidenced in dual confrontation assays.34 A recent report showed that inoculation of microbial consortium consisting of rhizobium, AMF, PPFM and Bacillus altitudinis – FD48 significantly improves the growth and yield of groundnut.174
Positive responses of a Methylobacterium application have also been experimented on wheat,79 barnyard millet (Echinochloa frumentacea Var. COKV 2),175 biodiesel plant Crambe abyssinica,176 Ginger,177 rice,16,178 cotton179,180 and cardamom.181 All these high-throughput studies have been undertaken to gather knowledge about consequences of interactions between Methylobacterium and their host plants. The combination of multiple plant growth promoting characteristics keeps PPFMs as an attractive microbial tool in agriculture. They are categorized as biosafety level one organisms as there is no report of Methylobacterium mediated pathogenicity in plants till now. Taking into account of their beneficial attributes to the host plants various Methylobacterium based biofertilizers have been launched to the markets. For instance, Newleaf Symbiotics, a company designing next generation of Agri biologicals as biocomplements to existing chemicals, developed a commercial product composed only of Methylobacterium to accelerate the growth of cotton, tomato, peanut, rice, corn, soybean, and wheat.182 Even though, many potential Methylobacterium strains have been described previously, relatively small number of products are currently available in the market.
Very recently, a seed coating technology using immobilized cells of plant growth promoting Methylorubrum aminovorans to improve seed quality of cotton was introduced by Pragathi et al.183 This novel technology uses microbial cells of Methylorubrum aminovorans immobilized in a composite nanofibre matrix composed of Chitosan and Poly Vinyl Alcohol (PVA) as an effective localized delivery system. Yet another similar study conducted by Mukiri et al.184 put forwarded a seed invigouration technique for groundnut plants that uses electrospun Polyvinyl alcohol (PVA) nanofibre containing immobilized microbial cells of Methylorubrum aminovorans. Application of encapsulated Methylorubrum aminovorans was successful in enhancing root colonization followed by improving seed germination, seedling vigor and growth of groundnut plants.
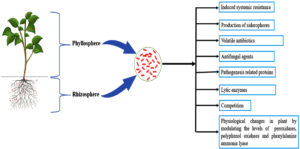
PPFM as Biological Control Agents of Plant Diseases
Biocontrol activity of PPFM on soil borne phytopathogens has been recorded in addition to its positive effect on plant growth14 (Table 2; Figure 2). PPFM have also been well elucidated for their induced systemic resistance (ISR) activity against various plant pathogens.14,121,185
Table (2):
Biological control of plant diseases by PPFM.
Methylotrophs |
Pathogen(s) |
Source |
References |
|---|---|---|---|
Methylobacterium sp. PPFM-Os-07 |
Rhizoctonia solani |
Rice phyllosphere |
14 |
Methylobacterium sp. |
Aspergillus niger, Sclerotium rolfsii |
Groundnut phyllosphere |
185 |
Methylobacterium sp. Co-47 Methylobacterium sp. MV-10 Methylobacterium sp. LE-1 Methylobacterium sp. AM-1 |
Macrophomina phaseolina, Phytophthora infestans, Fusarium oxysporum, Fusarium udum, Pythium aphanidermatum, Sclerotium rolfsii |
Rice phyllosphere |
187 |
Methylobacterium fujisawaense TNAU 14 |
Meloidogvne incognita |
Tomato rhizosphere |
191 |
Methylobacterium sp. Co-47 Methylobacterium sp. MV-10 Methylobacterium sp. LE-1 Methylobacterium sp. AM-1 |
Rhizoctonia solani |
Rice phyllosphere |
188 |
Methylobacterium rhodinum Methylobacterium aminovorans |
Rhizoctonia solani |
Soil |
198 |
Methylobacterium sp. GPPFM13 |
Macrophomina phaseolina, Sclerotium rolfsii, Pythium myriotylum, Colletotrichum gloeosporioides Fusarium oxysporum |
Ginger phyllosphere |
177 |
Methylobacterium populi |
Colletotrichum capsici |
Chilli phyllosphere |
190 |
Seed treatment or foliar spray of Methylobacterium on rice induced the pathogenesis related proteins which protected the plants against sheath blight pathogen Rhizoctonia solani under pot culture conditions.14 PPFMs bring about several physiological changes in plants, making the plants more resistant to pathogens. M. extorquens CO-47 induced the accumulation of peroxidase, polyphenol oxidase, phenylalanine lyase and phenols in plants and subsequently suppressed the pathogen Rhizoctonia solani.14 Methylobacterium spp. treated groundnut plants challenged with Aspergillus niger or Sclerotium rolfsii resulted in enhancement of seed germination and seedling vigour index. It also caused the increase in the activities of β-1,3- glucanase, phenylalanine ammonia lyase (PAL) and peroxidase (PO). Also, presence of five isozymes of polyphenol oxidase and PO were noticed in Methylobacterium treated plants when challenged with the pathogens.185 Application of Methylobacterium strains on tomato induced the defence response against the plant pathogen Ralstonia solanacearum.186 As already stated, Methylobacterium spp. synthesize anti-phytopathogen factors, such as siderophores.136 The antagonistic effect on the fungal pathogens tested may also be attributed to the salicylic acid, a type of siderophore, produced by PPFM isolates as already proved in Methylobacterium oryzae CBMB20 challenge inoculated with Pseudomonas syringae pv. tomato in tomato plants compared to control or M. oryzae treated plants under growth chamber and green-house conditions.121 According to Poorniammal et al.187 volatile antibiotics produced by Methylobacterium ceases mycelial growth of Sclerotium rolfsii, Fusarium udum, Fusarium oxysporum, Pythium aphanidermatum, Colletotrichum capsici, and Cercospora capsici, and also inhibit the growth of Xanthomonas campestris with various biocontrol efficacies under in vitro conditions. Methylobacteria isolate CO-47 hindered the mycelial growth of Rhizoctonia solani and the inhibition zone measured under in vitro conditions was 1.4 cm.188 Methylotrophs inhabiting mangrove sediment have been found to be powerful biocontrol agent against root rot pathogen Macrophomina phaseolina.189 As reported by Santosh et al.190 inoculation of PPFM isolates to chilli grown under field conditions remarkably reduces the anthracnose disease caused by Colletotrichum capsici.
Another interesting finding made was the biocontrol potential of M. fujisawaense against Meloidogyne incognita (Kofoid and White) chitwood race 3. M. fujisawaense filtrate was found to be highly effective in inhibiting egg hatching and reducing root penetration of M. incognita in tomato plants.191 Literature published on biocontrol activity of Methylobacterium to date are consistent and conclusive. Induced systemic resistance activity in plants on methylotrophic bacterial treatment suggests the possibility that PPFM bacteria could be used as a means for biological control of plant diseases.
