ISSN: 0973-7510
E-ISSN: 2581-690X
K. pneumoniae is known to cause hospital and community acquired infections. It is usually associated with upper & lower respiratory infections, septicaemia, urinary tract infection, wound infections, neonatal sepsis, meningitis, and endophthalmitis. The virulence factors play a role in its existence in different environmental conditions and therefore help in establishing Klebsiella pneumoniae infection in the human body. Multi drug resistant Klebsiella pneumoniae is an increasing threat to human health. Klebsiella pneumoniae is one of the species recognized as nosocomial pathogens that exhibit multidrug resistance and virulence in ESKAPE group as per WHO. The study was conducted to determine the various virulence factors & the antimicrobial pattern of Klebsiella pneumoniae isolates. A cross sectional observational study, conducted in Department of Microbiology of R.L. Jalappa Hospital and Research Centre, Kolar, Sample size of 150. All 150 Klebsiella pneumoniae isolates collected for the study, The Klebsiella pneumoniae isolates which were positive for various virulence factors were as follows on hemolysis 7(4.66%), capsule 150(100%), Hypermucoviscosity formation 66(44%), biofilm production 81(54%), siderophore production 110(73.33%), protease 135(90%), gelatinase 126(84%), lipase production 119(79.33%), lecithinase activity 82(54.66%). The drug resistance klebsiella pneumoniae were as follows: ESBL producers 24(16.67%), AmpC producers were 22(14.67%), MDR 116(74.20%), extensive drug resistant (XDR) 30(20%), pan drug resistant (PDR) 42(28%), Carbapenem resistance 65.33% reported. The increasing coexistence of virulence factors & antimicrobial resistance pattern is of particular concern. Hence active surveillance for antimicrobial resistance & virulence determinants is imperative now to implement effective control measures to prevent the rapid spread of drug resistance.
Klebsiella pneumoniae, Virulence Factors, Antimicrobial Resistance Pattern
Klebsiella pneumoniae is a gram negative, capsulated, non motile bacilli, lactose fermenting, facultative anaerobic bacteria belonging to family Enterobacteriaceae.1,
K. pneumoniae asymptomatically colonizes the skin, mouth, respiratory & gastrointestinal flora. K. pneumoniae is one of the known agents of hospital and community acquired infections.2 It is usually associated with lower respiratory infections (pneumonia), bacteremia, septicaemia, urinary tract infection, wound infections, intra abdominal infections neonatal septicemia, liver abscess, meningitis, endophthalmitis , catheter associated urinary tract infection (CAUTI), ventilator associated pneumonia (VAP) & central line associated bloodstream infection (CLABSI) cases.3
“The pathogenicity of K. pneumoniae mainly arise from various virulence factors which allow it to overcome innate host immunity and to maintain infection in a mammalian host”.4 These virulence factors play a role in its survival in different environmental conditions and therefore help in establishing infection in the human body.5 K. pneumoniae pathogenicity is attributed to several virulence factors like fimbrial adhesins, lipopolysaccharides, capsule and siderophores, biofilm formation, lipase, lecithinase, haemagglutination, protease, gelatinase, hemolysis and hypermucoviscosity. All these virulence factors have the potential to produce a wide variety of infections in hospitalized patients & in the community.6,7
It also produces hemolysin protein, an exotoxin (cytolyic toxin) which cause lysis of blood cells & therefore facilitate the dissemination of bacteria.8
There are 3 variants of Klebsiella pneumoniae – Classical K. pneumonia (cKP), Hypervirulent K. pneumoniae (hvKP) & Multidrugresistant hvKP (MDR-hvKP).9,10
K. pneumoniae infections are more common with classic K. pneumoniae (cKp) type”.8 These strains persist in hospital environments and cause infections in debilitated patients. These cKp strains are distinct from hypervirulent K.pneumoniae (hvKp) strains.11
K. pneumoniae has the ability to form biofilms, responsible for colonization of the gastrointestinal, respiratory, urinary tract and the development of invasive infections especially in immunocompromised patients Klebsiella pneumoniae, can adhere to medical devices forming biofilms, thus preventing the antibacterial factors. 12,13
In the antibiotic era, K. pneumoniae is a documented cause of healthcare-associated infections (HAI); these strains are naturally resistant to ampicillin, carbenicillin and ticarcillin because of production of a chromosomal penicillinase, sulfhydryl variable (SHV-1). The global rise of multidrug-resistant (MDR) K.p strains represent an increasing threat to human health with a high mortality rate.14
According to WHO in 2017 Klebsiella pneumoniae is one of the species recognized in ESKAPE group (Enterococcus faecium, Staphylococcus aureus, K. pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species), associated by their characteristic potential to escape or evade the action of antimicrobial agents.15
This was a Cross sectional observational study conducted in the Department of Microbiology at Central Diagnostics Laboratory Services (CDLS), R.L. Jalappa Hospital and Research Centre, Tamaka, Kolar. The study was done from October 2019 to May 2021, for about 1.6 years.
