ISSN: 0973-7510
E-ISSN: 2581-690X
Staphylococcus aureus (S.aureus) is a prevalent organism causing infections in the community and hospital. A variety of antibiotics are used, including the Macrolide-Lincosamide-StreptograminB (MLSB) family of antibiotics in which clindamycin is the preferred agent. Widespread use of these antibiotics leads to resistance to these MLSB antibiotics; a D-test can characterize the different MLSB phenotypes. This study was taken up with an objective to perform a double disc diffusion test for detecting different phenotypes in S.aureus with particular reference to inducible clindamycin resistance. Out of a total of 174(100%) strains of S.aureus, 98(56.32%) were MRSA, and 76(43.68%) were MSSA. All isolates were tested by D-test. A total of 47(27.01%) were of cMLSB phenotype, 31(17.82%) were of iMLSB phenotype, and 96(55.17%) were of MS phenotype. The majority of MRSA strains were cMLSB phenotype(76.60%) and iMLSB phenotype (64.52%) in comparison to MSSA isolates. Although iMLSB phenotypes are present in both MRSA and MSSA, iMLSB was more in MRSA isolates. Appropriate susceptibility data is essential for a clinician to start clindamycin therapy to prevent therapeutic failures with inducible MLSB resistance in S.aureus isolates. It will be appropriate for all the clinical laboratories to report inducible Clindamycin resistance in S.aureus strains (both MRSA and MSSA), for which D-test is a reliable testing method.
Staphylococcus aureus, MRSA, MSSA, D-test, inducible clindamycin resistance, MLSB phenotypes
Staphylococcus aureus (S.aureus) is a commonly encountered organism causing infections both in hospital and community settings1.The genus Staphylococcus contains 32 species, of which 16 species are found in humans. S.aureus is one of the most virulent species having many virulence factors like surface proteins, capsular polysaccharides, cytotoxins, superantigens, enzymes responsible for producing an array of ailments from superficial infections to deep-seated and life-threatening infections2.
Treatment of S.aureus infections is usually with antibiotics like β lactams, glycopeptides, quinolones, oxazolidinone, etc. S.aureus has developed resistance to multiple antibiotics by various mechanisms like efflux of the drug, drug inactivation, target alteration, production of β lactamase, etc.3 Emergence of Methicillin-Resistant S.aureus(MRSA) strains which is a typical hospital acquired organism and acquiring multidrug resistance has still complicated the treatment . The Macrolide-Lincosamide-StreptograminB(MLSB) family of antibiotics are the agents used against such strains. MLSB includes Macrolide (Eg: Erythromycin, Azithromycin, Spiramycin), Lincosamides (Eg: Clindamycin, Lincomycin), and StreptograminB (Eg: Quinupristin, Dalfopristin). These agents are different chemically, but all of them act by inhibition of protein synthesis, among which clindamycin is the recommended agent due to its pharmacokinetics, and its ability to reach various tissues, including bones4.
Widespread use of the MLSB group of antibiotics leads to an increase in S.aureus strains becoming resistant to these drugs, which can be due to any of the following mechanisms:
- erm, a gene of S.aureus produces rRNA methylase, which brings about changes in the antibiotic binding site. The production of the erm gene can be either constitutive or inducible, leading to cMLSB or iMLSB phenotypes, respectively.
- Efflux of antibiotics by msrA gene, which is called MS phenotype.
- Inactivation of lincosamide by chemical alteration by the inuA gene5.
Organisms develop resistance to these groups of antibiotics by acquiring genes called erm genes responsible for producing methylases. S.aureus strains harbor the genes like erm A, B, C & Y in their plasmids, conferring resistance to MLSB antibiotics. The resistance can be inducible resistance where the strains exhibiting this type of resistance don’t encode for methylases but become active only in the presence of antimicrobial agents like erythromycin, which is an inducer of erm genes. Another type of resistance exhibited by organisms is called constitutive resistance, in which methylases are produced even in the absence of inducer like erythromycin6.
The isolates having the inducible erm gene exhibit resistance to agents like erythromycin, which are the inducer but will appear to be susceptible to the lincosamide and the non-inducer macrolides. Hence, using antibiotics like clindamycin will lead to the selection of constitutive mutants leading to treatment failures7.