Pink Pigmented Facultative Methylotrophs (PPFMs) are generally regarded as ubiquitous colonizers of several, if not all plants. Their beneficial interactions with host plants and the symbiotic nature, and other valuable attributes are attracting greater attention. Methylotrophic bacteria with plant growth promotion ability are widely accepted as efficient and potential microbes for enhancing agricultural yield, and hence can be developed into a reliable component in sustainable agricultural systems. Multiple mechanisms of plant growth promotion, such as phytohormone production, nutrient acquisition and biocontrol activities emphasize the potential of the PPFMs in crop production. Since PPFMs can promote plant growth as well as prevent infection by phytopathogens, they can be used as better alternatives to chemical fertilizers and fungicides in sustainable agriculture. The importance of plant microbiome in plant and crop health is getting realized of late. The innovations focusing on core microbiome understanding of crop plants could help integrate PPFM as a component in synthetic microbiome development which would open up environmentally-friendly opportunities to take care of the current crop production challenges.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Fall R, Benson A. Leaf methanol- the simplest natural product from plants. Trends Plant Sci. 1996;1(9):296-301.
Crossref - Nemecek-marshall M, MacDonald RC, Franzen JJ, Wojciechowski CL, Fall R. Methanol emission from leaves: enzymatic detection of gas-phase methanol and relation of methanol fluxes to stomatal conductance and leaf development. Plant Physiol. 1995;108(4):1359-1368.
Crossref - Huve K, Christ MM, Kleist E, et al. Simultaneous growth and emission measurements demonstrate an interactive control of methanol release by leaf expansion and stomata. J Exp Bot. 2007;58(7):1783-1793.
Crossref - Anthony C. Bacterial oxidation of methane and methanol. Adv Microbial Physiol. 1986;27:113-210.
Crossref - Carvajal TM, Tan RL, Lee AC. Pink Pigmented Facultative Methylotrophic (PPFM) bacteria from the human oral cavity. Philippine J Syst Biol. 2011;5:1-9.
- Green PN, Bousifield IJ. A taxonomic study of some Gram- negative facultatively methylotrophic bacteria. J Gen Microbiol. 1989;128(3):623-625.
Crossref - Bassalik K. Uber die Verarbeitung der oxalsaure dursch Bacillus extorquens nov. sp. Jahrb. wiss. Bot. 1913;53:255-302.
- Kuono K, Ozaki A. Distribution and identification of methanol utilizing bacteria. In G Tenui et al (eds.), Microbial Growth on C1 Compounds. Society of Fermentation Technology, Japan Publisher. 1975:11-21.
- Patt TE, Cole GC, Hanson RS. Methylobacterium, a new genus of facultatively methylotrophic bacteria. Int J Syst Bacteriol. 1976;26(2):226-229.
Crossref - Green PN, Bousfield IJ. Emendation of Methylobacterium Patt, Cole, and Hanson 1976; Methylobacterium rhodinum (Heumann 1962) comb. nov. corrig.; Methylobacterium radiotolerans (Ito and Iizuka 1971) comb. nov. corrig.; and Methylobacterium mesophilicum (Austin and Goodfellow 1979) comb. nov. Int J Syst Evol Microbiol. 1983;33(4):875-877.
Crossref - Anthony C. The quinoprotein dehydrogenases for methanol and glucose. Arch. Biochem. Biophys. 2004;428(1):2-9.
Crossref - Keltjens JT, Pol A, Reimann J, Op den Camp HJ. PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl Microbiol Biotechnol. 2014;98(14):6163-6183.
Crossref - Alessa O, Ogura Y, Fujitani Y, et al. Comprehensive comparative genomics and phenotyping of Methylobacterium species. Front Microbiol. 2021;12:740610.
Crossref - Madhaiyan M, Poonguzhali S, Senthilkumar M, et al. Growth promotion and induction of systemic resistance in rice cultivar Co-47 (Oryza sativa L.) by Methylobacterium spp. Bot Bull Acad Sin. 2004;45:315-325.
- Lee HS, Madhaiyan M, Kim CW, Choi SJ, Chung KY, Sa T. Physiological enhancement of early growth of rice seedlings (Oryza sativa L.) by production of phytohormone of N2-fixing methylotrophic isolates. Biol Fert Soils. 2006;42(5):402-408.
Crossref - Lai K, Nguyen NT, Miwa H, Yasuda M, Nguyen HH, Okazaki S. Diversity of Methylobacterium spp. in the rice of the Vietnamese Mekong Delta. Microbes Environ.2020;35(1):ME19111.
Crossref - Madhaiyan M, Kim B, Poonguzhali S, et al. Methylobacterium oryzae sp. nov., an aerobic, pink-pigmented, facultatively methylotrophic, 1-aminocyclopropane-1-carboxylate deaminase producing bacterium isolated from rice. Int J Syst Evol Microbiol. 2007;57(2):326-331.
Crossref - Renier A, Jourand P, Rapior S, Poinsot V, Sy A, Dreyfus B, Moulin L. Symbiotic properties of Methylobacterium nodulans ORS 2060T: A classic process for an atypical symbiont. Soil Biol Biochem.2008;40(6):1404-1412.
Crossref - Ardley JK, O’Hara GW, Reeve WG, et al. Root nodule bacteria isolated from South African Lotononis bainesii, L. listii and L. solitudinis are species of Methylobacterium that are unable to utilize methanol. Arch Microbiol. 2009;191(4):311-318.
Crossref - Senthilkumar M, Madhaiyan M, Sundaram SP, Kannaiyan S. Intercellular colonization and growth promoting effects of Methylobacterium sp. with plant-growth regulators on rice (Oryza sativa L. Cv CO-43). Microbiol Res.2009;164(1):92-104.
Crossref - Cao YR, Wang Q, Jin RX, Tang SK, Jiang Y, He WX, Lai HX, Xu LH, Jiang CL. Methylobacterium soli sp. nov. a methanol-utilizing bacterium isolated from the forest soil. Antonie van Leeuwenhoek. 2011;99(3):629-634.
Crossref - Salam LB, Obayori OS, Raji SA. Biodegradation of used engine oil by a methylotrophic bacterium, Methylobacterium mesophilicum isolated from tropical hydrocarbon-contaminated soil. Petroleum Sci Technol. 2015;33(2):186-195.
Crossref - Kim J, Chhetri G, Kim I, Kim MK, Seo, T. Methylobacterium durans sp. nov., a radiation-resistant bacterium isolated from gamma ray-irradiated soil. Antonie van Leeuwenhoek. 2020;113(2):211-220.
Crossref - Ten LN, Li W, Elderiny NS, Kim MK, Lee SY, Rooney AP, Jung HY. Methylobacterium segetis sp. nov., a novel member of the family Methylobacteriaceae isolated from soil on Jeju Island. Arch Microbiol. 2020;202(4):747-754.
Crossref - Ito M, Shimizu T, Nakamura A. Complete genome sequence of Kaistia sp. strain 32K, isolated from soil as a mixed single colony with Methylobacterium sp. strain ME121. Microbiol Resour Announcements. 2021;10(10):e00019-21.
Crossref - Maeng S, Kim DU, Lim S, et al. Methylobacterium radioduranssp. nov., a novel radiation-resistant Methylobacterium. Arch Microbiol. 2021;203(6):3435-3442.