Sample Size
150 Klebsiella pneumoniae isolates
Inclusion Criteria
All Klebsiella pneumoniae species isolated from various clinical samples were included in our study.
Exclusion Criteria
Patient who refused to give the informed consent for the study.
Klebsiella pneumoniae isolated from stool samples were excluded from the study. A total of 150 Klebsiella pneumoniae species isolated from different clinical samples: Pus sample (n = 51), respiratory samples were as follows – sputum (n = 22) & ET samples (n = 43),Blood culture (n = 15), Urine (n = 10) and Body Fluids (n = 5), miscellaneous samples (n = 3) like vaginal sample, central line sample, ear/eye samples as shown in Table 1. The samples were cultured on Blood agar media and Mac conkey agar then were incubated at 37°C for 24 hours. K. pneumoniae was identified by standard biochemical tests including morphology of colony, Gram staining, oxidase, triple sugar iron medium (TSI), Methyl Red, indole test VogesProskauer (MR-VP), urease, lysine decarboxylase, arginine dihydrolase and Simmons citrate agar.16 The clinical isolates, were determined for the following virulence factors as mentioned below:
Hemolysis, Capsule Formation, Hypermucoviscosity, Biofilm Formation, Siderophore Production, Protease, Gelatinase, Haemagglutination Assay, Lipase & Lecithinase Activity (Table 2).
Table (1):
Type of samples |
No. of isolates (%) |
|---|---|
Pus sample |
51 (34 %) |
Endotracheal tube (ET) sample |
43 (28.67 %) |
Sputum |
22 (14.66 %) |
Blood culture |
15 (10.0 %) |
Urine |
10 (6.67 %) |
Fluids (CSF, pleural, Peritoneal, Synovial fluids) |
5 (3.33 %) |
Miscellaneous (Ear, eye swab, vaginal sample) |
4 (2.67 %) |
Table (2):
Distribution of Klebsiella pneumoniae isolates positive for various virulence factors.
No. |
Detection of various virulence factors by phenotypic methods |
K. pneumonia isolates |
|---|---|---|
1. |
Hemolysis |
7 (4.66 %) |
2. |
Capsule |
150 (100.0 %) |
3. |
Hypermucoviscosity (HMV) |
66 (44.0 %) |
4. |
Biofilm Formation |
81 (54.0 %) |
5. |
Siderophore Production |
110 (73.33 %) |
6. |
Protease |
135 (90.0 %) |
7. |
Gelatinase Test |
126 (84.0 %) |
8. |
Lipase |
119 (79.33 %) |
9. |
Lecithinase Activity |
82 (54.66 %) |
Methodology For Detection of Virulence Factors: Hemolysis
Klebsiella pneumoniae colonies were inoculated on routine sheep blood agar at 37°C & observed for hemolysis.6
Detection of Capsule
Overnight incubated Klebsiella pneumoniae colony was taken on a clean slide, smear stained with methylene blue for 2 mins, then washed with tap water & observed for hemolysis. Observed for capsule around the organism.17
Hyper Mucoviscosity [Hv] Modified String Test: Klebsiella pneumoniae isolate was inoculated on routine sheep blood agar media, incubated at 37°C for 24 hours. Then a standard inoculation loop was used to demonstrate the string test When the formed string stretched >5 mm in length, it indicated HMV phenotype.18
Biofilm Formation
96 well Microtiter plate method is commonly used to demonstrate the biofilm formation by Klebsiella pneumoniae isolates in various samples as shown in Figure 1.