So while testing in vitro, interpretation of different phenotypes has to be done. cMLSB phenotypes are resistant to macrolides like erythromycin and lincosamides like clindamycin. iMLSB phenotypes are resistant to erythromycin and appear sensitive to clindamycin when tested without an inducer. But, in the presence of inducer of erm gene like erythromycin, they are resistant to clindamycin with a D-shaped zone of inhibition. MS phenotypes are sensitive to clindamycin without a D zone and resistant to erythromycin due to drug efflux mechanisms4.
Determination of inducible clindamycin resistance by double disc diffusion test is advisable to avoid false sensitive reporting of clindamycin. The use of clindamycin in iMLSB phenotypes can lead to treatment failure because of the selection of cMLSB phenotypic strains. D-test which is an induction test useful in distinguishing S.aureus isolates which have inducible erm mediated resistance, i.e., iMLSB phenotypes from those with resistance due to drug efflux mechanism, i.e., MS phenotypes, and it is essential to test in vitro to differentiate iMLSB and MS phenotype strains to avoid clinical therapeutic failure8,9.
Inducible Clindamycin resistance can be tested phenotypically by double disc diffusion test (D-test) or genotypically by molecular methods like Polymerase Chain Reaction(PCR) for detecting erm gene.10 Though molecular techniques like PCR are more sensitive, its cost, requirement for technical expertise, and non-availability at all testing facilities make it less preferable than simple, easy to perform D-test.
This is a descriptive cross-sectional study conducted in the microbiology department at Dr. Pinnamaneni Siddhartha Institute of Medical Sciences and Research Foundation, Andhra Pradesh, India, after the Institutional Ethics Committee approval for a period of two years, i.e., from January 2018 to December 2019.
A total of 339 Staphylococcus aureus isolates obtained from various clinical samples were incorporated in the study and were characterized by conventional tests, including Gram’s staining, culture, and standard biochemical tests. Antibiotic sensitivity testing of all the isolates was done by Kirby Bauer disc diffusion method on Mueller Hinton agar (MHA) by using antibiotic discs (obtained from HIMEDIA lab Mumbai)of Penicillin (10units), Cefoxitin (30mcg), Ciprofloxacin (5mcg), Linezolid (30mcg), Erythromycin (15mcg) and Clindamycin (2mcg); interpreted as sensitive, intermediate and resistant as per CLSI guidelines. Vancomycin was reported by performing E-test. Identification of methicillin sensitive S.aureus (MSSA) and MRSA strains were according to CLSI guidelines10. Double disc diffusion test was done for all the isolates by placing Clindamycin(2mcg) and Erythromycin(15mcg) discs 15mm apart.
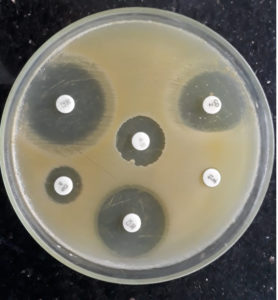
Flattening of the zone of inhibition around the clindamycin disc facing the Erythromycin disc was considered D-test positive, indicating inducible clindamycin resistance (Fig. 1). All such isolates were reported as clindamycin resistant.
The strains were interpreted as constitutive MLSB phenotype if resistant to erythromycin with zone size ≤13mm and clindamycin with zone size ≤14mm, and those strains that were resistant to erythromycin with zone size ≤13mm and sensitive to clindamycin with zone size ≥21mm without D-zone was interpreted as MS phenotype11.(Table 1)
Table (1):
Showing the interpretation of the D-test.
D-test phenotype |
Erythromycin(E) Zone size |
Clindamycin(CD) |
D-test interpretation |
|---|---|---|---|
Constitutive MLSB(cMLSB) |
R(≤13mm) |
R(≤14mm) |
Growth up to CD and E discs |
Inducible MLSB(iMLSB) |
R(≤13mm) |
S(≥21mm) |
Flattened D shaped zone of inhibition around CD adjacent to E disc |
MS phenotype |
R(≤13mm) |
S(≥21mm) |
No D-zone, clear zone around Clindamycin disc |
S: Sensitive; R: Resistant; CD: Clindamycin; E: Erythromycin
S.aureus ATCC 25923 was used as a control strain.
Results tabulated and analyzed statistically.
Of the total 339 S.aureus isolates,165 were sensitive to both erythromycin and clindamycin. D-test further characterized the remaining 174 isolates resistant to either erythromycin or clindamycin, or both.