Crossref - Zhao C, Wan Y, Cao X, Zhang H, Bao X. Comparative genomics and analysis of the mechanism of PQQ overproduction in Methylobacterium.World J.Microbiol Biotechnol. 2021;37(6):1-10.
Crossref - Anda M, Ikeda S, Eda S, Okubo T, Sato S, Tabata S, Mitsui H, Minamisawa K. Isolation and genetic characterization of Aurantimonas and Methylobacterium strains from stems of hypernodulated soybeans. Microbes Environ. 2011;26(2):172-180.
Crossref - Mizuno M, Yurimoto H, Yoshida N, Iguchi H, Sakai Y. Distribution of Pink pigmented facultative methylotrophs on leaves of vegetables. Biosci Biotechnol Biochem. 2012;76(3):578-580.
Crossref - Eevers N, Van Hamme JD, Bottos EM, Weyens N, Vangronsveld J. Draft genome sequence of Methylobacterium radiotolerans, a DDE-degrading and plant growth-promoting strain isolated from Cucurbita pepo. Genome Announc. 2015;3(3):e00488-15.
Crossref - Chaudhry V, Baindara P, Pal VK, Chawla N, Patil PB, Korpole S. Methylobacterium indicum sp. nov., a facultative methylotrophic bacterium isolated from rice seed. Syst Appl Microbiol.2016;39(1):25-32.
Crossref - Roodi D, Millner JP, McGill C, Johnson RD, Jauregui R, Card SD. Methylobacterium, a major component of the culturable bacterial endophyte community of wild Brassica seed. Peer J. 2020;8:e9514.
Crossref - Photolo MM, Mavumengwana V, Sitole L, Tlou MG. Antimicrobial and antioxidant properties of a bacterial endophyte, Methylobacterium radiotolerans MAMP 4754, isolated from Combretum erythrophyllum seeds. Int J Microbiol. 2020;9483670.
Crossref - Grossi CEM, Fantino E, Serral F, et al. Methylobacterium sp. 2A is a plant growth-promoting rhizobacteria that has the potential to improve potato crop yield under adverse conditions. Front Plant Sci. 2020;11:71.
Crossref - Senthilkumar M, Pushpakanth P, Jose AP, Krishnamoorthy R, Anandham R. Diversity and functional characterization of endophytic Methylobacterium isolated from banana cultivars of South India and its impact on early growth of tissue culture banana plantlets. J Appl Microbiol. 2021;131(5):2448-2465.
Crossref - Rahim AA, Ibrahim NA, Ishak FN, Mean LJ, Ayub NAM, Fazilah NN. Investigation of newly isolated Methylobacterium sp. as potential biofertilizer. In IOP Conference Series: Earth Environ Sci. 2021;765(1):012063.
Crossref - Ishak FN, Rahim AA, Mean LJ, Ayub NAM, Fazilah NN. Preliminary analysis of endophytic plant growth promoting (pgp) Methylobacterium sp. isolated from palm oil (Elaeis guineensis) leaves. In IOP Conference Series: Earth Environ Sci. 2021;765(1):012071.
Crossref - Kosmiatin M, Husni A, Salma S. In vitro growth response of Patchouli (Pogostemon cablin) cultured in medium containing Methylobacterium spp. filtrate. IOP Conference Series: Earth Environ Sci. 2021;762(1):012076.
Crossref - Christian N, Basurto BE, Toussaint A, et al. Elevated CO2 reduces a common soybean leaf endophyte. bioRxiv. 2021.
Crossref - Kutschera U. Plant-associated methylobacteria as co-evolved phytosymbionts: a hypothesis. Plant Signaling & Behavior.2(2):74-78.
- Austin B, Goodfellow M, Dickinson CH. Numerical taxonomy of phylloplane bacteria isolated from Lolium perenne. J Gen Microbiol. 1978;104(1):139-155.
Crossref - Basile DV, Slade LL, Corpe WA. An association between a bacterium and a liverwort, Scapania nemorosa. Bull Torrey Bot Club. 1969;96(6):711-714.
- Austin B, Goodfellow M. Pseudomonas mesophilica, a new species of pink bacteria isolated from leaf surfaces. Int J Syst Bacteriol. 1979;29(4):373-378.
Crossref - Corpe WA, Basile DV. Methanol- utilizing bacteria associated with green plants. Dev Ind Microbiol. 1982;23:483-493.
- Corpe WA. A method for detecting Methylotrophic bacteria on solid surfaces. J Microbiol.1985;3(3-4):215-221.
Crossref - Jorge GL, Kisiala A, Morrison E, Aoki M, Nogueira APO, Emery RJN. Endosymbiotic Methylobacterium oryzaemitigates the impact of limited water availability in lentil (Lens culinaris Medik.) by increasing plant cytokinin levels. Environ Exp Bot. 2019;162:525-540.
Crossref - Pirttila AM, Laukkanen H, Pospiech H, Myllyla R, Hohtola A. Detection of intracellular bacteria in the buds of scotch pine (Pinus sylvestrisL.) by in situ hybridization. Appl Environ Microbiol. 2000;66(7):3073-3077.
Crossref - Poonguzhali S, Madhaiyan M, Yim WJ, Kim KA, Sa TM. Colonization pattern of plant root and leaf surfaces visualized by use of green-fluorescent-marked strain of Methylobacterium suomiense and its persistence in rhizosphere. Appl Microbiol Biotechnol. 2008;78(6):1033-1043.
Crossref - Jia LJ, Zhang KS, Tang K, Meng JY, Zheng C, Feng FY. Methylobacterium crusticola sp. nov., isolated from biological soil crusts. Int J Syst Evol Microbiol.2020;70(3):2089-2095.
Crossref - Jiang L, An D, Wang X, et al. Methylobacterium planium sp. nov., isolated from a lichen sample. Arch Microbiol. 2020;202(7):1709-1715.
Crossref - Pascual JA, Ros M, Martinez J, et al. Methylobacterium symbioticum sp. nov., a new species isolated from spores of Glomus iranicum var. tenuihypharum.Curr Microbiol. 2020;77(9):2031-2041.
Crossref - Feng GD, Chen W, Zhang XJ, Zhang J, Wang SN, Zhu H. Methylobacterium nonmethylotrophicum sp. nov., isolated from tungsten mine tailing. Int J Syst Evol Microbiol.2020;70(4):2867-2872.
Crossref - dela Rosa CJO, Lee AC, Rivera WL. Pink pigmented facultative methylotrophic bacteria isolated from fermented Philippine shrimp paste. Trop Life Sci Res. 2021;32(2):147-161
- Balachandar D, Raja P, Sundaram SP. Genetic and metabolic diversity of pink pigmented facultative methylotrophs in phyllosphere of tropical plants. Braz J Microbiol. 2008;39(1):68-73.
Crossref - Raja P, Balachandar D, Sundaram SP. Genetic diversity and phylogeny of pink-pigmented facultative methylotrophic bacteria isolated from the phyllosphere of tropical crop plants. Biol Fertile Soils. 2008;45(1):45-53.