Method
200µl of overnight luria broth (LB) culture was transferred to a 96 well microtiter plate in triplets. Isolates sub cultured on Lysogeny Broth was incubated for 24 hr at 37°C. Further 25µl of 1% crystal violet was added to each well and incubated for 15 mins at room temperature. Wells were washed thrice with phosphate buffer solution (PBS) and ethanol was added to dissolve the strain.19
Interpretation of Biofilm Production
After testing the isolates, microtiter plate was assessed: The mean OD492 of the six wells was calculated (ODT). The cut-off OD (ODc) was defined as three standard deviations above the mean OD of the negative control wells. The level of the formed biofilm was asserted as follows: (i) Nonadherent: ODT ≤ ODC (ii)Weakly adherent: ODC < ODT ≤ 2ODC (iii)Moderately adherent: 2ODC < ODT ≤ 4ODC (iv)Strongly adherent: 4ODC < ODT.20
Siderophores
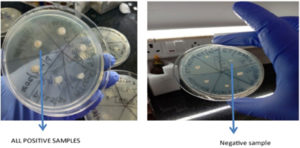
Nutrient agar supplemented with 200 mM of 2.2 –dipyridyl Klebsiella pneumoniae isolates will be streaked on agar plates & then incubated at 37°C for 24 hrs. Any bacterial growth will be considered positive for Siderophore production as shown in Figure 2.21
Protease
Klebsiella pneumoniae was inoculated on freshly prepared milk agar and incubated at 37°C for 72 hrs, formation of turbidity around the colonies – demonstrate protease production.7
Interpretation of the Test
The Development of any growth at stab inoculated sites were considered as positive & no growth at the inoculation were considered as negative for the test.22,23
Gelatinase
Nutrient agar plate with 3% gelatin was prepared, colony was inoculated & incubated for 16 hrs at 37°C, then extra 5 hrs in refrigeration (at 4°C). The gelatinase producing colonies were surrounded by a clear zone once mercuric chloride was poured on plates while the medium became opaque.
Interpretation of the Test
The Development of any growth at stab inoculation sites were considered as positive & no growth at the inoculation were considered as negative for the test as shown in Figure 3.24
Haemagglutination Assay
Mannose sensitive haemagglutination was confirmed by absence of haemagglutination, and Mannose resistant haemagglutination: was confirmed by presence of haemagglutination (with 3% O -blood group human RBC in presence of 2% mannose).25
Lipase Activity
Klebsiella pneumoniae isolate grown on blood agar was re-inoculated on egg-yolk agar & was incubated at 37°C for one day. Then plates were flooded with copper sulphate solution for 20 mins, removed the excess solution and plate was allowed to dry. Observed the changes.
Lecithinase Activity
Klebsiella pneumoniae isolated from nutrient agar was re-inoculated on egg-yolk agar, incubated for 37°C for 24 hrs then the plates were sprayed with copper sulphate solution for 20 mins, drain off the excess solution & plate was allowed to dry.