Out of the 174 isolates, 98(56.32%) were MRSA, and 76 isolates were MSSA(43.68%), as shown in Fig. 2.
All the 174 isolates of S.aureus were subjected to D-test to characterize as cMLSB, iMLSB, or MS phenotype. Among the 174 isolates tested, 47(27.01%) strains were of cMLSB phenotype, 31(17.82%) strains were of iMLSB phenotype, and 96(55.17%) strains were of MS phenotype, as shown in Table 2.
Table (2):
Showing different S.aureus phenotypes.
Phenotype |
No. of isolates |
|---|---|
cMLSB |
47(27.01%) |
iMLSB |
31(17.82%) |
MS phenotype |
96(55.17%) |
Total |
174(100%) |
Out of the total 98 isolates of MRSA 36(76.60%) were cMLSB,20 (64.52%) iMLSB and 42(43.75%) MS phenotype.Out of total 76 MSSA isolates 11(23.40%) were cMLSB, 11(35.48%)iMLSB and 54(56.25%)MS phenotype as shown in Table 3.
Table (3):
Showing phenotypes among MRSA and MSSA isolates.
| Phenotype | Methicillin Resistance | Total | |
|---|---|---|---|
| MRSA | MSSA | ||
| cMLSB | 36(76.60%) | 11(23.40%) | 47 |
| iMLSB | 20(64.52%) | 11(35.48%) | 31 |
| MS phenotype | 42(43.75%) | 54(56.25%) | 96 |
| Total | 98 | 76 | 174 |
In the present study, the iMLSB phenotype was more in MRSA isolates (64.52%) than in MSSA isolates(35.48%).
Clindamycin is an excellent and preferred agent to treat superficial infections with S.aureus and a preferred antibiotic in patients allergic to penicillin12. Resistance to clindamycin in S.aureus strains with inducible phenotype may be reported as sensitive if not tested by D-test giving a false sensitive report which could result in treatment failure and also the emergence of constitutive erm mutants13.
The incidence of iMLSB in our study was 17.82% which was comparable with Toleti S et al.14 (18%), Lall M et al.9 (20.3%), and Adaleti R et al.15(22%). Bingo S et al.16 had reported an incidence of iMLSB to be 28.5% which is higher than in our study. Prabhu K et al.17 had reported 10.52% of iMLSB, which was less compared to our study. 31(17.82%) isolates of Staphylococci would have been reported as sensitive if not tested with a D-test, conveying a false report to the treating clinician.
In the present study, among MRSA isolates, cMLSB phenotypes were 36(76.60%), iMLSB phenotypes 20(64.52%), and MS phenotypes were 42(43.75%), and in MSSA isolates, cMLSB phenotypes are 11(23.40%) iMLSB are 11(35.48%), and MS phenotype are 54(56.25%). In our study, both cMLSB and iMLSB phenotypes are more in MRSA isolates compared to MSSA isolates.
According to Toleti S et al.14, the prevalence of iMLSB phenotype was 22.72% in MRSA isolates and 11.11% in MSSA isolates, and Bingo S et al. 16 found that iMLSB phenotypes in MRSA were 91.9%. In MSSA, it was 8.1%, and according to Lall M et al. 9 MRSA isolates showing iMLSB phenotype are 37.1%, and MSSA was 6%. According to Prabhu K et al.17 iMLSB phenotypes in MRSA was 20% and MSSA was 6.5%, and these results were similar to our study showing iMLSB phenotypes more in MRSA isolates than in MSSA isolates.
In a study done by Adaleti R et al. 15, The iMLSB phenotypes in MRSA were 18.2%, and MSSA was 40% which can be considered because the frequency of iMLSB phenotypes varies widely, ranging from 7 to 94%18 and there are also few other studies showing higher percentage in MSSA isolates than MRSA isolates19.
Clindamycin is a preferred antibiotic in superficial Staphylococcal infections and an alternative in penicillin-allergic patients.
False sensitive reports can lead to Clindamycin therapy failures and the selection of a constitutive resistant mutant in an iMLSB strain. So it will be appropriate that all clinical laboratories test and report inducible clindamycin resistance in both MRSA & MSSA by double disc diffusion test, which is a straight forward method to identify iMLSB phenotypes.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All listed author(s) have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Yilmaz G, Aydin K, Iskender S, Caylan R, Koksal I. Detection and prevalence of inducible clindamycin resistance in Staphylococci. J Med Microbiol. 2007;56(Pt 3):342-345.