Crossref - Dourado MN, Ferreira A, Araujo WL, Azevedo JL, Lacava PT. The diversity of endophytic methylotrophic bacteria in an oil-contaminated and an oil-free mangrove ecosystem and their tolerance to heavy metals. Biotechnol Res Int. 2012;759865.
Crossref - Kaur GJ, Kumar M, Tomar RS. Genetic diversity of epiphytic pink pigmented facultative methylotrophs from leaf phyllosphere of crop plants. Artic Int J Pharma Bio Sci. 2016;220-229.
- de Andrade PAM, Dias ACF, Cotta SR, et al. Differential niche occupation and the biotechnological potential of Methylobacterium species associated with sugarcane plants. Afr J Microbiol Res.2018;12(25):595-605.
Crossref - Balachandar D, Raja P, Nirmala K, Rithyl TR, Sundaram SP. Impact of transgenic Bt-cotton on the diversity of pink-pigmented facultative methylotrophs. World J Microbiol Biotechnol. 2008;24(10):2087-2095.
Crossref - Schweiggert RM, Carle R. Carotenoid production by bacteria, microalgae, and fungi. In Baranska,M, Kaczor A, (eds.), Carotenoids: Nutrition, Analysis and Technology, Ist Ed. John Wiley and Sons, Ltd. Chichester, UK. 2016;217-240.
- Ram S, Mitra M, Shah F, Tirkey SR, Mishra S. Bacteria as an alternate biofactory for carotenoid production: A review of its applications, opportunities and challenges. J Funct Foods.2020;67:103867.
Crossref - Yabuzaki J. Carotenoids database: Structures, chemical fingerprints and distribution among organisms. Database. 2017;2017:bax004.
Crossref - Urakami T, Araki H, Suzuki K, Komogata K. Further studies of the genus Methylobacterium and description of Methylobacterium aminovorans sp. nov. Int. J Syst Bacteriol. 1993;43(3):504-513.
Crossref - Konovalova HM, Shylin SO, Rokytko PV. Characteristics of carotenoids of methylotrophic bacteria of Methylobacterium genus. Mikrobiolohichnyi Zhurnal. 2007;69(1):35-41.
- Krinsky NI. Carotenoids as antioxidants. Nutrition. 2001;17(10):815-817.
Crossref - Subhaswaraj P, Jobina R, Parasuraman P, Siddhardha B. Plant growth promoting activity of Pink Pigmented Facultative Methylotroph-Methylobacterium extorquens MM2 on Lycopersicon esculentum L. J Appl Biol Biotechnol. 2017;5(1):0-4.
Crossref - Trotsenko YA, Ivanova EG, Doronina NV. Aerobic methylotrophic bacteria as phytosymbionts. Microbiol. 2001;70:623-632.
Crossref - Godinho A, Bhosle S. Carotenes produced by alkaliphilic orange-pigmented strain of Microbacterium arborescens-AGSB isolated from coastal sand dunes. Indian J Mar Sci. 2008;37(3):307-312. http://nopr.niscpr.res.in/handle/123456789/2053
- Ye RW, Yao H, Stead K, et al. Construction of the astaxanthin biosynthetic pathway in a methanotrophic bacterium Methylomonas sp. strain 16a. J Ind Microbiol Biotechnol. 2007;34(4):289-299.
Crossref - Peel D, Quayle JR. Microbial growth on C-1 compounds. 1. Isolation and characterization of Pseudomonas AM1. Biochem J. 1961;81(3):465-469.
Crossref - Klaui, H, Bauernfeind JC. Carotenoids as food colors, In Bauernfeind JC (ed), Carotenoids as Food Colorants and Vitamin A Precursors. Academic Press, New York. 1981:48-317.
- Marusich WL Bauernfeind JC. Oxycarotenoids in poultry feed. In Bauernfeind JC (ed.), Carotenoids as Food Colorants and Vitamin A Precursors. Academic Press, New York. 1981:319-462.
Crossref - Simpson KL, Katayama W, Chlchester CO. In Bauernfeind JC (ed.), Carotenoids as Food Colorants and Vitamin A Precursors. Academic Press, New York. 1981:463-538.
Crossref - Shimzu S, Sato K, Hiraoka M, Yamashita F, Kobabayashi T. Carotenoid formation by a facultative methylotroph, Protaminobacter tuber. J Ferment Technol. 1982;60:163-166.
- Nelis HJ, De Leenheer AP. Profiling and quantitation of bacterial carotenoids by liquid chromatography and photodiode array detection. Appl Environ Microbiol. 1989;55(12):3065-3071.
Crossref - Laxmi J, Palanichamy V, Reddy N, Rajsekaran C, Kumari NV, Bhaskar M. Standardization of cultivation parameters for the extraction of carotenoid from pink pigmented facultative methylotrophic (PPFM) bacteria. Asian J Pharma Clin Res. 2012;5:187-192.
- Omer ZS, Tombolini R, Broberg A, Gerhardson B. Indole-3-acetic acid production by pink-pigmented facultative methylotrophic bacteria. Plant Growth Reg. 2004;43(1):93-96.
Crossref - Anitha KG. Enhancing seed germination of mono and dicotyledons through IAA production of PPFM. Trends Soil Sci Plant Nutr J. 2010;1:14-18.
- Meena KK, Kumar M, Kalyuzhnaya MG, et al. Epiphytic pink-pigmented methylotrophic bacteria enhance germination and seedling growth of wheat (Triticum aestivum) by producing phytohormone. Antonie Van Leeuwenhoek. 2012;101(4):777-786.
Crossref - Kalyaeva MA, Zacharchenko NS, Doronina NV. Plant growth and morphogenesis in vitro is promoted by associative methylotrophic bacteria. Russian J Plant Physiol. 2001;48(4):514-517.
Crossref - Santosh S, Santosh HB, Sreenivasa MN. Assessment of native pink pigmented facultative methylotrophs of chilli (Capsicum annuum L.) for their plant growth promotional abilities. Int. J. Curr. Microbiol. App. Sci. 2019;8(1):1196-205.
Crossref - Madhaiyan M, Poonguzhali S, Ryu J, Sa T. Regulation of ethylene levels in canola (Brassica campestris) by 1-aminocyclopropane-1-carboxylate deaminase – containing Methylobacterium fujisawaense. Planta. 2006;224(2):268-278.
Crossref - Sy A, Giraud E, Jourand P, et al. Methylotrophic Methylobacterium bacteria nodulate and fix nitrogen in symbiosis with legumes. J Bacteriol. 2001;183(1):214-220.
Crossref - Agafonova NV, Kaparullina EN, Doronina NV, Trotsenko YA. Phosphate-solubilizing activity of aerobic Methylobacteria. Microbiol. 2013;82:864-867.
- Ryu J, Madhaiyan M, Poonguzhali S, et al. Plant growth substances produced by Methylobacterium spp. and their effect on tomato (Lycopersicon esculentum L.) and red pepper (Capsicum annuum L.) growth. J Microbiol Biotechnol. 2006;16(10):1622-1628.