Interpretation of Lipase & Lecithinase Test
If there is clearance/halo around the colonies it was considered as positive for this test. For lipase production – development of bluish green type of colonies even after removing excess copper sulphate were considered as positive test results.26
Methodology for Detection of Antimicrobial Resistance
For each pure isolate an antimicrobial sensitivity testing was performed by disk diffusion technique was done on Mueller Hinton Agar [MHA] and antibiotic discs will be placed and incubated for 18- 24 hours. Based on the zone size, the isolates will be classified as sensitive, moderately sensitive and resistance as per the CLSI [Clinical and Laboratory Standards Institute] guidelines.27
The gram negative antibiotic panel for routine antibiotic susceptible testing will be as follows:
- Piperacillin, Piperacillin-Tazobactam, Ampicillin, Amoxicillin-Clavulanate, Gentamicin, Amikacin, Tobramycin, Cefotaxime, Ceftriaxone, Ceftazidime, Ceftazidime-Clavulanic Acid, Cefoxitin, Ciprofloxacin, Levofloxacin, Imipenem, Meropenem, Ertapenem, TrimethoprimSulfamethoxazole (COT), Chloramphenicol, Doxycycline, Nitrofurantoin & Norfloxacin [For Urine Sample Only].
KPC Positive Samples
Detected by KPC Hi Media agar, when blue-green colonies are produced on culture plate it will be considered as positive for Carbapenem resistance.28
Table (3):
Types of antimicrobial pattern in our study.
Types of antimicrobial pattern in our study |
No of isolates (%) |
|---|---|
ESBL type |
25 (16.67 %) |
AmpC producers |
22 (14.67 %) |
MDR |
116 (74.20 %) |
XDR |
30 (20.0 %) |
PDR |
42 (28.0 %) |
Imipenem (IMP) resistance |
95 (63.33 %) |
Meropenem (MRP) resistance |
105 (70.0 %) |
Ertapenem (ETP) resistance |
94 (62.66 %) |
All 150 Klebsiella pneumonia isolates collected in our study were positive for various virulence factors as follows: hemolysis 7(4.66%), capsule 150(100%), Hypermucoviscosity formation 66(44%), biofilm production 81(54%), siderophore production 110(73.33%), protease 135(90%), gelatinase 126(84%), lipase production 119(79.33%), lecithinase activity 82(54.66%) as depicted in Table 2.
The drug resistance pattern of Klebsiella pneumoniae isolates were as follows ESBLs, 25 (16.67%), AmpC producers were 22 (14.67%), MDR 116 (74.20%), extensive drug resistant (XDR) 30 (20%), pan drug resistant (PDR) 42 (28%), Carbapenem resistance 65.33% reported as depicted in Table 3.
K. pneumoniae strains produce a variety of infections and employs various virulence factors to colonize and spread in the human body. The exploding antimicrobial resistance of this bacteria in recent years is of great concern to the scientific world.29
Pathogenicity of K. pneumoniae is the result of production of many virulence factors that help these bacteria overcome the immune system and cause various diseases.30
In our study, we collected 150 clinical K. pneumoniae samples from pus sample, respiratory samples, blood culture, urine and body fluids.
The following 10 virulence factors were analysed in our study – Hemolysis, Capsule Formation, Hypermucoviscosity, Biofilm Formation, Siderophore Production, Protease, Gelatinase, Haemagglutination Assay, Lipase & Lecithinase Activity.
The Klebsiella pneumoniae isolates from our study producing capsule was 100%, this is in concordance with other study findings – Aljanaby et al – 100%, Imtiaz et al – 99%31,32 and Klebsiella pneumoniae has thick polysaccharide capsule this provides protection to bacteria against opsonization and phagocytosis by macrophages and neutrophils, due to blockade of Toll like receptors (TLR-4) & inhibit the expression of IL -8.33
High virulence is typical for K. pneumoniae strains that cause community-acquired infections, LPS protects against humoral defences and activate immune system.