Crossref - Cheung AL, Projan SJ, Gresham H. The genomic aspect of virulence, sepsis, and resistance to killing mechanisms in Staphylococcus aureus. Curr Infect Dis Rep. 2002;4(5):400-410.
Crossref - Kesah C, Ben Redjeb S, Odugbemi TO, et al. Prevalence of methicillin resistant Staphylococcus aureus in eight African hospitals and Malta. Clin Microbiol Infect. 2003;9(2):153-156.
Crossref - Fiebelkorn KR, Crawford SA, McElmeel ML, Jorgensen JH. Practical disk diffusion method for Detection of inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci. J Clin Microbiol. 2003;41(10):4740-4744.
Crossref - Brisson-Noel A, Delrieu P, Samain D, Courvalin P. Inactivation of lincosamide antibiotics in Staphylococcus. Identification of lincosamide O-nucleotidyl transferases and comparison of the corresponding resistance genes. J Biol Chem. 1988;263(31):15880-15887.
Crossref - Leclercq R. Mechanism of resistance to macrolides and lincosamides. Nature of the resistance elements and their clinical implications. Clin Infect Dis. 2002;34(4):482-492.
Crossref - Watanakunakorn C. Clindamycin therapy of Staphylococcus aureus endocarditis. Clinical relapse and level of resistance to clindamycin, lincomycin, and erythromycin. Am J Med. 1976;60(3):419-425.
Crossref - Steward CD, Raney PM, Morrell AK, et al. Testing for induction of clindamycin resistance in erythromycin resistant isolates of Staphylococcus aureus. J Clin Microbiol. 2005;43(4):1716-1721.
Crossref - Lall M, Sahni AK. Prevalence of inducible clindamycin resistance in Staphylococcus aureus isolated from clinical samples. Med J Armed Forces India. 2014;70(1):43-47.
Crossref - Clinical and Laboratory Standards Institute.Performance standards for antimicrobial susceptibility testing; Twenty-Seventh Informational Supplement. CLSI document M100-S27. Wayne, PA: CLSI;2017.
- Hamilton-Miller JMT, Shah. Patterns of phenotypic resistance to the macrolide-lincosamide-ketolide-streptogramin group of antibiotics in staphylococci. J Antimicrob Chemother. 2000;46(6):941-949.
Crossref - Drinkovic D, Fuller ER, Shore KP, Holland DJ, Ellis-Pegler R. Clindamycin treatment of Staphylococcus aureus expressing inducible clindamycin resistance. J Antimicrob Chemother. 2001;48(2):315-316.
Crossref - Deotale V, Mendiratta DK, Raut V, Narang P. Inducible clindamycin resistance in Staphylococcus aureus isolated from clinical samples. Indian J Med Microbiol. 2010;28(2):124-126.
Crossref - Toleti S, Bobbillapati JR, Kollipaka SR, Myneni RB. Detection of inducible clindamycin resistance and susceptibilities to other antimicrobial agents in clinical isolates of Staphylococcus aureus. Int J Res Med Sci. 2015;3(3):612-616.
Crossref - Adaleti R, Nakipogulu Y, Ceran N, TasdemirC, Kaya F, Tasdemir S. Prevalence of phenotypic resistance of Staphylococcus aureus isolates to macrolide, lincosamide, streptogramin B, ketolide, and linezolid resistance in Turkey. Braz J Infect Dis. 2010;14(1):11-14.
Crossref - Bingo S, Basak S, Das S, Kaushik P. Inducible clindamycin resistance in Staphylococcus aureus and its therapeutic implications. Int J Curr Res. 2017;9(10):59930-59933.
- Prabhu K, Rao S, Rao V. Inducible Clindamycin Resistance in Staphylococcus aureus Isolated from Clinical Samples. J Lab Physicians. 2011;3(1):25-27.
Crossref - Patel M, Waites KB, Moser SA, Cloud GA, Hoesley CJ. Prevalence of inducible clindamycin resistance among community- and hospital-associated Staphylococcus aureus isolates. J Clin Microbiol. 2006;44(7):2481-2484.
Crossref - Schreckenberger PC, Ilendo E, Ristow KL. Incidence of constitutive and inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci in a community and a tertiary care hospital. J Clin Microbiol. 2004;42(6):2777-2779.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.