- Patten CL, Glick BR. Bacterial biosynthesis of indole-3–acetic acid. Can J Microbiol. 1996;42(3):207-220.
Crossref - Ivanova EG, doronina NV, Trotsenko YA. Aerobic Methylobacteria are capable of synthesizing auxins. Mikrobiologiya. 2001;70(4):392-397.
Crossref - Holland MA, Polacco JC. Urease-null and hydrogenase-null phenotypes of a phylloplane bacterium reveal altered nickel metabolism in two soybean mutants. Plant Physiol. 1992;98(3):942-948.
Crossref - Holland MA, Polacco JC. PPFMs and other covert contaminants: Is there more to plant physiology than just plant. Annu Rev Plant Physiol Plant Mol Biol. 1994;45(1):197-209.
Crossref - Holland MA. Methylobacterium and plants. Rec Res Dev Plant Physiol. 1997;1:207-213.
- Lee KH, Madhaiyan M, Kim CW, Lee HS, Poonguzhali S, Sa T. Isolation and characterization of the IAA producing Methylotrophic bacteria from phyllosphere of rice cultivars (Oryza sativa L.). Korean J Soil Sci Fertil. 2004;37:235-244.
- Thangamani G, Sundaram SP. Potential of facultative methylotrophs in increasing the yield of tomato crop. In Proceedings of the ICAR National Symposium on Biotechnological Intervention for Improvement of Horticultural Crops: Issues and Strategies in Symbiohort, 10-12 January, 2005, Thrissur. Centre for Plant Biotechnology and Molecular Biology, College of Horticulture, Kerala Agricultural University, Thrissur, Kerala. 2005:362-365.
- Radha TK. Studies on methylotrophs and their beneficial effects on soybean (Glycine max Merrill.). M. Sc. (Ag) thesis, University of Agricultural Science, Dharwad, 139p. 2007.
- Jones NP. Molecular diversity of Arbuscular mycorrhizal fungi and pink pigmented facultative methylotrophic bacteria and their influence on grapevine (Vitis vinifera). Ph.D. (Ag) Thesis, University of Agricultural Science, Dharwad. 2010:156p.
- Phillips DA, Torrey JG. Cytokinin production by Rhizobium japonicum. Physiologia Plantarum. 1970;23(6):1057-1063.
Crossref - Tien TM, Gaskins MH, Hubbell D. Plant growth substances produced by Azospirillum brasilense and their effect on the growth of pearl millet (Pennisetum americanum L.). Appl Environ Microbiol. 1979;37(5):1016-1024.
Crossref - Cacciari I, Lippi D, Pietrosanti T, Pietrosanti W. Phytohormone-like substances produced by single and mixed diazotrophic cultures of Azospirillum and Arthrobacter. Plant Soil. 1989;115:151-153.
Crossref - Nieto KF, Frankenberger Jr WT. Biosynthesis of cytokinins by Azotobacter chroococcum. Soil Biol Biochem. 1989;21(7):967-972.
Crossref - Taller BJ, Wong TY. Cytokinins in Azotobacter vinelandii culture medium. Appl Environ Microbiol. 1989;55(1):266-267.
Crossref - Anwar H, Shahida H. Cytokinin production by some bacteria: its impact on cell division in cucumber cotyledons. African J Microbiol Res. 2009;3(11):704-712.
- Hayat R, Ahmed I, Sheirdil RA. An overview of plant growth promoting rhizobacteria (PGPR) for sustainable agriculture. In Ashraf M, Ozturk, M, Ahmad M, Aksoy A (eds.), Crop Production for Agricultural Improvement. Springer, Dordrecht. 2012:557-559.
- GroBkinsky DK, Tafner R, Moreno MV, et al. Cytokinin production by Pseudomonas fluorescens G20-18 determines biocontrol activity against Pseudomonas syringae in Arabidopsis. Sci Rep. 2016;6(1):23310.
Crossref - McDonald IR, Murrell JC. The methanol dehydrogenase structural gene mxaF and its use as a functional gene probe for methanotrophs and methylotrophs. Appl Environ Microbiol. 1997;63(8):3218-3224.
Crossref - Ivanova EG, Doroniana NY, Shepelyakovskaya AO, Laman AG, Brovkof A, Trotsenko A. Facultative and obligate aerobic Methylobacteria synthesise cytokinins. Microbiol. 2000;69(6):646-651.
Crossref - Koenig RL, Morris RO, Polacco JC. tRNA is the source of low level trans-zeatin production in Methylobacterium spp. J Bacteriol. 2002;184(7):1832-1842.
Crossref - Madhaiyan M, Poonguzhali S, Lee HS, Hari K, Sundaram SP, Sa TM. Pink-pigmented facultative methylotrophic bacteria accelerate germination, growth and yield of sugarcane clone Co86032 (Saccharum officinarum L.). Biol Fertil Soils. 2005;41(5):350-358.
Crossref - Madhaiyan M, Poonguzhali S, Sundaram S, Sa T. A new insight into foliar applied methanol influencing phylloplane methylotrophic dynamics and growth promotion of cotton (Gossypium hirsutum L.) and sugarcane (Saccharum officinarum L.). Environ Exp Bot. 2006;57(1-2):168-176.
Crossref - El-Gawad HGAE, Ibrahim MFM, El-Hafez AA, El- Yazied A. Contribution of pink pigmented facultative Methylotrophic bacteria in promoting antioxidant enzymes, growth and yield of Snap Bean. Am Eurasian J Agric Environ Sci. 2015;15:1331-1345.
Crossref - Palberg D, Kisiala A, Jorge GL, Emery RJ. A survey of Methylobacterium species and strains reveals widespread production and varying profiles of cytokinin phytohormones. BMC Microbiol. 2022;22(1):49.
Crossref - Saleem M, Asghar HN, Khan MY, Zahir ZA. Gibberellic acid in combination with pressmud enhances the growth of sunflower and stabilizes chromium (VI)-contaminated soil. Environ Sci Pollut Res. 2015;22(14):10610-10617.
Crossref - Ullah S, Anwar S, Rehman M, et al. Interactive effect of gibberellic acid and NPK fertilizer combinations on ramie yield and bast fibre quality. Sci Rep. 2017;7(1):10647.
Crossref - Dinler BS, Cetinkaya H, Akgun M, Korkmaz K. Simultaneous treatment of different gibberellic acid doses induces ion accumulation and response mechanisms to salt damage in maize roots. J Plant Biochem Physiol. 2021;9(3):258.
- Yanni YG, Rizk RY, Abd El-Fattah FK, Squartini A, Corich V, Giacomini A. The beneficial plant growth-promoting association of Rhizobium leguminosarum bv. trifolii with rice roots. Funct Plant Biol. 2001;28(9):845-870.
Crossref - Joo GJ, Kin YM, Kim JT, Rhee IK, Kim JH, Lee IJ. Gibberellins-producing rhizobacteria increase endogenous gibberellins content and promote growth of red peppers. J Microbiol. 2005;43(6):510-515.