The Klebsiella pneumoniae isolates producing siderophore was 73.33% is our study results, (Siderophore production is more from endotracheal & pus samples in our study). This is in contrast to other studies which reported 100% in – Aljanaby et al.31,34
It has been shown that the development of invasive forms of the disease is associated with the production of several siderophores by K. pneumoniae strains. Hv-KP strains produce more siderophore molecules and in their more active form, than c-KP.35
The Klebsiella pneumoniae isolates producing Hypermucoviscosity (HMV) detected by positive string test was 44% in our study. The study conducted by other investigators on HMV produced from Klebsiella pneumoniae varies from 7 – 62%, Aljanaby et al reported 62%, Lee et al studied extensively on HMV – showed 14.8% from hospital acquired Klebsiella pneumoniae infections & 41.5% from community acquired Klebsiella pneumoniae infections. But in contrast Rama Krishnan et al study done in 2019 reported only 7% and Imtiaz et al reported 22%.6,31,32,36
Detection of the rmp A gene in K. pneumoniae isolates / Hv-KP are responsible for invasive forms of infection in previously healthy adults (liver abscesses, meningitis, and endophthalmitis), hv-KP strains are more resistant to the complement and phagocytosis.37
The Klebsiella pneumoniae isolates from our study producing lipase was 79.33%, this is in contrast to Rama Krishnan et al study showed 58%.6
In our study, the Klebsiella pneumoniae isolates producing biofilm are 54% (n = 81), our study findings are in contrast to Aljanaby et al reported 100%, Imtiaz et al – showed 94% results, & Rama Krishnan et al study showed 79%. In contrast, a study Dougnan et al. reported with only 3.34% of biofilm formation in Klebsiella pneumoniae isolates.6,31,32,38
Our findings on hemolysis was 4.66%; this is in concordance with other findings of Imtiaz et al. – showed 8%.32 Our findings are in contrast to Dougnan et al which reports 20% for hemolysis.38
In our study, the Klebsiella pneumoniae isolates producing lecithinase enzyme as virulence factor 54.66% (n =82), this is in concordance with Rama Krishnan et al study showed 55%. Our study is in contrast to findings of Dougnan et al which showed 3.34%.6,38
In our study, the Klebsiella pneumoniae isolates producing protease enzyme was 90% (n =135), this is in contrast to earlier studies like Rama Krishnan et al study showed 44%.6 In our study the Klebsiella pneumoniae isolates producing gelatinase enzyme on gelatin media was 84% (n = 126), this findings are in contrast to Rama Krishnan et al study showed 41% & Imtiaz et al – showed 12% only. 6,32
The antibiotic resistance of K. pneumoniae strains is associated mainly with the production of ESBL. In 2017 the World Health Organization declared K. pneumoniae as one of the most dangerous superbugs along with Acinetobacter baumanii and Pseudomonas aeruginosa. This is of great concern because some antibiotic resistance are carried by mobile genetic elements (plasmids & transposons), which facilitate horizontal genetic transfer and promote the spread of antimicrobial resistance within and between species.39
Compared to the multicenter study carried out in Garcia Fernandez et al, our study findings out of 150 K. pneumoniae isolates showed: ESBL K. pneumoniae 16.67% (n=25 isolates), AmpC producer type 14.67% (n=22), multi drug resistance (MDR) – 74.20% (n=116), extensive drug resistant (XDR) – 20% (n=30), pan drug resistant (PDR) – 28% (n=42).40
In our study MDR isolates reported was 74.20%, this is in contrast to a meta analysis done by Asri et al on MDR K. pneumoniae isolates was 32.8%, Our study is in concordance with Ferreira et al study based on genotypic analysis reported 84% of MDR Klebsiella pneumoniae in their study.41,42
In our study the XDR isolates were 20.0%. Our findings are in concordance with Wenzi Bi et al showed 26.71. In their study XDR was resistant to all antibiotics except Tigecycline & Polymyxin B. In our study we have reported as resistance to all antimicrobials except chloramphenicol.43
Our study findings on CRKP isolates were as follows: Imipenem resistance 63.33%, meropenem resistance 70% & ertapenem resistance 62.66%; our study results are in contrast to studies Lombardi et al which reported 35% of CRKP isolates & Kahraman EP et al showed Imipenem resistance of 5.1% & meropenem resistance of 3.4% respectively.44,45
The reasons for antimicrobial drug resistance in Klebsiella pneumoniae could be due to diminished expression/loss of porin channels, alteration in outer membrane proteins (OMP) permeability, or efflux pump overexpression & carbapenem hydrolysis.46
KPCs also inactivate all beta-lactam antibiotics and are only partially inhibited by beta-lactamase inhibitors like clavulanic acid, tazobactam and boronic acid.47
The presence of several virulence factors accompanied by antimicrobial resistance had made Klebsiella pneumoniae an important infectious agent of global concern and lead to treatment failure. The increasing coexistence of the virulence factors & antimicrobial is a major concern as it can lead to untreatable/invasive K. pneumoniae infections. Active surveillance is needed for antimicrobial resistance and also for virulence determinants, to avoid the transmission and spread of multidrug resistant strains. In our study, a lot of MDR K. pneumoniae strains were observed which warrants the role of antimicrobial stewardship programme, as It is vital & plays an important role in prevention & control of MDR K. pneumoniae strains.
ACKNOWLEDGMENTS
The authors would like to thank patients who willingly participated in this study by giving informed consent & clinical samples to be processed accordingly.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
MSH helped in collection & processing of samples, data collection, analysis of data & writing. AN helped in design of study, analysis of data & manuscript editing. PNS helped in revision of intellectual content & manuscript editing.
FUNDING
None.
DATA AVAILABILITY
All datasets generated/analysed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee of Sri Devaraj Urs Medical College, Tamaka, Kolar [No: SDUMC/KLR/IEC/113/2019-20, Dated 11 -10- 2019].
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- Ashurst JV, Dawson A. Klebsiella Pneumonia. StatPearls [Inernet]. 2022. https://www.ncbi.nlm.nih.gov/books/NBK519004/
- Martin RM, Bachman MA. Colonization, Infection, and the Accessory Genome of Klebsiella pneumoniae. Front Cell Infect Microbiol. 2018;8:4.
Crossref - Ko WC, Paterson DL, Sagnimeni AJ, et al. Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerg Infect Dis. 2002;8(2):160-166.
Crossref - Bengoechea JA, Sa Pessoa J. Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS Microbiol Rev. 2019;43(2):123-144.
Crossref - Paczosa MK, Mecsas J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol Mol Biol Rev. 2016;80(3):629-661.
Crossref - Ramakrishnan K, JenithaJohnsi P, Seetha KS. Clinical isolates of Klebsiella pneumoniae and its virulence factors from a tertiary carehospital. Indian J Microbiol Res. 2019;6(2):109-112.
- Piperaki ET, Syrogiannopoulos GA, Tzouvelekis LS and Daikos GL. Klebsiella pneumoniae: Virulence, Biofilm and Antimicrobial Resistance. The Pediatric Infect Dis Jl .2017:36 (10):1002-1005.
- Esmaeel JR, Sadeq JN. Hemolysin gene detection in some isolates of Klebsiella pneumonia by PCR. Al-Qadisiyah Journal of Veterinary Medicine Sciences. 2018;17(2):49-52.
Crossref - Zhu J, Wang T, Chen L, Du H. Virulence Factors in Hypervirulent Klebsiella pneumoniae. Front Microbiol. 2021;12:642484.
Crossref - Lin Z, Zheng J, Bai B, et al. Characteristics of Hypervirulent Klebsiella pneumoniae: Does Low Expression of rmpA Contribute to the Absence of Hypervirulence? Front Microbiol. 2020;11:436.
Crossref - Choby JE, Howard-Anderson J, Weiss DS. Hypervirulent Klebsiella pneumoniae – clinical and molecular perspectives. J Intern Med. 2020;287(3):283-300.
Crossref - Alcantar-Curiel MD, Blackburn D, Saldana Z, et al. Multi-functional analysis of Klebsiella pneumoniae fimbrial types in adherence and biofilm formation. Virulence. 2013;4(2):129-138.