- Kang S, Joo GJ, Hamayun M. Gibberellin production and phosphate solubilization by newly isolated strain of Acinetobacter calcoaceticus and its effect on plant growth. Biotechnol Lett. 2009;31(2):277-281.
Crossref - Sgroy V, Cassan F, Masciarelli O, Del Papa MF, Lagares A, Luna V. Isolation and characterization of endophytic plant growth-promoting (PGPB) or stress homeostasis-regulating (PSHB) bacteria associated to the halophyte Prosopis strombulifera. Appl Microbiol Biotechnol. 2009;85(2):371-381.
Crossref - Rajan S, Devi TS, Maina C, Prathibaha K, Sundaram S. Methylotrophs-a novel bioinoculant for sustainable agriculture. The Ecosan. 2012;6(3,4):141-144.
- Sheela RP, Jones PN, Jagadeesh KS, Venugopal CK, Gundlur SS. Isolation and characterization of methylotrophs isolated from Coleus forskohlii. Bioinfolet. 2013;10:59-61.
- Savitha P, Sreenivasa MN, Nirmalnath JP. Production of plant growth hormones by pink pigmented facultative methylotrophs. J Pure Appl Microbiol. 2013;7(2):981-985.
- Braun V. Avoidance of iron toxicity through regulation of bacterial iron transport. Biol Chem. 1997;378(4):779-786.
- Indiragandhi P, Anandham R, Kim K, Yim W, Madhaiyan M, Sa T. Induction of defense responses in tomato against Pseudomonas syringae pv. tomato by regulating the stress ethylene level with Methylobacterium oryzae CBMB20 containing 1-aminocyclopropane-1-carboxylate deaminase. World J Microbiol Biotechnol. 2008;24(7):1037-1045.
Crossref - Simionato AVC, Simo CC, Ifuentes A, et al. Capillary electrophoresis-mass spectrometry of citrus endophytic bacteria siderophores. Electrophoresis. 2006;27(13):2567-2574.
Crossref - Silva-Stenico ME, Pacheco FTH, Rodriques JLM, Carrilho E, Tsai SM. Growth and siderophore production of Xylella fastidiosa under iron-limited conditions. Microbiol Res. 2005;160(4):429-436.
Crossref - Lacava PT, Stenico SME, Araujo WL, Simionato AVC, Carrilho E, Tsai SM, Azevedo JL. Detection of siderophores in endophytic bacteria Methylobacterium spp. associated with Xylella fastidiosa subsp. Pauca. Pesq Agropec Bras. 2008;43(4):521-528.
Crossref - Gholizadeh A, Kohnehrouz BB. A molecular evidence for the presence of methylobacterial-type Fe siderophore receptor in Celosia cristata. Plant Soil Environ. 2010;56(3):133-138.
- Vaidehi K, Sekar C. Amino acid conjugated hydroxamate type of siderophore production in Methylobacterium phyllosphaerae MB-5. CIBtech J Microbiol. 2012;1(1):25-30. https://www.cibtech.org/J-Microbiology/PUBLICATIONS/2012/Vol-1-No-1/04…005%20Seran…Amino…Methylobacterium…24-30.pdf
- Juma PO, Fujitani Y, Alessa O, et al. Siderophore for Lanthanide and Iron uptake for methylotrophy and lant growth promotion in Methylobacterium aquaticum strain 22A. Front Microbiol. 2022;13: 921635.
Crossref - Maheswari M, Murthy ANG, Shanker AK . 12 – Nitrogen Nutrition in Crops and Its Importance in Crop Quality. The Indian Nitrogen Assessment. 2017:175-186.
Crossref - Taiz L, Zeiger E, Moller IM, Murphy A. Plant Physiology and Development, 6th Ed. Sinauer Associates, Sunderland, CT. 2015.
- Glick BR. The enhancement of plant growth by free-living bacteria. Can J Microbiol. 1995;41(2):109-117.
Crossref - Raja P, Uma S, Sundaram S. Non-nodulating pink pigmented facultative Methylobacterium sp. with a functional nifH gene. World J Microbiol Biotechnol. 2006;22(12):1381-1384.
Crossref - Lidstrom ME, Chistoserdova L. Plants in the pink: cytokinin production by Methylobacterium. J Bacteriol. 2002;184(7):1818-1818.
Crossref - Rekadwad BN. Growth promotion of crop plants by Methylobacterium organophilum: Efficient bioinoculant and bio-fertilizer isolated from mud. Res Biotechnol. 2014;5(5):1-6.
- Madhaiyan M, Alex THH, Ngoh ST, Prithiviraj B, Ji L. Leaf-residing Methylobacterium species fix nitrogen and promote biomass and seed production in Jatropha curcas. Biotechnol Biofuels. 2015;8:222.
Crossref - Khan MS, Zaidi A, Ahemad M, Oves M, Wani PA. Plant growth promotion by phosphate solubilising fungi – Current perspective. Arch Agron Soil Sci. 2010;56(1):73-98.
Crossref - Jha CK, Saraf M. Plant growth promoting rhizobacteria (PGPR): A review. J Agric Res Dev. 2015;5:108-119.
Crossref - Goswami D, Thakker JN, Dhandhukia PC. Portraying mechanics of Plant Growth Promoting Rhizobacteria (PGPR). Cogent Food Agric. 2016;2(1):1-19.
Crossref - Dipta B, Bhardwaj S, Kaushal M, Kirti S, Sharma R. Obliteration of phosphorus deficiency in plants by microbial interceded approach. Symbiosis.2019;78(2):163-176.
Crossref - Vazquez P, Holguin G, Puente ME, Lopez-Cortes A, Bashan Y. Phosphate-solubilizing microorganisms associated with the rhizosphere of mangroves in a semiarid coastal lagoon. Biol Fertil Soils. 2000;30(5):460-468.
Crossref - Kaur H, Kaur J, Gera R. Plant growth promoting rhizobacteria: A boon to agriculture. Int J Cell Sci Biotechnol. 2016;5:17-22.
- Rodrıiguez H, Fraga R, Gonzalez T, Bashan Y. Genetics of phosphate solubilization and its potential applications for improving plant growth-promoting bacteria. Plant Soil. 2006;287(1):15-21.
Crossref - Weyens N, Truyens S, Dupae J, et al. Potential of Pseudomonas putida W619-TCE to reduce TCE phytotoxicity and evapotranspiration in poplar cuttings. Environ Pollut. 2010;158(9):2915-2919.
Crossref - Jayashree S, Vadivukkarasi P, Anand K, Kato Y, Seshadri S. Evaluation of pink-pigmented facultative methylotrophic bacteria for phosphate solubilization. Arch Microbiol.2011;193(8):543-552.
Crossref - Kumari A. Molecular characterization of mineral phosphate solubilization in Serratia marcescens and Methylobacterium sp. Ph.D dissertation thesis, University of Agricultural Sciences, Dharwad. 2011:69.
- Kwak MJ, Jeong H, Madhaiyan M, et al. Genome information of Methylobacterium oryzae, a plant-probiotic methylotroph in the phyllosphere. Plos One. 2014;9(3):1-14.