Crossref - Guerra MES, Destro G, Vieira B, et al. Klebsiella pneumoniae Biofilms and Their Role in Disease Pathogenesis. Front Cell Infect Microbiol. 2022;12:877995.
Crossref - Effah CY, Sun T, Liu S, Wu Y. Klebsiella pneumoniae: an increasing threat to public health. Ann Clin Microbiol Antimicrob. 2020;19(1):1.
Crossref - Mulani MS, Kamble EE, Kumkar SN, Tawre MS, Pardesi KR. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front Microbiol. 2019;10:539.
Crossref - Tille PM. Bailey and Scott`s Diagnostic Microbiology. 14th Ed. Missouri: Elsevier publishers (p) Ltd; 2017:336-354.
- Standard Operating Procedures Bacteriology Antimicrobial Resistance Surveillance and Research Network 2nd Edition, 2019.Indian Council of Medical Research New Delhi, India
- Lin Y-C, Lu M-C, Tang H-L, et al. Assessment of hypermucoviscosity as a virulence factor for experimental Klebsiella pneumoniae infections: Comparative virulence analysis with hypermucoviscosity-negative strain. BMC microbiology. 2011;11:50.
Crossref - Stepanovic S, Vukovic D, Dakic I, Savic B, Svabio-Vlahovic M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods. 2000;40(2),175–179.
Crossref - Deka N. Comparison of Tissue Culture plate method, Tube Method and Congo Red Agar Method for the detection of biofilm formation by Coagulase Negative Staphylococcus isolated from Non-clinical Isolates. Int J Curr Microbiol App Sci. 2014:3(10);810-815.
- Namikawa H, Niki M, Niki M, et al. Siderophore production as a biomarker for Klebsiella pneumoniae strains that cause sepsis: A pilot study. J Formos Med Assoc. 2022;121(4):848-855.
- Taher NA, Baqir BAA, Abdul FR. A Study on the Effect of Ethidium Bromide on Virulence Factors (Protease and Biofilm Formation) by Klebsiella Pneamoniae isolated from Different Clinical Sources. Historical Res Lett. 2016;(30):1-6.
- Alam M, Imran M. Screening and Potential of Gram Negative Bacterial Isolates for their Extracellular Enzymatic Activities Isolated from the Hospital Aquatic Environment. J Basic Clin Pharma. 2018;9:41-45.
- Cevahir N, Demir M, Kaleli I, Gurbuz M, Tikvesli S. Evaluation of biofilm production, gelatinase activity, and mannose resistant hemagglutination in Acinetobacter baumannii strains. J Microbiol Imunnol Infect. 2008;41(6):513-518.
- Mishra M, Thakar YS, Pathak AA. Haemagglutination, haemolysin production and serum resistance of Proteus and related species isolated from clinical sources. Indian J Med Microbiol. 2001;19(2):5-11.
- Esselmann TM, Liu PV. Lecithinase production by Gram negative bacteria. J Bacteriol. 1961;81(6):939-945.
- CLSI. Performance standards for Antimicrobial Susceptibility testing. 29th ed.CLSI supplement M100. Wayne P A: Clinical and Laboratory Standards Institute. 2019
- Mahapatra A, Nikitha K, Rath S, Behera B, Gupta K. Evaluation of HiCrome KPC Agar for Screening of Carbapenem-Resistant Enterobacterales Colonization. J Lab Physicians. 2021;13(4):358–361.
Crossref - Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11(4):589-603.
Crossref - Opoku-Temeng C, Malachowa N, Kobayashi SD, DeLeo F, R: Innate Host Defense against Klebsiella pneumoniae and the Outlook for Development of Immunotherapies. J Innate Immun. 2022;14(3):167-181.
Crossref - Aljanaby AAJ, Alhasani AHA. Virulence factors and antibiotic susceptibility patterns of multidrug resistance Klebsiella pneumoniae isolated from different clinical infections. Afr J Microbiol Res. 2016;10(22):829-843.
Crossref - Imtiaz W, Syed Z, Rafaque Z, Andrews SC, Dasti JI. Analysis of Antibiotic Resistance and Virulence Traits Genetic and Phenotypic in Klebsiella pneumoniae Clinical Isolates from Pakistan: Identification of significant levels of Carbapenem and Colistin Resistance. Infect Drug Resist. 2021;14:227-236.
- Ali S, Alam M, Hasan G, Hassan M. Potential therapeutic targets of Klebsiella pneumoniae: a multi-omics review perspective. Briefings in Functional Genomics. 2022;21(2):63-77.
Crossref - Naga I, Ghazal A, Alomari S, Gaballah A. Detection of Biofilm and Siderophore Encoding Genes Implicated in the Pathogenesis of Klebsiella pneumoniae Isolated from Different Clinical Specimens. Egypt J Med Microbiol. 2021;30:101-108.
Crossref - Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. 2019;32(3):e00001-19.
Crossref - Lee HC, Chuang YC, Yu WL, et al. Clinical implications of hypermucoviscosity phenotype in Klebsiella pneumoniae isolates: association with invasive syndrome in patients with Community Acquired bacteremia. J Intern Med. 2006;259(6):606-614.
Crossref - Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4(2):107-118.
Crossref - Dougnon V, Assogba P, Mohammed J, et al. Urinary Tract Infections in Benin: Exploring the Virulence Factors and Antibiotic Resistance and Virulence Genes among Bacterial Isolates. International Journal of Pathogen Research, 2021;7(1):28-36.
Crossref - WHO publishes list of bacteria for which new antibiotics are urgently needed. https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed
- Garcıa-Fernandez S, Garcia-Castillo M, Bou G, et al. Activity of Ceftolozane/Tazobactam Against Pseudomonas aeruginosa and Enterobacterales Isolates Recovered From Intensive Care Unit Patients in Spain: The Superior Multicentre Study. Int J Antimicrob Agents. 2019;53(5):682-688.
Crossref - Mohd Asri NA, Ahmad S, Mohamud R, et al. Global Prevalence of Nosocomial Multidrug-Resistant Klebsiella pneumoniae: A Systematic Review and Meta-Analysis. Antibiotics. 2021;10(12):1508.
Crossref - Ferreira RL, da Silva BCM, Rezende GS, et al. High Prevalence of Multidrug-Resistant Klebsiella pneumoniae Harboring Several Virulence and β-Lactamase Encoding Genes in a Brazilian Intensive Care Unit. Front Microbiol. 2019;9:3198.
Crossref - Bi W, Liu H, Dunstan RA, et al. Extensively Drug-Resistant Klebsiella pneumoniae Causing Nosocomial Bloodstream Infections in China: Molecular Investigation of Antibiotic Resistance Determinants, Informing Therapy, and Clinical Outcomes. Front Microbiol. 2017;8:1230.
Crossref - Lombardi F, Gaia P, Valaperta R, et al. Emergence of Carbapenem-Resistant Klebsiella pneumoniae: Progressive Spread and Four-Year Period of Observation in a Cardiac Surgery Division. BioMed Res Int. 2015;215:871947.
Crossref - Kahraman EP, Ciftcia IH. The Antibiotic Resistance Patterns of Klebsiella pneumoniae Clinic Isolates: A Comprehensive Meta-Analysis. Open J Bac. 2017;1(1):021-026.
- Brunson DN, Maldosevic E, Velez A, Figgins E, Ellis TN. Porin loss in Klebsiella pneumoniae clinical isolates impacts production of virulence factors and survival within macrophages. Int J Med Microbiol. 2019;309(3-4):213-224.
Crossref - Meletis G. Carbapenem resistance: overview of the problem and future perspectives. Ther Adv Infect Dis. 2016;3(1):15-21.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.