- Bhattacharyya PN, Jha DK. Plant growth-promoting rhizobacteria (PGPR): Emergence in agriculture. World. J Microbiol Biotech. 2012;28(4):1327-1350.
Crossref - Li Q, Saleh-Lakha S, Glick BR. The effect of native and ACC deaminase-containing Azospirillum brasilense Cd1843 on the rooting of carnation cuttings. Can J Microbiol. 2005;51(6):511-514.
Crossref - Saleem M, Arshad M, Hussain S, Bhatti AS. Perspective of plant growth promoting rhizobacteria (PGPR) containing ACC deaminase in stress agriculture. J Ind Microbiol Biotechnol. 2007;34(10):635-648.
Crossref - Etesami H, Beattie GA. Mining halophytes for plant growth-promoting halotolerant acteria to enhance the salinity tolerance of non-halophytic crops. Front Microbiol.2018;9:148.
Crossref - Martinez-Viveros O, Jorquera M, Crowley DE, Gajardo G, Mora ML. Mechanisms and practical considerations involved in plant growth promotion by rhizobacteria. J Soil Sci Plant Nutr. 2010;10(3):293-319.
Crossref - Glickmann E, Gardan L, Jacquet S, Hussain S, Elasri M, Petit A, Dessaux Y. Auxin production is a common feature of most pathovars of Pseudomonas syringae. Mol Plant Microbe Interaction. 1998;11(2):156-162.
Crossref - Honma M, Shimomura T. Metabolism of 1-aminocyclopropane-1-carboxylic acid. Agric Biol Chem. 1978;42(10):1825-1831. doi: https://academic.oup.com/bbb/article-abstract/42/10/1825/5972028#no-access-message
- Boyd M, Nkongolo KK. Nickel induces changes in expression of genes encoding 1-aminocyclopropane-1-carboxylic acid deaminase and glutathione reductase in Picea glauca. Chem Ecol. 2021;37(7):589-602.
Crossref - Nadeem SM, Ahmad M, Tufail MA, Asghar HN, Nazli F, Zahir ZA. Appraising the potential of EPS-producing rhizobacteria with ACC-deaminase activity to improve growth and physiology of maize under drought stress. Physiologia Plantarum. 2021;172(2):463-476.
Crossref - Sofy MR, Aboseidah AA, Heneidak SA, Ahmed HR. ACC deaminase containing endophytic bacteria ameliorate salt stress in Pisum sativum through reduced oxidative damage and induction of antioxidative defense systems. Environ Sci Pollut Res. 2021;28(30):40971-40991.
Crossref - Han L, Zhang H, Xu Y, Li Y, Zhou J. Biological characteristics and salt-tolerant plant growth-promoting effects of an ACC deaminase-producing Burkholderia pyrrocinia strain isolated from the tea rhizosphere. Arch Microbiol. 2021;203(5):1-12.
Crossref - Ahmad J. 1-aminocyclopropane 1-carboxylic acid deaminase producing bacteria inoculation for improving the maize seed germination and seedling growth. Agric Sci J. 2021;3(1):46-55.
Crossref - Katiyar P, Dubey RC, Maheshwari DK. ACC deaminase-producing Ensifer adhaerensKS23 enhances proximate nutrient of Pisum sativum L. cultivated in high altitude. Arch Microbiol. 2021;203(3):2689-2698.
Crossref - Alemneh AA, Zhou Y, Ryder MH, Denton MD.Large-scale screening of rhizobacteria to enhance the chickpea-Mesorhizobium symbiosis using a plant-based strategy. Rhizosphere.2021;18:100361.
Crossref - Kumar A, Maleva M, Bruno LB, Rajkumar M. Synergistic effect of ACC deaminase producing Pseudomonas sp. TR15a and siderophore producing Bacillus aerophilus TR15c for enhanced growth and copper accumulation in Helianthus annuus L. Chemosphere. 2021;276:130038.
Crossref - Liu C.H, Siew W, Hung YT, Jiang YT, Huang CH. 1-aminocyclopropane-1-carboxylate (ACC) deaminase gene in Pseudomonas azotoformans is associated with the amelioration of salinity stress in tomato. J Agric Food Chem.2021;69(3):913-921.
Crossref - Wang C, Wang H, Li Y, et al. Plant growth-promoting rhizobacteria isolation from rhizosphere of submerged macrophytes and their growth-promoting effect on Vallisneria natans under high sediment organic matter load. Microbial Biotechnol. 2021;14(2):726-736.
Crossref - Jin K, Li H, Li X, et al. Rhizosphere bacteria containing ACC deaminase decrease root ethylene emission and improve maize root growth with localized nutrient supply. Food Energy Security. 2021;10(2):275-284.
Crossref - Chinnadurai C, Balachandar D, Sundaram SP. Characterization of 1-aminocyclopropane-1-carboxylate deaminase producing Methylobacteria from phyllosphere of rice and their role in ethylene regulation. World J Microbiol Biotechnol.2009:25(8):403-1411.
Crossref - Yim WJ, Chauhan PS, Madhaiyan M, Tipayno SC, Sa T. Plant growth promontory attributes by 1-aminocyclopropane-1-carboxylate (ACC) deaminase producing Methylobacterium oryzae strains isolated from rice. In 19th World Congress of Soil Science, Soil Solutions for a Changing World. Brisbane, Australia: International Union of Soil Sciences. 2010:96-99.
- Yim W, Woo S, Kim K, Sa T. Regulation of ethylene emission in tomato (Lycopersicon esculentum Mill.) and red pepper (Capsicum annuum L.) inoculated with ACC deaminase producing Methylobacterium spp. Korean J Soil Sci Fert. 2012;45(1):37-42.
Crossref - Ekimova GA, Fedorov DN, Doronina N.V. Khmelenina VN, Mustakhimov II. AcdR protein is an activator of transcription of 1-aminocyclopropane-1-carboxylate deaminase in Methylobacterium radiotoleransJCM 2831. Antonie van Leeuwenhoek. 2022;115(9):1165-1176.
Crossref - Madhaiyan M. Molecular aspects, diversity and plant interaction of facultative methylotrophs occurring in tropical plants. Ph.D. dissertation thesis, Tamilnadu Agricultural University, Coimbatore, Tamilnadu, India. 2003:126.
- Radhika S, Velayutham K, Thavaprakaash N. Effect of nutrients and plant growth regulators on the quality parameters and yield of baby corn (Zea mays L.). Int J Trop Agric. 2008;26:255-258.
- Meenakshi BC, Savalgi VP. The effect of co-inoculation of Methylobacterium and B. japonicum on plant growth, dry matter content and enzyme activities in soybean. Karnataka J Agric Sci. 2009;22:344-348.
- Pattanashetti SR. Isolation and characterization of pink pigmented facultative methylotrophs from Coleus forskohlii and their influence on growth and tuber yield. M.Sc. (Ag) thesis, University of Agricultural Sciences, Dharwad. 2012:73.
- Boruah HPD, Chauhan PS, Yim WJ, Han GH, Sa TM. Comparison of plant growth promoting Methylobacterium spp. and exogenous indole-3-acetic acid application on red pepper and tomato seedling development. Korean J Soil Sci Fertil. 2010;43(1):96-104.
- Pattnaik S, Rajkumari J, Paramanandham P, Busi S. Indole acetic acid production and growth-promoting activity of Methylobacterium extorquens MP1 and Methylobacterium zatmanii MS4 in tomato. Int J Veg Sci. 2017;23(4):321-330.
Crossref - Saravanan K, Gurusamy A, Chelviramessh, Kannan P. Effect of land configuration and microbial consortium on growth and yield of groundnut (Arachis hypogaea L.). Pharma Innovation J. 2022;11(8):1209-1212.
- Poorniammal R, Senthilkumar M, Prabhu S, Anandhi K. Effect of Methylobacterium on seed germination, growth and yield of Barnyard Millet (Echinochloa frumentacea Var. COKV 2) under Rainfed Condition. J Pharmacognosy Phytochemistry. 2020;9(2):1675-1677.
Crossref - de Aquino GS, Ventura MU, Alexandrino RP, et al. Plant-promoting rhizobacteria Methylobacterium komagatae increases crambe yields, root system and plant height. Ind Crops Products.2018;121:277-281.
Crossref - Vadivukkarasi P, Bhai RS. Phyllosphere-associated Methylobacterium: a potential biostimulant for ginger (Zingiber officinale Rosc.) cultivation. Arch Microbiol.2020;202(2):369-375.
Crossref - Nysanth NS, Meenakumari KS, Syriac EK, Beena R. Screening of pink pigmented facultative methylotrophs for growth enhancement in paddy. Biocatalysis Agric Biotechnol.2019;18:101055.
Crossref - Ragadevi K, Jeyakumar P, Djanaguiraman M, et al. Influence of microbial priming on germination and seedling growth traits of compact cotton CO17. Int J Plant Soil Sci. 2021;33(24):530-540.
Crossref - Selvakumar S, Thiruppathi M, Immanuel RR, Sivaraman A, Chakravarthy GA. Impact of foliar fertilization of nutrients and biofertilizer on the growth and yield of hybrid cotton (Gossypium hirsutum L.). Crop Res. 2022;57(4):275-280.
Crossref - Soundarajan S, Marimuthu R, Arunkumar K, Dhanaphal K, Venkatesan T, Sivakumar G. Pink pigmented facultative methylotrophic bacteria (PPFMs) as microbial farmers in small cardamom plantation. Pharma Innovation J. 2022;11(4):607-610.
- Laurita T, Kerovuo J. Plant microbiome innovation: M-trophs. Ind Biotechnol. 2018;14(3):129-133.
Crossref - Pragathi G, Raja K, Jerlin R, Chinna Mukiri and Ramalakshmi A. Methylorubrum aminovorans infused Chitosan/PVA composite nanofibre seed coating to improve germination and seedling vigour in cotton. Pharma Innovation J. 2022;11(7):2606-2611.
- Mukiri C, Raja K, Senthilkumar M, et al. Immobilization of beneficial microbe Methylobacterium aminovorans in electrospun nanofibre as potential seed coatings for improving germination and growth of groundnut Arachis hypogaea. Plant Growth Reg. 2022;97(2):419-427.
Crossref - Madhaiyan M, Reddy BVS, Anandham R, Senthikumar M, Poonguzhali S, Sundaram S, Sa T. Plant growth promoting Methylobacterium induces defense responses in groundnut (Arachis hypogaea L.) compared with rot pathogens. Curr Microbiol. 2006;53(4):270-276.
Crossref - Yim W, Seshadri S, Kim K, Lee G, Sa T. Ethylene emission and PR protein synthesis in ACC deaminase producing Methylobacterium spp. inoculated tomato plants (Lycopersicon esculentum Mill.) challenged with Ralstonia solanacearum under greenhouse conditions. Plant Physiol Biochem. 2013;67(9):5-104.
Crossref - Poorniammal R, Sundaram SP, Kumutha K. In vitro biocontrol activity of Methylobacterium extorquens against fungal pathogens. Int J Plant Prot. 2009;2:59-62.
- Poorniammal R, Sundaram SP, Kumutha K. Induced systemic resistance by Methylobacterium extorquens against Rhizoctonia solani in Cotton. Int J Plant Prot. 2010;2(2):199-204.
- Kumar M, Srivastava AK, Pandey AK. Biocontrol activity of some potent Methylotrophs isolated from Bhitarkanika mangrove sediment. Int J Curr Res Biosci Plant Biol. 2015;2:101-106.
- Santosh S, Sreenivasa MN. Field evaluation of native pink pigmented facultative methylotrophs for growth promotion and anthracnose management in chilli. Int J Curr Microbiol App Sci. 2020;9(3):718-7261.
Crossref - Prabhu S, Kumar S, Subramanian S, Sundaram SP. Suppressive effect of Methylobacterium fujisawaense against root-knot nematode, Meloidogyne incognita.Indian J Nematol. 2009;39(2):165-169.
- Lee MK, Lee GS, Yim WJ, et al. Inoculation effect of Methylobacterium suomiense on growth of red pepper under different levels of organic and chemical fertilizers. Korean J Soil Sci Fertil. 2009;42(4):266-273.
- Beneduzi A, Moreira F, Costa PB, et al. Diversity and plant growth promoting evaluation abilities of bacteria isolated from sugarcane cultivated in the South of Brazil. Appl Soil Ecol. 2013;63:94-104.
Crossref - Chauhan PS, Lee GS, Lee MK, Yim WJ, Lee GJ, Kim YS, Chung JB, Sa, TM. Effect of Methylobacterium oryzae CBMB20 inoculation and methanol spray on growth of red pepper (Capsicum annuum L.) at different fertilizer levels. Korean J Soil Sci Fertil. 2010;43(4):514-521.
- Tani A, Sahin N, Fujitani Y, Kato A, Sato K, Kimbara K. Methylobacterium species promoting rice and barley growth and interaction specificity revealed with whole-cell matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF/MS) analysis. PLoS ONE. 2015;10(6):e0129509.
Crossref - Photolo MM, Sitole L, Mavumengwana V, Tlou MG. Genomic and Physiological investigation of heavy metal resistance from plant endophytic Methylobacterium radiotolerans MAMP 4754, isolated from Combretum erythrophyllum. Int J Environ Rese Public Health.2021;18(3):997.
Crossref - Vannucchi F, Imperato V, Saran A, et al. Inoculated seed endophytes modify the Poplar responses to trace elements in polluted soil. Agronomy. 2021;11(10):1987.
Crossref - Kassem MM, Hauka FI, Afify AH, Nour El-Din M, Omara AA. Isolation and identification of some PPFMs bacterial isolates and their potentiality as biofertilizers and biocontrol agents to Rhizoctonia solani. J Agric Chem Biotechnol. 2013;4(2):79-92.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.